Abstract
DNA microarrays were used to analyze Candida glabrata oropharyngeal isolates from seven hematopoietic stem cell transplant recipients whose isolates developed azole resistance while the recipients received fluconazole prophylaxis. Transcriptional profiling of the paired isolates revealed 19 genes upregulated in the majority of resistant isolates compared to their paired susceptible isolates. All seven resistant isolates had greater than 2-fold upregulation of C. glabrata PDR1 (CgPDR1), a master transcriptional regulator of the pleiotropic drug resistance (PDR) network, and all seven resistant isolates showed upregulation of known CgPDR1 target genes. The altered transcriptome can be explained in part by the observation that all seven resistant isolates had acquired a single nonsynonymous mutation in their CgPDR1 open reading frame. Four mutations occurred in the regulatory domain (L280P, L344S, G348A, and S391L) and one in the activation domain (G943S), while two mutations (N764I and R772I) occurred in an undefined region. Association of azole resistance and the CgPDR1 mutations was investigated in the same genetic background by introducing the CgPDR1 sequences from one sensitive isolate and five resistant isolates into a laboratory azole-hypersusceptible strain (Cgpdr1 strain) via integrative transformation. The Cgpdr1 strain was restored to wild-type fluconazole susceptibility when transformed with CgPDR1 from the susceptible isolate but became resistant when transformed with CgPDR1 from the resistant isolates. However, despite the identical genetic backgrounds, upregulation of CgPDR1 and CgPDR1 target genes varied between the five transformants, independent of the domain locations in which the mutations occurred. In summary, gain-of-function mutations in CgPDR1 contributed to the clinical azole resistance, but different mutations had various degrees of impact on the CgPDR1 target genes.
Candida glabrata is a haploid yeast and closely related to Saccharomyces cerevisiae. To date, second to Candida albicans, C. glabrata has emerged as the most common cause of bloodstream infection (candidemia) in many countries (15, 17). Ergosterol is an important component of fungal cell membranes, and the ergosterol biosynthetic pathway has been a primary target of antifungal drugs, including azoles (e.g., fluconazole) and allylamines (terbinafine). ERG11 encodes a cytochrome P-450-dependent C14 lanosterol demethylase (Erg11p) and is essential in ergosterol biosynthesis. Azole antifungals inhibit Erg11p activity and lead to the depletion of ergosterol. However, C. glabrata possesses intrinsically low susceptibility to fluconazole compared to C. albicans and frequently further develops resistance during prolonged treatment with fluconazole (4, 16, 18, 19, 27).
Azole resistance in pathogenic fungi has been reviewed recently (14). Drug efflux due to ATP-binding-cassette (ABC) transporters has been found to be a major contributor to azole resistance in several species, including C. glabrata. C. glabrata Pdr1p (CgPdr1p), a master transcriptional regulator of pleiotropic drug resistance (PDR), contributes to azole resistance by regulating gene expression of various transporters and plays a central role in fluconazole resistance acquired by C. glabrata (8, 25, 28-30). Gain-of-function (GOF) mutations in the transcriptional regulator, CgPdr1p, have been found in C. glabrata clinical isolates (8, 28) and in a laboratory strain (30). The mutations have been accompanied by an increased expression of drug efflux pumps and other target genes involved in the response to xenobiotics. The resistant isolates have varied in their regulation of the three ABC transporter genes most important for azole resistance CgCDR1, PDH1 (CgCDR2), and CgSNQ2 (8, 26).
The relationship between the CgPdr1p protein domain and downstream effects of these mutations in C. glabrata appeared worthy of further analysis. We selected seven pairs of isolates from patients receiving fluconazole prophylaxis following hematopoietic stem cell transplantation. Pairs from the same patient had the same contour-clamped homogeneous electric field (CHEF) gel patterns but differed in fluconazole susceptibility (4). We identified the nonsynonymous mutations in CgPDR1 of the seven clinical pairs and analyzed the transcriptome of each clinical pair by DNA microarray analysis. To eliminate the possibility that differences within the clinical pairs were due to mutations other than those in CgPDR1, we expressed the CgPDR1 gene from one susceptible isolate and five resistant isolates in the same Cgpdr1 host. The impact of CgPDR1 GOF mutations on the transcription of CgPDR1 and four of the CgPDR1 target genes was determined by quantitative real-time PCR (qRT-PCR). Despite expression in the same host, the GOF mutations differed in the upregulation of CgPDR1 and in the upregulation of its four target genes.
MATERIALS AND METHODS
Strains and culture conditions.
Plasmids were maintained in Escherichia coli XL1-Blue (Stratagene, La Jolla, CA), TOP10 (Invitrogen, Carlsbad, CA), or TOP10F′ (Invitrogen) host cells grown in LB with 50 μg/ml ampicillin, 50 μg/ml kanamycin, or 12.5 μg/ml chloramphenicol, depending on the plasmids.
Candida glabrata strains, including four strains from a previous study (Table 1), were cultured on either yeast extract-peptone-dextrose (YPD) containing 1% Bacto yeast extract (Difco Laboratories, Detroit, MI), 2% Bacto peptone (Difco Laboratories), and 2% glucose (Sigma, St. Louis, MO) or minimum medium (MIN) containing 0.67% yeast nitrogen base without amino acids (Difco Laboratories) plus 2% glucose.
TABLE 1.
