Abstract
The trimethoprim resistance gene dfrK was found to be part of the novel Tn554-related transposon Tn559 integrated in the chromosomal radC gene of a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. While Tn559 and Tn554 had similar arrangements of the transposase genes tnpA, tnpB, and tnpC, the Tn554-associated resistance genes erm(A) and spc were replaced by dfrK in Tn559. Circular forms of Tn559 were detected and suggest the functional activity of this transposon.
The trimethoprim resistance gene dfrK was recently detected on plasmid pKKS2187 in a porcine methicillin-resistant Staphylococcus aureus (MRSA) strain of the multilocus sequence type 398 (ST398) (6). In this plasmid, the gene dfrK was physically linked to the tetracycline resistance gene tet(L). Further analysis of plasmids from porcine MRSA ST398 strains identified the tet(L)-dfrK genes in close proximity to the kanamycin/neomycin resistance gene aadD on plasmid pKKS825 (7) or to the macrolide-lincosamide-streptogramin B (MLSB) resistance gene erm(T) on plasmid pKKS25 (8). While dfrK was usually plasmid borne and linked to tet(L), a recent survey on the presence of dfrK among German coagulase-positive staphylococci from animals (3, 5, 19) identified the porcine S. aureus strain 2171 in which dfrK was neither located on a plasmid nor linked to tet(L).
This strain was obtained from a sow suffering from a genital tract infection (19). Susceptibility testing by broth microdilution (2) revealed susceptibility to oxacillin (MIC, 0.5 μg/ml) but resistance to penicillins (MICs, 8 μg/ml [penicillin] and 4 μg/ml [ampicillin]), macrolides and lincosamides (MICs, ≥64 μg/ml [erythromycin] and ≥128 μg/ml [clindamycin]), tetracycline (MIC, 32 μg/ml), enrofloxacin (MIC, 8 μg/ml), and trimethoprim (MIC, ≥128 μg/ml). Specific PCR assays (3, 13, 18) (Table 1) revealed the presence of the β-lactamase gene blaZ, the MLSB resistance gene erm(B), the tetracycline resistance gene tet(M), and the trimethoprim resistance gene dfrK. S. aureus 2171 proved to be negative for mecA (11). Multilocus sequence typing and spa typing (9) revealed the sequence type 398 and spa type t011. Plasmid analysis showed the presence of three plasmids of sizes below 10 kb, none of which conferred trimethoprim resistance in repeated protoplast and electrotransformation experiments nor showed the presence of dfrK in Southern blot hybridization experiments. The dfrK probe consisted of the internal PCR-generated 214-bp fragment of the dfrK gene (Table 1). A single band of ca. 3.7 kb hybridizing with the dfrK probe was seen in whole-cell DNA digested with EcoRI. This observation suggested that a single copy of dfrK was most likely present in the chromosomal DNA of S. aureus 2171.
TABLE 1.
PCR primers used in this study
| Primer designation | Primer sequence (5′→3′) | Annealing temp (°C) | Amplicon size (in bp) | Reference |
|---|---|---|---|---|
| blaZ_fw | TGCTTCAACTTCAAAAGCGATA | 55 | 563 | This study |
| blaZ_rv | GTTCAGATTGGCCCTTAGGA | |||
| dfrK_fw | GCTGCGATGGATAAGAACAG | 50 | 214 | 3 |
| dfrK_rv | GGACGATTTCACAACCATTAAAGC | |||
| dfrK_inv1 | CGGAAGAGCATTACCTGGAA | 55 | 3,642 | This study |
| dfrK_inv2 | AATTGGATATCCCTTTGTAGTATTTTT | |||
| Tn559_circ-fw | TCCATGAACTCGTACAGCAA | 55 | 778 | This study |
| Tn559_circ-rv | TGGTTGTGAAATTGTCCATTC | |||
| radC_fw | GGAAAGGATGGGGAGAAGAG | 55 | 2,305 | This study |
| tnpB_rv | TGCTTCAATTTCCACTCTCG |
To gain information about the dfrK flanking regions, the EcoRI fragments were religated and subjected to an inverse PCR using the primers dfrKinv1 and dfrKinv2 (Table 1). These primers are located 29 bp apart from each other within the dfrK gene. The resulting amplicon was cloned into pCR-Blunt II-Topo (Invitrogen, Karlsruhe, Germany) with subsequent transformation into chemically competent Escherichia coli TOP10 and sequenced using the M13 universal and reverse primers as well as with the dfrKinv1 and dfrKinv2 primers. Completion of the sequence was done by primer walking using primers derived from the sequence obtained with the abovementioned primers. Sequence analysis revealed similarities to part of tnpB and tnpC of transposon Tn554 in the dfrK upstream region and to the 3′ end of the chromosomal radC gene in the dfrK downstream region. Database searches identified a Tn554 element integrated into the radC reading frame in the whole-genome sequence of S. aureus N315 (12). Assuming that there is a similar situation in the genome of S. aureus 2171, we designed a PCR assay using one primer located in the 5′-terminal part of radC and the other from the S. aureus 2171-specific tnpB sequence (Table 1). An amplicon of 2,305 bp was obtained and completely sequenced by primer walking starting with the radC-fw and tnpB-rev primers.
