Abstract
We have developed a simple PCR-based high-resolution melt curve analysis for identification of the quinolone resistance gene aac(6′)-Ib-cr through regions encompassing the two defining single nucleotide mutations. Dissociation curves showed 100% concordance with DNA sequencing, including the identification of a strain where aac(6′)-Ib and aac(6′)-Ib-cr coexist.
The cr variant of aac(6′)-Ib encodes an aminoglycoside acetyltransferase that confers reduced susceptibility to ciprofloxacin and norfloxacin by N acetylation of their piperazinyl amines (8). aac(6′)-Ib-cr belongs to the group of plasmid-mediated quinolone resistance genes that determine small increases in the MICs that are sufficient to facilitate the selection of higher-level-resistance mutants (10). However, this low-level quinolone resistance is below the CLSI breakpoint for nonsusceptibility and is not detected in the clinical laboratory. Development of efficient techniques for detection of clinical isolates carrying aac(6′)-Ib-cr may improve optimization of antibiotic treatment. The resistance phenotype of AAC(6′)-Ib-cr is dependent on the effects of the individual mutations. Asp181Tyr (coded by G541T) produces a partial-resistance phenotype and Trp104Arg (coded by either T310C or T310A) no detectable resistance, but together, the two mutations confer the full resistance phenotype (the nucleotide positions correspond to GenBank accession number AF479774; see also the cr variant under GenBank accession number AY259086) (8). We previously showed that the gap-ligase chain reaction (LCR) is an inexpensive technique suited to large-scale surveys, albeit time-consuming (13).
Improved real-time PCR machines and software analysis packages in recent years have enhanced the resolution of melting temperature (Tm) differences between amplicons from 2°C to 0.01°C in modern instruments (4, 7). Such resolution is now sufficient for identification of transitions or transversions that involve A ↔ C or G ↔ T and can be applied in high-resolution melting curve analysis (HRMA) to identify a single nucleotide change inside a full-length amplicon (12). Melt analysis and its more sophisticated high-resolution variant are being increasingly applied to identify quinolone resistance mutations in type II topoisomerases of Haemophilus influenzae (5) and Neisseria gonorrhoeae (11), to genotype Mycoplasma pneumoniae isolates (9), or to identify multidrug-resistant Mycobacterium tuberculosis (6). We developed and validated a real-time PCR-based HRMA using SYBR green I to rapidly detect aac(6′)-Ib-cr and distinguish it from aac(6′)-Ib.
A homology search in GenBank identified 181 sequences described as aac(6′)-Ib, of which 22 corresponded to the aac(6′)-Ib-cr variant. Alignment of all sequences was used to design two pairs of primers (Geneious Pro 4) (2). The pair comprising aac6-5′278 (GTCGTACGTTGCTCTTGGAA) and aac6-5′352 (GGTCTATTCCGCGTACTCCT) and the pair comprising aac6-3′508 (GGGTTTGAGAGGCAAG GTA) and aac6-3′582 (GAATGCCTGGCGTGTTTG) amplified 73- and 74-bp products, designated the 5′ region and the 3′ region, respectively, that corresponded to nucleotides 278 to 352 and 508 to 582, respectively, in aac(6′)-Ib (GenBank accession number AF479774).
In developing the HRMA method, we used four different alleles of aac(6′)-Ib that we had previously generated by site-directed mutagenesis (8): aac0, encoding wild-type aac(6′)-Ib; aac1, encoding aac(6′)-Ib-cr; and aac2 and aac3, encoding aac(6′)-Ib with single mutations T310C and G541T, respectively. The assay was validated on nine aac(6′)-ib-cr-positive and 10 wild-type strains from a collection of clinical isolates already screened for aac(6′)-Ib-cr by gap-LCR and verified by sequencing (13).
Plasmid DNA from control strains was extracted using a QIAamp DNA minikit (Qiagen, Valencia, CA) in accordance with the manufacturer's instructions. Colonies were transferred to Tris-HCl (pH 7.4) in a 2-ml screw-cap tube and heated for 2 min at 98°C to prepare DNA templates from tested strains.