Candida glabrata strains used in this study
| Strain | Parental strain | Genotype or description | Reference(s) or source |
|---|---|---|---|
| NCCLS84 | Wild-type strain 84 | ATCC 90030a | |
| 84u | NCCLS84 | ura3 | 10 |
| CgB4 | 84u | ura3 cgpdr1::Tn5<Cm URA3> | 28 |
| Cg1S | Clinical susceptible isolate, Cg12581, pair 1b | 4, 28 | |
| Cg2R | Clinical resistant isolate, Cg13928, pair 1b | 4, 28 | |
| Cg3S | Clinical susceptible isolate, pair 2b | 4 | |
| Cg4R | Clinical resistant isolate, pair 2b | 4 | |
| Cg5S | Clinical susceptible isolate, pair 3b | 4 | |
| Cg6R | Clinical resistant isolate, pair 3b | 4 | |
| Cg7S | Clinical susceptible isolate, Cg1660, pair 4b | 4, 28 | |
| Cg8R | Clinical resistant isolate, Cg4672, pair 4b | 4, 28 | |
| Cg11S | Clinical susceptible isolate, pair 6b | 4 | |
| Cg12R | Clinical resistant isolate, pair 6b | 4 | |
| Cg13S | Clinical susceptible isolate, pair 7b | 4 | |
| Cg14R | Clinical resistant isolate, pair 7b | 4 | |
| Cg15S | Clinical susceptible isolate, pair 8b | 4 | |
| Cg16R | Clinical resistant isolate, pair 8b | 4 | |
| Cg17S | Clinical susceptible isolate, pair 9b | 4 | |
| Cg18R | Clinical resistant isolate, pair 9b | 4 | |
| Cg21S | Clinical susceptible isolate, pair 11b | 4 | |
| Cg22R | Clinical resistant isolate, pair 11b | 4 | |
| C1Sac | CgB4 | ura3 CgPDR1-Cg1S | This study |
| C1Sbc | CgB4 | ura3 CgPDR1-Cg1S | This study |
| C4Rac | CgB4 | ura3 CgPDR1-Cg4R | This study |
| C4Rbc | CgB4 | ura3 CgPDR1-Cg4R | This study |
| C6Rac | CgB4 | ura3 CgPDR1-Cg6R | This study |
| C6Rbc | CgB4 | ura3 CgPDR1-Cg6R | This study |
| C14Rac | CgB4 | ura3 CgPDR1-Cg14R | This study |
| C14Rbc | CgB4 | ura3 CgPDR1-Cg14R | This study |
| C16Rac | CgB4 | ura3 CgPDR1-Cg16R | This study |
| C16Rbc | CgB4 | ura3 CgPDR1-Cg16R | This study |
| C18Rac | CgB4 | ura3 CgPDR1-Cg18R | This study |
| C18Rbc | CgB4 | ura3 CgPDR1-Cg18R | This study |
American Type Culture Collection, Manassas, VA.
Comparsion of the more susceptible and more resistant isolates within each paired clinical isolate.
Complementation by integrative transformation. Two independent complemented transformants were selected for each CgPDR1 GOF mutation, labeled “a” and “b.”
The seven pairs of oropharyngeal sequential isolates were chosen for study because each pair came from an individual hematopoietic stem cell transplant recipient receiving fluconazole (FHCRC protocol number 954). The more resistant isolate of each pair acquired increased fluconazole resistance during therapy, while the karyotype remained unchanged from its paired more-susceptible isolate (4).
Drug sensitivity assay.
MIC of fluconazole (courtesy of Pfizer, Sandwich, United Kingdom) was determined with the CSLI (formerly NCCLS) microtiter test by using the MIC producing 80% growth reduction (MIC80) as the MIC; the test was modified by addition of 2% glucose to the buffered RPMI medium (Sigma) and incubation at 37°C for 48 h with 250 cells of inoculum per well. In the case of the ura3 mutant, the RPMI medium was supplemented with 20 μg/ml of uracil (Sigma).
Microarray analysis.
DNA microarray analysis was used to identify genes with altered expression in the resistant clinical isolates and the CgPDR1-complemented strains. Total RNA was isolated from the log phase culture of C. glabrata grown in YPD by using Trizol (Invitrogen) and the RNeasy MiniElute cleanup kit (Qiagen, Valencia, CA). Pin-spotted 70-mer oligonucleotide in-house arrays fabricated at the NIAID were used for analysis of clinical pairs initially, but later, Agilent custom arrays (Agilent Technologies, Santa Clara, CA) were used for analysis of the CgPDR1-complemented strains, as the in-house printing of arrays was discontinued.
For the in-house microarrays, a total of 5,908 70-mer oligonucleotides were purchased from Institut Pasteur (Paris, France) and were used for microarray printing at the NIAID Microarray Research Facility. Expression of each open reading frame (ORF) is measured by hybridization to a specific 70-mer oligonucleotide (7, 12). Thirty micrograms of total RNA from sensitive isolates and resistant isolates was reversed transcribed to cDNA to incorporate the fluorescent Cy3-dUTP and Cy5-dUTP (GE Health Care, Piscataway, NJ), respectively. The labeled cDNA of paired sensitive/resistant isolates was combined and used for microarray hybridization. Each group consisted of a sensitive/resistant pair with five microarrays, including one or two with reciprocal labeling. The microarrays were prehybridized at 42°C in prehybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% bovine serum albumin [BSA], 0.1% SDS) for 30 to 60 min and then hybridized to the labeled cDNA in 50 μl of hybridization buffer (25% formamide, 5× SSC, 0.2% SDS, 20 μg/ml poly[dA]40-60, 200 μg/ml Cot-1 DNA [Invitrogen], 80 μg/ml yeast tRNA) overnight at 42°C. The microarrays were washed three times in wash buffer A (1× SSC, 0.05% SDS) and washing buffer B (0.1× SSC). The in-house arrays were scanned with a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA). All microarray data archive and analyses were done in the Web-based mAdb system provided by the Bioinformatics and Molecular Analysis group (BIMAS) at the Center for Information Technology (CIT), NIH. The data were filtered with the parameters that included genes present in three or more arrays per group and each array with 80% or more genes present. The data set of each of the paired isolates was then analyzed by Student's t test. The genes with P values less than 0.001 and with at least 2-fold altered gene expression were then selected. The final data set included all the genes with altered expression in at least one clinical pair.
