Abstract
Core-shell nanostructures with nonionic amphiphilic shells and ionic cores encapsulating gentamicin were designed for therapy against intracellular pathogens, including Salmonella and Listeria. Flow cytometry and confocal microscopy showed that their uptake into J774A.1 macrophages proceeded mainly by fluid-phase endocytosis and clathrin-mediated pathways. The nanostructures were nontoxic in vitro at doses of 50 to 250 μg/ml, and they significantly reduced the amounts of intracellular Salmonella (0.53 log) and Listeria (3.16 log), thereby suggesting effective transport into the cells.
Gentamicin is an aminoglycoside with antimicrobial activity against Gram-positive and Gram-negative bacteria (3). Its efficacy against these bacteria is established, yet its poor cell membrane permeability limits its clinical use, particularly against intracellular bacterial infections (4). We previously reported core-shell nanostructures encapsulating gentamicin that had poly(sodium acrylate-b-ethylene oxide-b-propylene oxide-b-ethylene oxide-b-sodium acrylate) (PAA−+Na-b-PEO-b-PPO-b-PEO-b-PAA−+Na) with PEO-b-PPO-b-PEO amphiphilic shells and block lengths of ∼3,600, 1,800, and 3,600 g/mole, respectively (designated ∼3600-b-1800-b-3600 g/mole) (from Pluronic F68) (8). PAA−+Na homopolymers were also incorporated into the nanostructure cores to increase their sizes and charge, thus facilitating high loadings of the polycationic gentamicin in the cores. The stabilities of those nanostructures in the presence of phosphate salts, however, were relatively poor. In the present study, we aimed to improve the stability of the nanostructures in physiological media to enhance delivery of gentamicin into macrophages. The strategy was to incorporate a higher-molecular-weight hydrophobic PPO in the shells, and more PPO relative to the hydrophilic PEO, so that enhanced hydrophobic interactions would contribute to their stabilities in physiological media. Thus, a PAA−+Na-b-PEO-b-PPO-b-PEO-b-PAA−+Na copolymer (nonionic portion derived from Pluronic P85) with block lengths of 2160-b-1100-b-2300-b-1100-b-2160 g/mole was blended with a hydrophilic PEO-b-PAA−+Na copolymer with block lengths of 2000-b-7200 g/mole, and these copolymers were condensed with gentamicin to afford core-shell nanostructures. These nanostructures were investigated in vitro to establish transport into the macrophages and determine their activities against vacuolar Salmonella and cytoplasm-resident Listeria (1, 11).
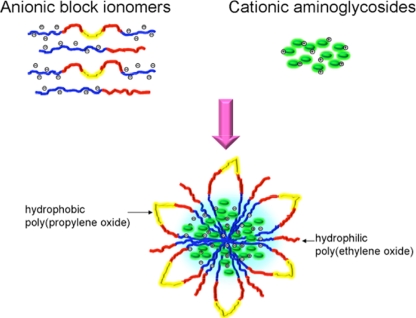
A scheme for preparing the nanostructures is shown in Fig. 1. PAA−+Na-b-PEO-b-PPO-b-PEO-b-PAA−+Na (25 mg; 1.7 × 10−4 equivalents [eq] of carboxylates) and PEO-b-PAA−+Na (25 mg; 2.7 × 10−4 eq of carboxylates) were dissolved in 25 ml of phosphate-buffered saline (PBS). The solution was sonicated while 5 ml of gentamicin sulfate solution (13.3 mg ml−1 gentamicin sulfate, equal to 40 mg gentamicin; 4.2 × 10−4 eq of cations) was added to form a turbid dispersion of nanostructures in PBS. Dynamic light scattering (DLS; Malvern Zetasizer NanoZS particle analyzer; Malvern Instruments, Ltd., Malvern, United Kingdom) at 37 ± 0.1°C and pH 7.4 in PBS showed a monomodal peak with an intensity average diameter of 548 ± 53 nm, thus indicating that a relatively homogeneous distribution of nanostructure sizes are produced with this procedure. This size distribution was in contrast to that of the previously reported nanostructures prepared with less of the hydrophobic PPO component in the nanostructure shells (8), for which DLS showed three peaks for the intensity average diameters centered around 11, 287, and 4,373 nm. The nanostructures were collected by centrifugation at 20,000 rpm for 15 min (Optima L-XP Ultracentrifuge; Beckman Coulter), washed twice with sterile deionized (DI) water and freeze-dried. The gentamicin concentration in the nanostructures was assayed (9) and determined to be 25% gentamicin by weight. The nomenclature describing these nanostructures is PN (PNF when labeled with fluorescein). The methodology to synthesize PNFs was reported previously (8). For flow cytometry, 1.25 × 105 J774A.1 macrophage cells/ml were preincubated with inhibitors of clathrin-mediated transport (10 μg/ml of chlorpromazine), caveolar-mediated transport (5 μg/ml of filipin), or fluid-phase endocytosis (450 mM sucrose) for 30 min (6). The cells were then incubated with 15 μg/ml of PNFs for 2 h, washed twice with Hanks balanced salt solution (HBBS) buffer, and the fluorescence intensities were analyzed by fluorescence-activated cell sorter (FACS) flow cytometry (BD FACS Aria) with an excitation wavelength of 488 nm and a 530/30-nm emission filter. Five thousand single-cell events were counted to compute the geometric mean fluorescence intensities of the cells from triplicate samples. Intracellular trafficking of PNFs were also analyzed by confocal microscopy (8). Cytotoxicity was evaluated using MTS assays (Promega) by incubating J774A.1 cells with gentamicin sulfate, Pluronic P85 (precursor polymer for the nanostructure shells), the copolymers, PNs, and untreated controls at a dose range of 50 to 250 μg/well for 24 h, and absorbance at 490 nm was measured. The ability of the PNs to kill intracellular bacteria at a dose of 25 μg/ml of gentamicin (100 μg/ml of PNs) was evaluated with an intracellular infection model using J774A.1 cells infected with Salmonella enterica serovar Typhimurium strain LT2 (6) or Listeria monocytogenes at a multiplicity of infection (MOI) of 10 bacteria per macrophage (5). Treatment groups were compared for differences in mean CFU using analysis of variance (ANOVA) followed by Tukey's procedure for multiple comparisons.