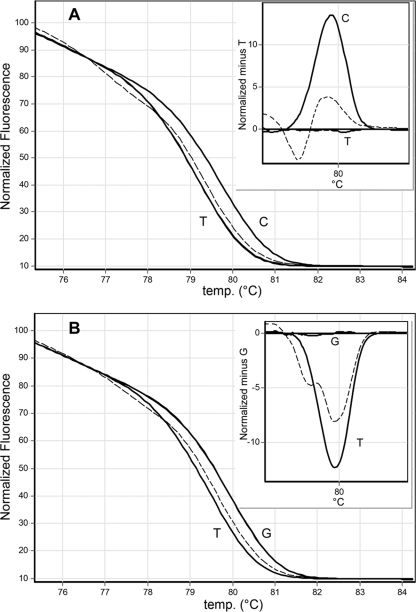
Real-time PCR and HRMA were performed using a Rotor-Gene 6000 apparatus (Corbett Life Science, Australia) in a total volume of 20 μl; the run consisted of 30 cycles at 93°C for 10 s, followed by 58°C for 10 s and 72°C for 6 s. The high-resolution-melt (HRM) conditions were 2 s at 95°C followed by 90 s at 55°C premelt, with an HRM ramp from 76°C to 86°C, rising by 0.04°C each step and holding for 2 s on each step. Gain optimization before the melt on all tubes was selected. SYBR green I (DyNAmo Flash SYBR green quantitative PCR [qPCR] kit; Finnzymes) was used with an excitation wavelength at 470 nm and detection at 510 nm. For normalization, the temperature ranges were 75.34°C to 77.51°C for the leading range and 82.39°C to 84.30°C for the trailing range. Calculations were done using the Rotor-Gene software program (version 1.7). A confidence value is provided as an integrity check of autocalled results. Serial 10-fold dilutions of extracted DNA from the control strains were amplified and subjected to HRMA. The distinctive typing that resulted from the type-specific melt profiles of the 5′ region (Fig. 1 A) and the 3′ region (Fig. 1B) showed that all amplicons were reliably sorted into one of the two distinct groups within each region.
FIG. 1.
Dissociation curves. Normalized and temperature-shifted difference plots for mutant discrimination by HRMA. (A) Normalized melt curve plot of the 5′ region showing T → C transition and (as shown in the nested graph) the normalized temperature minus the temperature shift for the same amplicon (T). (B) Normalized melt curve plot of the 3′ region, showing G → T transversion and (as shown in the nested graph) the normalized temperature minus the temperature shift for the same amplicon (G). Corresponding nucleotides (C, G, and T) are depicted next to each curve. The nucleotide present in aac(6′)-Ib was used to normalize each temperature shift graph. Dotted lines correspond to a heterozygote.
The estimated error rates for genotyping homozygotes as a function of their Tm varied from instrument to instrument, and this rate was found to be less than 0.01 at 0.5°C for an amplicon of 110 bp with the use of the Rotor-Gene 3000 instrument (4). The intra-assay variation was calculated for two replicas of four 10-fold dilutions of each mutant. The standard deviations (SD) of the Tm varied from 0.01 (with a Tm of 79.20°C for nucleotide T in the 5′ region and a 73-bp amplicon) to 0.055 (for nucleotide T in the 3′ region with a Tm of 79.26°C and an amplicon of 74 bp). The differences in Tm (ΔTm) between amplicons for the 5′ and 3′ regions were 0.62°C and 0.71°C, respectively, with a confidence level above 97% in all cases.
As expected, no PCR products were obtained from dozens of clinical strains lacking aac(6′)-Ib and its variant. We then assayed 43 carbapenemase-producing Enterobacteriaceae with an unknown aac(6′)-Ib genotype isolated from wounds, sputa, and urine samples. One and 41 isolates were positive for aac(6′)-Ib-cr and aac(6′)-Ib, respectively; the HRMA results for these amplicons were sorted in the expected group, with confidence averages of 97.9% for the 5′ region (SD = 2.3) and 97.1% for the 3′ region (SD = 2.1). For one strain, a dissociation curve was interpreted as having variations (less than 85% confidence) in both the 5′ and the 3′ regions (Fig. 1). The distinct Tm plot of this amplicon is visible on the normalized graphics (Fig. 1, nested graphics). Sequencing demonstrated double peaks corresponding to the nucleotides cytosine and thymine at position 310 and guanine and thymine at position 541 in aac(6′)-Ib-cr.
PCR products were obtained for every strain used for validation, thus supporting the utility of the boiling extraction method as a reliable, fast, and inexpensive method for obtaining whole-cell DNA as a template for this PCR.