For Agilent custom microarrays, the array probes were designed against all NCBI Reference Sequence (RefSeq) mRNA sequences available for C. glabrata CBS138 as of September 2008. Sixty-base DNA sequences were selected using the e-Array software (Agilent Technologies), specifying one “best probe” per transcript, “base composition method,” and “3-prime bias.” Custom microarrays with 5,125 unique probes (one for each target transcript), replicated to eight spot features per probe, were manufactured by Agilent in the 4×44K format. Ten micrograms of total RNA was used for each fluorescent labeling. Each group constituted of four microarrays, including one with reciprocal labeling. Microarrays were hybridized using the Tecan HS Pro 4800 hybridization station with Agilent 2× gene expression hybridization HI-RPM buffer and 10× blocking reagent at 65°C for 17 h and washed with Agilent gene expression wash buffer 1 at room temperature and gene expression wash buffer 2 at 37°C. Then slides were dried under nitrogen gas for 3 min at 30°C. The slides were imaged using Agilent high-resolution DNA microarray scanner G2505C at 5-μm resolution with both 100% and 10% photomultiplier tubes. Agilent Feature Extraction software was used for image analysis. Statistical calculations were performed on the “processed signal” data by using the mAdb analysis system provided by the BIMAS group at the CIT, NIH. Data were filtered with the parameters that included genes present in three or more arrays per group and each array with 80% or more genes present.
DNA sequence analysis of CgPDR1.
Genomic DNA from the seven pairs of oropharyngeal isolates was used as the templates for PCR to amplify the CgPDR1 ORF as well as its 2.5-kb promoter region. PfuUltra High-Fidelity DNA polymerase (Stratagene) was used for PCR amplification for reducing the generation of mutations during PCR amplification. Two independent PCR amplifications were performed for each isolate to obtain the DNA for sequencing. Primer set PDR8S and PDR5AS (Table 2) was used for amplifying the ORF region with the following parameters: 95°C for 2 min; 30 cycles of 95°C for 30 s, 53°C for 30 s, 72°C for 5 min, with an extension on the last cycle at 72°C for 10 min. Primer set PDR9S and PDR17AS was used for amplifying the promoter region with the same parameters as described above. The PCR products were then sequenced and analyzed.
TABLE 2.
Primers and TaqMan probes used in this study
| Primer or probe | Sequence (5′-3′) |
|---|---|
| PCR and sequencing primers | |
| CgPDR1AS | GGACAGAAATTGGAACATCG |
| CgPDR2S | TATCCTAAGTATGGACAACG |
| CgPDR4AS | GATTCCTTAAGCCCGATAAG |
| CgPDR5AS | GGTTACACCACTACTAGTTGa |
| CgPDR8S | GGTGGAGCTCTTTAGCTACGTTATTGAGa |
| CgPDR9S | TGAGATGAAAGCAATAACTG |
| CgPDR10S | TCAGTACTACACCTGAGTTG |
| CgPDR15AS | AATCGTTGTCCATACTTAGG |
| CgPDR16AS | ACACTCTCAATAAACGGTTG |
| CgPDR17AS | GTCAATGGATGATTTTATCG |
| CgPDR18AS | ACAAGGTTTTAGCCCATTAC |
| CgPDR19AS | TAATACCTAGTTTTACCCAC |
| CgPDR21AS | AGTATTCCCAACAGTATGAG |
| CgPDR22AS | ATGCTTAGTCTCTGCTCAC |
| CgPDR24S | ATGTCCTTATCACTAGGTC |
| qRT-PCR probes | |
| CgACT1P | CCACGTTGTTCCAATTTACGCCGG |
| CgPDR1P | TCGAATATTATGCACCATCATGTCTGTGTTTAGCT |
| CgCDR1P | TTATCTGCTGCGATGGTTCCTGCTTCC |
| PDH1P | CAGGCTCACATGCAAACCAAGACTACCAT |
| CgSNQ2P | CCGATGGTGACGATGCGCACAG |
| CgYOR1P | CTCGCCGGTGCAGGATTACGATCTAGA |
| qRT-PCR primers | |
| CgACT1F | TTGGACTCTGGTGACGGTGTTA |
| CgACT1R | AAAATAGCGTGTGGCAAAGAGAA |
| CgPDR1F | AACGATTATTCAATTGCAACAACG |
| CgPDR1R | CCTCACAATAAGGAAAGTCTGCG |
| CgCDR1F | AGATGTGTTGGTTCTGTCTCAAAGAC |
| CgCDR1R | CCGGAATACATTGACAAACCAAG |
| PDH1F | AATGGATGTTAGAAGTAGTTGGAGCAG |
| PDH1R | TGTTCGGAATTTCTCCACACCT |
| CgSNQ2F | GCGGAAGATCGCACGAAG |
| CgSNQ2R | GGCGCGAGCGGGATA |
| CgYOR1F | CGCTGGGAAGGCCAAGA |
| CgYOR1R | CTCCCCGGACGTCAGAATAG |
Underlined bases are the restriction sites.
Plasmid construction.
The plasmid pCgACU-P2F5 carrying the CgPDR1 gene on a 8-kb KpnI DNA fragment from the clinical isolate Cg8R (Cg4672) was used as the backbone vector (11). For the cloning of CgPDR1 from several clinical isolates, the CgPDR1 ORFs from one susceptible isolate (Cg1S) and two resistant isolates (Cg4R and Cg18R) with mutations in the regulatory domain (RD) were obtained by PCR using the primers PDR8S and PDR5AS as described above. The PCR products were digested with DraIII, and the 0.9-kb DraIII DNA fragments were then cloned into the DraIII site of pCgACU-P2F5 to give pCgACU-Cg1S, pCgACU-Cg4R, and pCgACU-Cg18R, respectively. The CgPDR1 ORFs from the resistant isolates with mutations in the activation domain (Cg6R) or undefined region (Cg14R and Cg16R) were obtained by PCR using the primer set PDR8S and PDR5AS and the parameters described above. The PCR products were digested with HpaI and PacI. The 1.2-kb HpaI-PacI DNA fragment containing the activation domain and the undefined region was then cloned into the HpaI-PacI site of pCgACU-1S to give the plasmids pCgACU-Cg14R, pCgACU-Cg16R, and pCgACU-Cg6R, respectively.