FIG. 1.
Preparation of core-shell nanostructures encapsulating gentamicin.
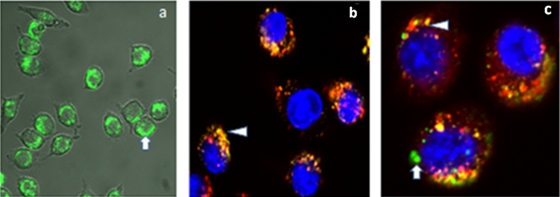
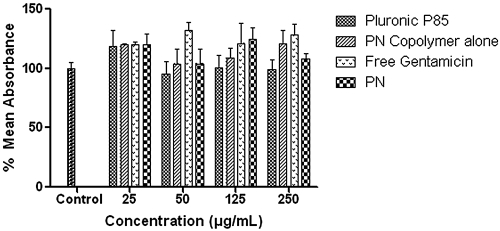
Based on the fluorescence of cells containing PNFs, flow cytometry suggested that PNF uptake was significantly reduced with chlorpromazine (∼46%) and sucrose (∼40%) compared to that with PNFs alone. It has been reported that the sucrose inhibitor of fluid-phase endocytosis can also inhibit clathrin-dependent receptor internalization by blocking clathrin-coated pit formation (12). Thus, inhibition by sucrose is regarded as somewhat nonspecific and could have contributed to reductions in uptake by both fluid-phase and clathrin-mediated pathways (7). In contrast, filipin did not significantly inhibit particle uptake. Confocal microscopy results agreed with those of flow cytometry. PNFs were taken up by the cells (Fig. 2 a). Macrophages were labeled positive for endosomes with Alexa Fluor 488 (Fig. 2b), and colocalization studies suggested that the PNFs resided in endosomes/lysosomes and also in the cytoplasm (Fig. 2c). Thus, transport of the PNFs into the cells could be an interplay of delivery by fluid-phase endocytosis into the cytosol or by a clathrin-mediated pathway involving endosomes, as reported elsewhere (2). For the dose range studied with the MTS assays, absorbance was comparable or higher than that of the untreated control, indicating a lack of toxicity (Fig. 3). Higher absorbance signified higher mitochondrial activity and an enhanced metabolic state of the cells. In vitro treatment studies showed that free gentamicin, the copolymers alone, and the infected control did not significantly clear intracellular Salmonella or Listeria. In contrast, PNs resulted in significant reductions of intracellular Salmonella (0.53 log) and Listeria (3.16 log) (P ≤ 0.05) (Table 1). Almost quantitative reduction of Listeria suggested that their subcellular localization may influence the capacity of the PNs to reach their intracellular bacterial targets. Plausibly, the vacuolar-resident Salmonella may not have been exposed to a high dose of the antimicrobial due to membrane barriers around the Salmonella within the cells. In contrast, cytoplasm-resident Listeria may directly interact with gentamicin, favoring efficient clearance (5). It should be noted that escape of Salmonella from the intracellular vacuoles partially depends on the duration of infection (10) and that the observed reduction in Salmonella could be due to escape of flagellated bacilli from the vacuoles and direct exposure to gentamicin in the cytoplasm. Development of drug delivery systems which release drugs in a time-dependent manner in the cell cytosol may be highly valuable for such treatments. In summary, these studies showed that the higher hydrophobic content in the PNs improves stability in physiological solutions over that in previously reported nanostructures (8) and that the efficacies of these nanostructures appear to depend on subcellular localization of the bacteria. Future studies aiming to target different intracellular niches of the bacterium may help develop an effective clearance strategy.
FIG. 2.