HRMA may detect other mutations that are not the target of the screening within the amplified region. In addition, because the melting temperature is the same, the HRMA might not have detected a T310A mutation. Recently, a report on the detection of aac(6′)-Ib-cr through its T310C or T310A mutations by the use of an asymmetric concentration of primers to promote amplification of the DNA strand complementary to an unlabeled and 3′ phosphorylated probe for HRMA was published (1). An evolutionary step for aac(6′)-Ib with a single mutation in either position 310 or position 541 (not investigated in that study) is plausible (1, 3), and its identification would be an important contribution to the understanding of the evolution of aac(6′)-Ib-cr and the tracking of its epidemiology. By analyzing the two aac(6′)-Ib-cr-characterizing regions, we ensured the accuracy of detection of the cr variant, even indicating whether this variant had been determined by a mutation at position 541. We have developed a simple and rapid real-time PCR-based HRMA that is able to detect aac(6′)-Ib-cr and discriminate between the two aac(6′)-Ib single nucleotide mutations required for the ciprofloxacin resistance phenotype. This approach provides an improvement over laborious procedures such as gap-LCR or expensive sequencing methods. Further research is needed in applying rapid diagnostic procedures for the detection of additional plasmid-mediated quinolone resistance genes.
Acknowledgments
This work was supported in part by grant Morasha 1833/07 from the Israel Science Foundation to J.S.
Footnotes
Published ahead of print on 24 May 2010.
REFERENCES
- 1.Bell, J. M., J. D. Turnidge, and P. Andersson. 2010. aac(6′)-Ib-cr genotyping by simultaneous high-resolution melting analysis of an unlabeled probe and full-length amplicon. Antimicrob. Agents Chemother. 54:1378-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drummond, A. J., B. Ashton, M. Cheung, J. Heled, M. Kearse, R. Moir, S. Stones-Havas, T. Thierer, and A. Wilson. 2009. Geneious v4.7. Biomatters, Ltd., Auckland, New Zealand.
- 3.Guillard, T., V. Duval, H. Moret, L. Brasme, V. Vernet-Garnier, and C. de Champs. 2010. Rapid detection of quinolone resistance gene aac(6′)-Ib-cr by pyrosequencing. J. Clin. Microbiol. 48:286-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrmann, M. G., J. D. Durtschi, L. K. Bromley, C. T. Wittwer, and K. V. Voelkerding. 2006. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin. Chem. 52:494-503. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura, S., K. Yanagihara, Y. Morinaga, K. Izumikawa, M. Seki, H. Kakeya, Y. Yamamoto, S. Kamihira, and S. Kohno. 2009. Melting curve analysis for rapid detection of topoisomerase gene mutations in Haemophilus influenzae. J. Clin. Microbiol. 47:781-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietzka, A. T., A. Indra, A. Stoger, J. Zeinzinger, M. Konrad, P. Hasenberger, F. Allerberger, and W. Ruppitsch. 2009. Rapid identification of multidrug-resistant Mycobacterium tuberculosis isolates by rpoB gene scanning using high-resolution melting curve PCR analysis. J. Antimicrob. Chemother. 63:1121-1127. [DOI] [PubMed] [Google Scholar]
- 7.Ririe, K. M., R. P. Rasmussen, and C. T. Wittwer. 1997. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245:154-160. [DOI] [PubMed] [Google Scholar]
- 8.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz, S. B., K. A. Thurman, S. L. Mitchell, B. J. Wolff, and J. M. Winchell. 2009. Genotyping of Mycoplasma pneumoniae isolates using real-time PCR and high-resolution melt analysis. Clin. Microbiol. Infect. 15:756-762. [DOI] [PubMed] [Google Scholar]
- 10.Strahilevitz, J., G. A. Jacoby, D. C. Hooper, and A. Robicsek. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernel-Pauillac, F., T. R. Hogan, J. W. Tapsall, and C. Goarant. 2009. Quinolone resistance in Neisseria gonorrhoeae: rapid genotyping of quinolone resistance-determining regions in gyrA and parC genes by melting curve analysis predicts susceptibility. Antimicrob. Agents Chemother. 53:1264-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vossen, R. H., E. Aten, A. Roos, and J. T. den Dunnen. 2009. High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum. Mutat. 30:860-866. [DOI] [PubMed] [Google Scholar]
- 13.Warburg, G., M. Korem, A. Robicsek, D. Engelstein, A. E. Moses, C. Block, and J. Strahilevitz. 2009. Changes in aac(6′)-Ib-cr prevalence and fluoroquinolone resistance in nosocomial isolates of Escherichia coli collected from 1991 through 2005. Antimicrob. Agents Chemother. 53:1268-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]