CgPDR1 complementation.
The CgPDR1 of clinical isolates was introduced into a laboratory cgpdr1 mutant (CgB4) (28), which allowed us to determine the impact of the CgPDR1 mutations on fluconazole resistance in the same genetic background. To introduce the CgPDR1 into the Tn<Cm URA3>-disrupted cgpdr1 locus in CgB4, the constructs containing CgPDR1 from the susceptible isolate (pCgACU-Cg1S) and five resistant clinical isolates (pCgACU-Cg4R, pCgACU-Cg6R, pCgACU-Cg14R, pCgACU-Cg16R, and pCgACU-Cg18R) were digested with HindIII, and the 2.9-kb HindIII DNA fragments containing the partial CgPDR1 ORFs were used to transform the cgpdr1 mutant CgB4, which is highly susceptible to fluconazole. Putative transformants were obtained based on the restoration of wild-type fluconazole susceptibility at 50 μg/ml and resistance to fluoroorotic acid (FOA) (28). To screen for the replacement of Tn<Cm URA3> by CgPDR1, FOA-resistant transformants were obtained and analyzed by PCR using the primer set CgPDR2S and CgPDR4AS with the following parameters: 95°C for 2 min; 35 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 2 min; with extension on the last cycle at 72°C for 10 min. Southern hybridization and DNA sequence analysis were done to confirm the CgPDR1 gene replacement (data not shown).
qRT-PCR.
Total RNA was isolated from C. glabrata log phase cultures grown in MIN rather than those grown in YPD to increase RNA purity. The total RNA was treated with DNase to remove the minute contamination of genomic DNA prior the reverse transcription with a high-capacity cDNA archive kit (Applied Biosystems, Foster City, CA). The parallel amplification between CgACT1 and the gene of interest was confirmed for each with probe-primer sets. Quantitative real-time PCR (qRT-PCR) was used to determine the expression level of CgACT1, CgPDR1, CgCDR1, PDH1, CgSNQ2, and CgYOR1 in C. glabrata. The sequences of TaqMan probes and forward and reverse primers are listed in Table 2. CgACT1 was used as an internal control for normalization. The threshold cycle (2−ΔΔCT) method was used for calculating the differences in gene expression.
Techniques and reagents.
C. glabrata genomic DNA was isolated from overnight cultures grown in YPD by using the MasterPure yeast purification kit (Epicentre, Madison, WI). Purified DNA fragments were recovered using the Strataprep gel DNA extraction kit (Stratagene). Hybond-N nylon membranes (Amersham, Arlington Heights, IL) were used for Southern hybridization analyses. DNA probes were labeled with [α-32P]dCTP or [α-32P]dATP (MP Biomedical, Solon, OH) by using the Prime-It II kit (Stratagene). DNA cloning and hybridization analyses were done according to the standard protocol (20). DNA sequencing was done using the DNA sequencing kit with a dRhodamine dye terminator (Applied Biosystems) and an ABI automatic DNA sequencing system (Perkin-Elmer, Foster City, CA). For sequencing of PCR products, PfuUltra DNA polymerase (Stratagene) was used for PCR amplification to minimize the rate of PCR-introduced mutations. The PCR products were cleaned with the Strataprep PCR purification kit (Stratagene) and used as the templates for DNA sequencing.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the CgPDR1 DNA sequences are HM17911 to HM17924. The array layout and probe sequences have been uploaded to the NCBI GEO microarray repository. The GEO accession number for the Agilent Cgda array is GPL10325. The accession number for the in-house array Cgaa is GPL8174. The GEO accession number for the in-house Cgaa array data is GSE21352, and the GEO accession number for the Agilent Cgda array data is GSE21355.
RESULTS
All seven clinical azole-resistant isolates had single nonsynonymous mutations in CgPDR1.
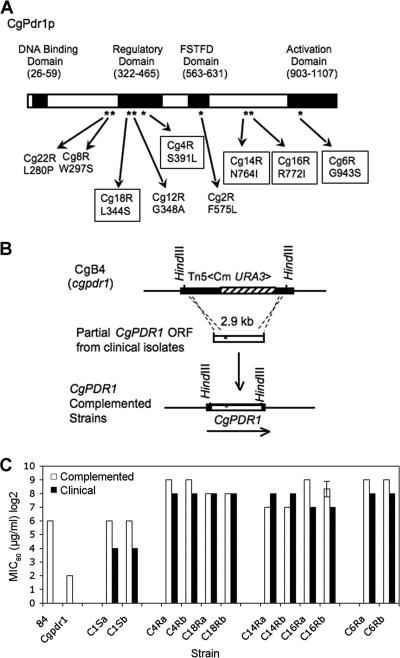
The fluconazole MIC80 of the more susceptible clinical isolates ranged from 16 to 64 μg/ml, while their paired more-resistant isolates ranged from 128 to 512 μg/ml (Table 3). To investigate whether CgPDR1 mutations contributed to the azole resistance in the oropharyngeal isolates of C. glabrata, the CgPDR1 ORFs (3.3 kb) of seven clinical azole-susceptible/azole-resistant pairs were sequenced along with their promoter regions (1.4 kb). DNA sequence analysis revealed that each of the seven clinical resistant isolates harbored a single nonsynonymous mutation at various regions of the CgPDR1 ORF compared to its paired azole-sensitive isolates (Table 3). No differences in the promoter sequences were found. All of the point mutations resulted in single amino acid substitutions. The majority of the amino acid substitutions resulted in changes in amino acid properties with the exception of the pair Cg12R/Cg11S, which retained a nonpolar aliphatic amino acid. Four putative functional domains (DNA binding, regulatory, fungus-specific transcriptional factor, and activation) were proposed in the CgPdr1p based on its similarity to S. cerevisiae Pdr1p. The majority of amino acid substitutions, four out of seven, were located in the regulatory domain (RD) (Cg22R, L280F; Cg18R, L344S; Cg12R, G348A; and Cg4R, S391L). While only one amino acid substitution occurred in the activation domain (Cg6R, G943S), there were two amino acid substitutions in an undefined region (Cg14R, N764; Cg16R, R772I), which is in the vicinity of a putative nuclear localization signal (NLS; amino acids 793 to 836) based on its similarity to the NLS of Pdr1p (amino acids 725 to 769) reported in S. cerevisiae (5). Together with the two mutations we identified previously in the regulatory and fungus-specific transcription factor domains (Cg8R, 297S; Cg2R, F575L) (28), a total of four domains/regions in CgPdr1p were identified as potentially being involved in the clinical azole resistance associated with the PDR network, with the regulatory domain being the predominant region for the mutations.