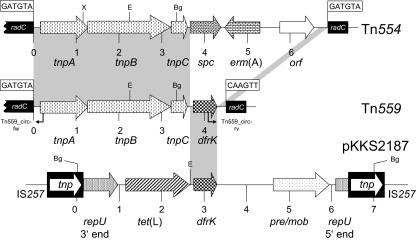
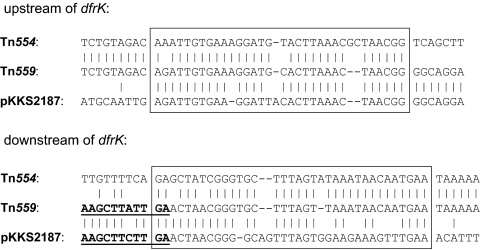
Analysis of the complete sequence revealed the presence of a transposon-like element of 4,289 bp, designated Tn559 (Fig. 1). Tn559 has three partly overlapping transposase genes, tnpA, tnpB, and tnpC. The 361-amino-acid (aa) TnpA protein of Tn559 showed 97.2% identity, the 630-aa TnpB protein showed 99.4% identity, and the 125-aa TnpC protein showed 99.2% identity to the same-sized proteins of Tn554 (1). Downstream of the tnpC gene, the reading frame for a 163-aa dihydrofolate reductase was detected. The gene revealed 97.4% nucleotide sequence identity to the dfrK genes, and the deduced protein showed 95.7% amino acid identity to the DfrK proteins known from staphylococcal plasmids pKKS2187, pKKS825, and pKKS25 (6-8). The sequences up- and downstream of the dfrK gene in Tn559 revealed two areas of homology which might have served for recombination of a plasmid-borne dfrK gene region with a Tn554 transposon (Fig. 2). Such a recombination resulted in the replacement of the former resistance gene region, consisting of the rRNA methylase gene erm(A), the spectinomycin resistance gene spc, and an open reading frame (ORF) of unknown function, by the dfrK gene.
FIG. 1.
Comparison of transposon Tn559 (FN677369) with transposon Tn554 (X03216) and the resistance gene region of plasmid pKKS2187 (FM207105). A distance scale in kb is given below each map. The position and orientation of the genes coding for transposition functions (tnpA, tnpB, and tnpC), antimicrobial resistance [erm(A), resistance to macrolides, lincosamides, and streptogramin B antibiotics; spc, spectinomycin resistance; dfrK, resistance to trimethoprim], plasmid replication (repU), plasmid recombination/mobilization (pre/mob), or unknown functions (orf) are indicated by arrows with the direction of transcription being shown by the arrowhead. The IS257 elements are shown as black boxes with the white arrow indicating the transposase gene tnp. Restriction endonuclease cleavage sites are abbreviated as follows: Bg (BglII), E (EcoRI), and X (XhoI). The positions of primers used for the detection of circular Tn559 forms are labeled Tn559_circ-fw and Tn559_circ-rv and indicated by arrows. The hexanucleotide sequences at the transposon junctions of the two transposons are shown in boxes. The regions of ≥97% nucleotide sequence identity between Tn554 and Tn559 as well as between Tn559 and pKKS2187 are marked by gray shading. The radC gene sequences up- and downstream of Tn554 and Tn559 are shown as black boxes. It should be noted that the radC sequence determined in S. aureus 2171 corresponds exactly to the primary target sequence of Tn554 known as att554 (14, 15).
FIG. 2.
Potential sites used for the development of Tn559 via recombination between a dfrK-carrying plasmid and a Tn554-like transposon. Vertical bars indicate identical bases compared to the Tn559 sequence. The recombination sites, where crossover is believed to have occurred, are boxed. The sequence displayed in bold underlined type in Tn559 and pKKS2187 represents the 3′ end of the dfrK gene.