Confocal microscopy. (a) Uptake of green PNFs in J774A.1 cells. (b) Nanostructures are shown by yellow-to-orange spots formed by green nanoparticles/dyes and red endosomes/lysosomes, showing that a majority of the Alexa Fluor appears to reside in endosomes (arrowhead). (c) PNFs are distributed throughout the cells. Colocalization of PNFs with endosomes/lysosomes after incubation for 2 h (arrowhead) and subcellular localization of PNFs (arrow).
FIG. 3.
MTS assays showing the percent mean absorbance at 490 nm after incubating J774A.1 cells with 50 to 250 μg ml−1 of PNs along with appropriate controls. Results are expressed as means from six measurements ± standard deviations (SD).
TABLE 1.
Killing of intracellular wild-type S. Typhimurium LT2 and L. monocytogenes in J774A.1 cellsa
| Group |
Salmonella |
Listeria |
||
|---|---|---|---|---|
| Log CFU (±SD) | Log CFU reduction | Log CFU (±SD) | Log CFU reduction | |
| Control | 4.06 (0.16) | 0.00 | 5.63 (0.35) | 0.00 |
| Free gentamicin | 3.98 (0.05) | 0.08 | 5.67 (0.14) | −0.04 |
| PN copolymers | 3.91 (0.08) | 0.15 | 5.50 (0.10) | 0.13 |
| PNs | 3.53 (0.20) | 0.53* | 2.46 (0.11) | 3.17* |
Shown are results for killing of intracellular wild-type S. Typhimurium LT2 and L. monocytogenes in J774A.1 cells incubated with free gentamicin, the copolymers alone, or the PNs at a dose of 25 μg gentamicin/ml for 6 h. Each result is the mean from triplicate assays performed together ± SD. Asterisks indicate values found to be significantly different (confidence level; t test, 0.05) from that for free gentamicin by statistical analysis (ANOVA followed by Tukey's procedure for multiple comparisons).
Acknowledgments
We appreciate the support from National Science Foundation grant DMR-0312046 and Virginia Tech's Institute for Critical Technology and Applied Science.
Footnotes
Published ahead of print on 1 June 2010.
REFERENCES
- 1.Bakowski, M. A., V. Braun, and J. H. Brumell. 2008. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic 9:2022-2031. [DOI] [PubMed] [Google Scholar]
- 2.Batrakova, E. V., and A. V. Kabanov. 2008. Pluronic block copolymers: evolution of drug delivery concept from inert nanocarriers to biological response modifiers. J. Control. Release 130:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos, M. G., H. R. Rawls, L. H. Innocentini-Mei, and N. Satsangi. 2009. In vitro gentamicin sustained and controlled release from chitosan cross-linked films. J. Mater. Sci. Mater. Med. 20:537-542. [DOI] [PubMed] [Google Scholar]
- 4.Gamazo, C., M. C. Lecaroz, S. Prior, A. I. Vitas, M. A. Campanero, J. M. Irache, and M. J. Blanco-Prieto. 2006. Chemical and biological factors in the control of Brucella and brucellosis. Curr. Drug Deliv. 3:359-365. [DOI] [PubMed] [Google Scholar]
- 5.Lutwyche, P., C. Cordeiro, D. J. Wiseman, M. St-Louis, M. Uh, M. J. Hope, M. S. Webb, and B. B. Finlay. 1998. Intracellular delivery and antibacterial activity of gentamicin encapsulated in pH-sensitive liposomes. Antimicrob. Agents Chemother. 42:2511-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 7.Perumal, O. P., R. Inapagolla, S. Kannan, and R. M. Kannan. 2008. The effect of surface functionality on cellular trafficking of dendrimers. Biomaterials 29:3469-3476. [DOI] [PubMed] [Google Scholar]
- 8.Ranjan, A. P. N., M. N. Seleem, N. Sriranganathan, R. Kasimanickam, M. Makris, and J. S. Riffle. 2009. In vitro trafficking and efficacy of core-shell nanostructures for treating intracellular Salmonella infections. Antimicrob. Agents Chemother. 53:3985-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranjan, A., P. N. M. Seleem, N. Jain, N. Sriranganathan, J. S. Riffle, and R. Kasimanickam. 2009. Drug delivery using novel nanoplexes against a Salmonella mouse infection model. J. Nanopart. Res. 12:905-914. [Google Scholar]
- 10.Sano, G., Y. Takada, S. Goto, K. Maruyama, Y. Shindo, K. Oka, H. Matsui, and K. Matsuo. 2007. Flagella facilitate escape of Salmonella from oncotic macrophages. J. Bacteriol. 189:8224-8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia, S., X. P. Dun, P. S. Hu, S. Kjaer, K. Zheng, Y. Qian, C. Solen, T. Xu, B. Fredholm, T. Hokfelt, and Z. Q. Xu. 2008. Postendocytotic traffic of the galanin R1 receptor: a lysosomal signal motif on the cytoplasmic terminus. Proc. Natl. Acad. Sci. U. S. A. 105:5609-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]