TABLE 3.
CgPDR1 mutations and fluconazole susceptibilities of clinical isolates
| Domain/region | Isolate | Fluconazole susceptibilitya | MIC80 (μg/ml) | Codon | Amino acid substitution | Amino acid property |
|---|---|---|---|---|---|---|
| Regulatory | Cg21S | Susceptible | 32 | TTG | Nonpolar, aliphatic | |
| Cg22R | Resistant | 256 | TTT | L280F | Nonpolar, aromatic | |
| Cg7Sb | Susceptible | 32-64 | TGG | Nonpolar, aromatic | ||
| Cg8Rb | Resistant | 512 | TCG | W297S | Polar-neutral | |
| Cg17S | Susceptible | 32 | TTG | Nonpolar, aliphatic | ||
| Cg18R | Resistant | 256 | TCG | L344S | Polar-neutral | |
| Cg11S | Susceptible | 32 | GGT | Nonpolar, aliphatic | ||
| Cg12R | Resistant | 256 | GCT | G348A | Nonpolar, aliphatic | |
| Cg3S | Susceptible | 32 | TCG | Polar-neutral | ||
| Cg4R | Resistant | 256 | TTG | S391L | Nonpolar, aliphatic | |
| FSTFc | Cg1Sb | Susceptible | 16 | TTC | Nonpolar, aromatic | |
| Cg2Rb | Resistant | 128 | CTC | F575L | Nonpolar, aliphatic | |
| Undefined | Cg13S | Susceptible | 64 | AAT | Polar-neutral | |
| Cg14R | Resistant | 256 | ATT | N764I | Nonpolar, aliphatic | |
| Cg15S | Susceptible | 16 | AGA | Polar-basic | ||
| Cg16R | Resistant | 128 | ATA | R772I | Nonpolar, aliphatic | |
| Activation | Cg5S | Susceptible | 32 | GGT | Nonpolar, aliphatic | |
| Cg6R | Resistant | 256 | AGT | G943S | Polar-neutral |
Comparison of the more susceptible and more resistant isolates within each paired clinical isolate.
Reported previously.
FSTF, fungus-specific transcriptional factor.
Clinical pairs with the CgPDR1 mutations in the same domain exhibited different transcriptional profiles.
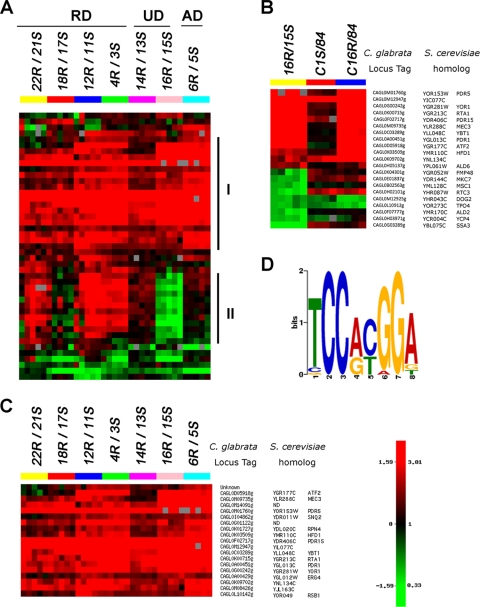
DNA microarray analysis was performed to determine the potential impact of different nonsynonymous CgPDR1 mutations on the transcriptional profiles of the clinical resistant isolates. Total RNA of the clinical pairs was reverse transcribed to incorporate Cy3-dUTP or Cy5-dUTP, which were combined and hybridized to the C. glabrata 70-mer oligonucleotide in-house microarrays. Figure 1 A provides the heat map of 45 genes with significant altered expression in at least one clinical pair. Five arrays for each of the seven clinical pairs and the expression ratios are shown in a log2 scale as either upregulation (in red) or downregulation (in green) of genes in the resistant isolates compared to its paired sensitive isolates.
FIG. 1.
Microarray analysis of clinical sensitive/resistant paired isolates. (A) Heat map of hierarchical gene clustering. For the seven pairs of azole-susceptible and -resistant isolates, five arrays for each pair and the expression ratio of resistant isolate over sensitive isolate are shown in log2 scale to show upregulation (in red) or downregulation (in green) of the genes in the resistant isolates. RD, regulatory domain; UD, undefined domain; AD, activation domain. Panel I, genes upregulated in the majority of seven groups; II, genes downregulated in 16R/15S. (B) Heat map of the genes with altered expression in 16R/15S compared to that of the complemented strains matched to the host wild-type strain 84. C1S, Cgpdr1 mutant complemented by CgPDR1 from Cg1S; C16R, Cgpdr1 mutant complemented by CgPDR1 from Cg16R. (C) Heat map and annotation of the 19 genes upregulated in a majority of the clinical resistant isolates (A, panel I). The C. glabrata locus tags of each ORF represented by the oligonucleotide as well as its S. cerevisiae homologs are listed on the right. ND, not determined. (D) Putative PDRE motif logo of CgPDR1. The 1-kb upstream sequences of 18 upregulated genes were used for the motif search with MEME version 4.3.0. The major motif was obtained with an E-value of 1.4 × 10−28.