As previously reported for Tn554 (14-17) and other members of this transposon family, such as Tn5406 (4) and Tn558 (10), Tn559 also neither contains inverted repeats at its ends nor generates a duplication of the target sequence at the integration site. Tn559 exhibited the hexanucleotide sequence 5′-GATGTA-3′ at the left-end junction and the sequence 5′-CAAGTT-3′ at the right-end junction. Studies of serial transposition of Tn554 into novel target sites revealed that the sequences at the junctions of Tn554 varied with respect to the target sites: with each new transposition event, the sequence originally present in the target site is found at the left end of Tn554, whereas the former left-end junction is now found at the right end and the former right-end junction is lost (14, 15, 17). Since transposition of Tn554 and its relatives includes the formation of circular forms which precede the integration of the transposon into a new target sequence (4, 14), inverse PCR assays using the primers Tn559_circ-fw and Tn559_circ-rv (Table 1) were conducted to detect such circular Tn559 intermediates. Amplicons of the expected size were obtained in repeated experiments from S. aureus 2171 and sequenced completely. In agreement with the transposition model (14, 15), this amplicon consisted of 322 bp of tnpA and its upstream region including the 6-bp core sequence 5′-GATGTA-3′ at the left end of Tn559, while the remaining 456 bp represented part of the dfrK gene and the right end of Tn559 up to—but not including—the sequence 5′-CAAGTT-3′. Evidence of the presence of circular Tn559 forms suggested the functional activity of this transposon in S. aureus 2171.
Nucleotide sequence accession number.
The sequence of Tn559 and its flanking regions has been deposited in the EMBL database under accession number FN677369.
Acknowledgments
We thank Kerstin Meyer for excellent technical assistance.
This study was financially supported by internal funding of the Friedrich-Loeffler-Institut.
Footnotes
Published ahead of print on 24 May 2010.
REFERENCES
- 1.Bastos, M. C., and E. Murphy. 1988. Transposon Tn554 encodes three products required for transposition. EMBO J. 7:2935-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CLSI. 2008. Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals; approved standard, 3rd ed. CLSI document M31-A3. CLSI, Wayne, PA.
- 3.Fessler, A., C. Scott, K. Kadlec, R. Ehricht, S. Monecke, and S. Schwarz. 2010. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65:619-625. [DOI] [PubMed] [Google Scholar]
- 4.Haroche, J., J. Allignet, and N. El Solh. 2002. Tn5406, a new staphylococcal transposon conferring resistance to streptogramin A and related compounds including dalfopristin. Antimicrob. Agents Chemother. 46:2337-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadlec, K., R. Ehricht, S. Monecke, U. Steinacker, H. Kaspar, J. Mankertz, and S. Schwarz. 2009. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 64:1156-1164. [DOI] [PubMed] [Google Scholar]
- 6.Kadlec, K., and S. Schwarz. 2009. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L). Antimicrob. Agents Chemother. 53:776-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadlec, K., and S. Schwarz. 2009. Identification of a novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 53:3589-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadlec, K., and S. Schwarz. 2010. Identification of a plasmid-borne resistance gene cluster comprising the resistance genes erm(T), dfrK, and tet(L) in a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 54:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kehrenberg, C., C. Cuny, B. Strommenger, S. Schwarz, and W. Witte. 2009. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53:779-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kehrenberg, C., and S. Schwarz. 2005. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob. Agents Chemother. 49:813-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 13.Lüthje, P., and S. Schwarz. 2006. Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide-lincosamide resistance phenotypes and genotypes. J. Antimicrob. Chemother. 57:966-969. [DOI] [PubMed] [Google Scholar]
- 14.Murphy, E. 1990. Properties of the site-specific transposable element Tn554, p. 123-135. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, NY.
- 15.Murphy, E. 1989. Transposable elements in gram-positive bacteria, p. 269-288. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, DC.
- 16.Murphy, E., L. Huwyler, and M. C. F. Bastos. 1985. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 4:3357-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy, E., and S. Löfdahl. 1984. Transposition of Tn554 does not generate a target duplication. Nature 307:292-294. [DOI] [PubMed] [Google Scholar]
- 18.Roberts, M. C., Y. Pang, D. E. Riley, S. L. Hillier, R. C. Berger, and J. N. Krieger. 1993. Detection of Tet M and Tet O tetracycline resistance genes by polymerase chain reaction. Mol. Cell. Probes 7:387-393. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz, S., E. Alešík, C. Werckenthin, M. Grobbel, A. Lübke-Becker, L. H. Wieler, and J. Wallmann. 2007. Antimicrobial susceptibility of coagulase-positive and coagulase-variable staphylococci from various indications of swine, dogs and cats as determined in the BfT-GermVet monitoring program 2004-2006. Berl. Münch. Tierärztl. Wochenschr. 120:372-379. [PubMed] [Google Scholar]