The hierarchical cluster I contained genes upregulated in a majority of the seven clinical resistant isolates (Fig. 1A, panel I). Differences in the transcriptional profiles among seven clinical pairs were also evidenced in the data set. Cluster II included many genes upregulated only in three groups, 22R/21S, 12R/11S, and 4R/3S (Fig. 1A, panel II). It was particularly striking for the pair 16R/15S, which exhibited many downregulated genes (Fig. 1A, panel II). This was not observed in the other clinical pairs analyzed. Both 14R/13S and 16R/15S had the mutations in the same region, but the transcriptional profile of 14R/13S in cluster II appeared more similar to that of 6R/5S than to that of 16R/15S. Similarly, the profile of 18R/17S was different from those of the other three RD pairs (22R/21S, 12R/11S, and 4R/3S), all of which had the mutations in the same domain as 18R/17S. In conclusion, the domain/region locations of CgPDR1 mutations did not show a direct correlation with the degree of similarity in their transcription profiles based on the microarray analysis.
Downregulation of genes in the clinical isolate Cg16R was unrelated to the GOF mutation of CgPDR1.
Because the clinical pair Cg16R/Cg15S shared a different gene expression pattern from the other pairs (Fig. 1A, panel II), we wished to determine if this was due to a unique effect of the R772I mutation or due to the genetic background in which the mutation occurred. Therefore, the CgPDR1 genes of Cg16R and Cg1S were introduced into the disrupted cgpdr1 locus of CgB4 (cgpdr1 mutant) via an integrative transformation for targeted gene replacement of the disrupted cgpdr1 gene (Fig. 2 B). Microarray analysis was performed to determine the potential impact of Cg16R nonsynonymous CgPDR1 mutation on the transcriptional profiles. Twelve genes upregulated in Cg16R were also upregulated in the complemented strains carrying the GOF CgPDR1 gene of Cg16R, while only three genes were upregulated in the complemented strain carrying the native CgPDR1 gene of Cg1S (Fig. 1B). This indicated that the upregulated gene expression was due to the CgPDR1 GOF mutation from Cg16R. In contrast, only three out of nine genes downregulated in Cg16R were downregulated in the complemented strains (C16R) carrying the GOF CgPDR1 gene of Cg16R. However, the three genes were also downregulated in the complemented strain (C1S) carrying the native CgPDR1 gene from Cg1S. Therefore, downregulation of genes in Cg16R was likely not related to the CgPDR1 GOF mutation.
FIG. 2.
CgPDR1 mutations and fluconazole susceptibilities. (A) Distribution of CgPDR1 mutations. The domains shown were based on the homology between S. cerevisiae Pdr1p and CgPdr1p. Asterisks indicate the locations of mutations. FSTFD is fungus-specific transcription factor domain. Five mutations (boxed) were selected to represent each of the domain/region groups for complementation analysis. (B) Targeted gene replacement via double-crossover homologous recombination; the 2.9-kb HindIII DNA fragment of the CgPDR1 ORF from each of clinical isolates, Cg1S, Cg4R, Cg18R, Cg14R, Cg16R, and Cg6R, was transformed into the Cgpdr1 mutant (CgB4) for targeted gene replacement. Two transformants, a and b, were selected from each complementation for analysis. The solid bar indicates the CgPDR1 ORF. (C) Fluconazole susceptibilities. Solid boxes, clinical isolates; open boxes, laboratory complemented strains. Strains: 84, wild-type strain; Cgpdr1, Cgpdr1 mutant (CgB4); Cg1Sa and Cg1Sb, Cgpdr1 mutant complemented by CgPDR1 from clinical sensitive isolate Cg1S; Cg4Ra and Cg4Rb, Cgpdr1 mutant complemented by CgPDR1 from Cg4R; Cg18Ra and Cg18Rb, Cgpdr1 mutant complemented by CgPDR1 from Cg18R; Cg14Ra and Cg14Rb, Cgpdr1 mutant complemented by CgPDR1 from Cg14R; Cg16Ra and Cg16Rb, Cgpdr1 mutant complemented by CgPDR1 from Cg16R; Cg6Ra and Cg6Rb, Cgpdr1 mutant complemented by CgPDR1 from Cg6R. All susceptibility tests were repeated in triplicate. The standard error of the geometric mean is shown for the isolate in which susceptibilities differed.
Pleiotropic drug resistance genes were upregulated in the majority of seven clinical resistant isolates in the absence of drug challenge.
Hierarchical clustering of the 45 genes according to their expression patterns also revealed that a cluster of 19 genes was upregulated in most of the seven clinical resistant isolates compared to in their paired more-susceptible isolates (Fig. 1A, panel I, and 1C). As C. glabrata and S. cerevisiae are closely related, gene names and annotations from S. cerevisiae were used to categorize the biological process and function of the annotated genes (Table 4) (http://www.yeastgenome.org/). The largest group was “transport,” which included six genes: PDR5 (CAGL0M01760g or CgCDR1), PDR15 (CAGL0F02717g or PDH1 or CgCDR2), SNQ2 (CAGL0I04862g), YOR1 (CAGL0G00242g), YBT1 (CAGL0C03289g), and RSB1 (CAGL0L10142g) (Fig. 1C and Table 4). The second group included ATF2 (CAGL0D05918g), HFD1 (CAGL0K03509g), RTA1 (CAGLK00715g), and ERG4 (CAGL0A00429g) and was categorized as “lipid, fatty acid, and sterol metabolism.” The third group, “transcription,” included three genes: PDR1 (CAGL0A00451g), RPN4 (CAGL0K01727g), and MEC3 (CAGL0M09735g). CgPDR1 is known to regulate the expression of CgCDR1 (PDR5) and PDH1 (PDR15) as well as CgSNQ2. CgCDR1 encodes the major fluconazole transporter in C. glabrata (21). PDH1 and CgSNQ2 have also been reported to be involved in the efflux of fluconazole in C. glabrata (10, 26). The last four genes included two genes homologous to uncharacterized genes in S. cerevisiae, YJL163C (CAGL0M08426g) and YIL077C (CAGL0M12947g), as well as two genes (CAGL0G01122g and CAGL0M14091g) which have no homolog in S. cerevisiae.
TABLE 4.
Annotated biological process and function of the 18 upregulated genes
| Group | Gene descriptiona |
|---|---|
| Transport | |
| PDR5 | ABC multidrug transporter involved in cellular detoxification, steroid transport, and cation resistance |
| PDR15 | ABC multidrug transporter involved in cellular detoxification |
| SNQ2 | ABC multidrug transporter involved in multidrug resistance |
| YOR1 | ABC multidrug transporter involved in cellular detoxification |
| YBT1 | ABC transporter involved in bile acid transport |
| RSB1 | Sphingolipid transporter |
| Lipid, fatty acid, and sterol metabolism | |
| ATF2 | Alcohol acetyltransferase and may be involved in steroid detoxification |
| HFD1 | Putative fatty acid aldehyde dehydrogenase |
| RTA1 | Overexpression confers 7-aminocholesterol resistance |
| ERG4 | C-24(28) sterol reductase, catalyzes the final step in ergosterol biosynthesis |
| Transcription | |
| PDR1 | Transcription factor of multidrug resistance |
| RPN4 | Transcription factor of proteasome genes and transcriptionally regulated by various stress responses |
| MEC3 | DNA damage and meiotic pachytene checkpoint protein, response to stress |
| Biological function unknown | |
| YJL163C | |
| YIL077C | |
| YNL134C | |
| Uncharacterized | |
| CAGL0G01122g | No S. cerevisiae homolog |
| CAGL0M14091g | No S. cerevisiae homolog |
Candida glabrata gene annotation by Génolevures based on homology with S. cerevisiae.
Multiple Em (expectation-maximization algorithm) for motif elicitation (MEME) (1) was used to analyze the upstream sequences of the 18 annotated genes. The motif analysis revealed that 17 out of 18 genes (Fig. 1B), except CgERG4, contained the putative pleiotropic drug response element (PDRE) motif (TCCRYGGA) in their 1-kb upstream regions. The canonical sequence “TCCACGGA” appeared at the highest frequency, and “TCCGTGGA” occurred the second most frequently (Table 5). A motif comparison using the TOMTOM tool (24) matched the motif with the PDRE motif of Pdr1p/Pdr3p in the S. cerevisiae promoter database (SCPD) (P value of 9.9 × 10−5), in which “TCCGCGGA” is the major PDRE motif. In summary, the upregulated expression of pleiotropic drug resistance genes in the clinical resistant isolates analyzed represented the critical elements of C. glabrata's response to xenobiotic stress. This response remained stable even when the cells were cultured in the absence of drug.
TABLE 5.
Putative pleiotropic drug response elements identified in the upstream region of the 17 genes
| C. glabrata gene | S. cerevisiae genea | Putative PDREb | Startc |
|---|---|---|---|
| CAGL0A00451g | PDR1 | TCCGTGGA | −558 |
| TCCACGGA | −702 | ||
| CAGL0C03289g | YBT1 | TCCACGGG | −451 |
| CAGL0D05918g | ATF2 | TCCGCGGA | −196 |
| TCCACGGA | −561 | ||
| TCCACGGA | −723 | ||
| CAGL0F02717g (PDH1) | PDR15 | TCCACGGA | −522 |
| TCCGTGGA | −558 | ||
| CAGL0G00242g | YOR1 | TCCGTGGA | −649 |
| CAGL0G01122g | TCCATGGA | −794 | |
| TCCATGGA | −804 | ||
| CAGL0I04862g | SNQ2 | TCCACGGA | −219 |
| TCCACGGG | −793 | ||
| CAGL0K00715g | RTA1 | TCCACGGA | −301 |
| TCCGCGGA | −380 | ||
| CAGL0K01727g | RPN4 | TCCGTGGA | −379 |
| TCCGTGGA | −395 | ||
| TCCACGGA | −553 | ||
| CAGL0K09702 | YNL134C | TCCACGGA | −610 |
| CAGL0K03509g | HFD1 | TCCGTGGA | −219 |
| CAGL0L10142g | RSB1 | TCCGTGGA | −882 |
| TCCACGGA | −986 | ||
| CAGL0M01760g (CgCDR1) | PDR5 | TCCACGGG | −134 |
| TCCACGGG | −228 | ||
| TCCACGGA | −388 | ||
| TCCACGGA | −516 | ||
| TCCACGGA | −970 | ||
| CAGL0M08426g | YIJ163C | TCCGTGGA | −451 |
| CAGL0M09735g | MEC3 | TCCGTGGA | −111 |
| TCCACGGA | −163 | ||
| CAGL0M12947g | YIL077C | TCCGTGGA | −473 |
| TCCACGGA | −503 | ||
| CAGL0M14091g | TCCACGGA | −245 | |
| TCCATGGA | −533 |
Candida glabrata gene annotation by Génolevures based on homology with S. cerevisiae.
P < 0.000045.
Nucleotide positions from translation initiation codon, ATG.
GOF CgPDR1 mutations led to increased fluconazole resistance in the same host.
Five resistant isolates and one susceptible isolate were selected for the gene replacement study to investigate whether the CgPDR1 mutations of various domains/regions contributed to the increased azole resistance. The CgPDR1 mutations were introduced into the disrupted cgpdr1 locus of CgB4 via an integrative transformation to replace the disrupted cgpdr1 gene (Fig. 2B). The clinical resistant isolates, Cg4R and Cg18R, were chosen to represent the group with mutations in the regulatory domain. The isolates Cg14R and Cg16R represented the group with the mutations in the undefined region, and the isolate Cg6R represented the group with the mutation in the activation domain. The clinical sensitive isolate, Cg1S, had no CgPdr1p amino acid substitution and served as the reference strain. The CgPDR1 mutations were introduced into the cgpdr1 locus by targeted gene replacement via a double-crossover homologous recombination. Two independent transformants, labeled “a” and “b,” were selected from each transformation for consistency confirmation, and the CgPDR1 mutations introduced were confirmed by DNA sequencing. The transformants were then subjected to fluconazole susceptibility analysis and compared to their corresponding clinical isolates (Fig. 2C). The two complemented strains, C1Sa and C1Sb, carrying the native copy of CgPDR1 from Cg1S had the fluconazole MIC80 restored to the level of the wild-type laboratory strain, NCCLS84, at 64 μg/ml. In contrast, the complemented strains carrying the CgPDR1 GOF mutations (Cg4Ra, Cg4Rb, Cg18Ra, Cg18Rb, Cg14Ra, Cg14Rb, Cg16Ra, Cg16Rb, Cg6Ra, and Cg6Rb) showed 2- to 8-fold increases in their resistance to fluconazole compared to the complemented strains without a GOF mutation: Cg1Sa and Cg1Sb. MICs of the resistant strains ranged from 128 to 512 μg/ml. All five GOF mutations resulted in increased fluconazole resistance. In conclusion, the increased fluconazole resistance in the complemented strains was due to the CgPDR1 GOF mutations.
CgPDR1 mutations differentially regulated the expression of the pleiotropic drug resistance genes.
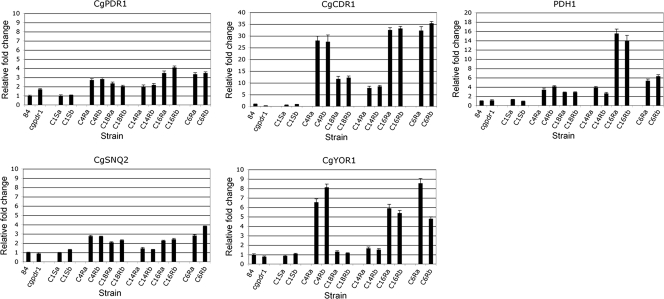
The expression of CgPDR1 and its target transporter genes, CgCDR1, PDH1, CgSNQ2, and CgYOR1, in the CgPDR1-complemented strains was determined by qRT-PCR. Microarray data showed that these genes were upregulated in majority of the clinical resistant isolates and likely to be the factors contributing to the increased fluconazole resistance in the complemented strains as well as in the clinical resistant isolates with the CgPDR1 mutations. The qRT-PCR results showed that the expression of CgPDR1, CgCDR1, and PDH1 was upregulated only in the complemented strains carrying the CgPDR1 GOF mutations and not in the complemented strains carrying the native CgPDR1 (Fig. 3). The wild-type strain, NCCLS84, was used as the reference, and the CgACT1 gene is used as the internal control. Disruption of CgPDR1 resulted in a slight increase in truncated cgpdr1 mRNA as observed previously (28). Integration of the native CgPDR1 copy restored and reduced the CgPDR1 expression level to that of the wild-type strain, NCCLS84. In contrast, integration of the CgPDR1 GOF mutations increased the CgPDR1 expression. Therefore, the expression level of CgPDR1 was modulated by the CgPDR1 mutations directly or indirectly. The expression of CgCDR1 was decreased in the Cgpdr1 mutant, as CgCDR1 is one of CgPDR1 primary downstream targets. Similar to the microarray analysis finding, the expression of both CgCDR1 and PDH1 was upregulated in all the complemented strains carrying the CgPDR1 GOF mutations. However, the upregulated expression levels of CgCDR1 and PDH1 were not parallel. The expression level of CgCDR1 correlated variably with the expression level of functional CgPDR1. The complemented strains with a higher expression of CgPDR1 seemed to always lead to higher expression levels of CgCDR1, which ranged from 8- to 35-fold increases compared to those of the wild-type strain, NCCLS84. In contrast to CgCDR1, the impact of CgPDR1 GOF mutations on PDH1 was much weaker in most of the cases, with increases no greater than 7-fold, with the exception of Cg16Ra and Cg16Rb, which had 14- to 16-fold increases. CgSNQ2 and CgYOR1, like CgCDR1 and PDH1, are also downstream targets of CgPDR1. However, not all the CgPDR1 GOF mutations led to increases in the expression of CgSNQ2 and CgYOR1. Similar to PDH1, the CgPDR1 GOF mutations had a weaker impact on the expression of CgSNQ2, which was no greater than a 4-fold increase, and only marginal increases were seen in Cg14Ra and Cg14Rb. The upregulated expression pattern of CgYOR1 seemed more parallel to that of CgCDR1 except for the marginal increases in Cg18Ra, Cg18Rb, Cg14Ra, and Cg14Rb. Therefore, the CgPDR1 GOF mutations regulated the expression of various pleiotropic drug resistance genes, CgCDR1, PDH1, and CgSNQ2 and CgYOR1, differently, and these differences in the gene expression did not correlate with the putative protein domains of GOF location and differed between the GOF mutations in the same domain.
FIG. 3.
qRT-PCR analysis of CgPDR1, CgCDR1, PDH1, CgSNQ2, and CgYOR1 expression. The expression of pleiotropic drug resistance genes was analyzed by qRT-PCR. CgACT1 was used as an internal control for normalization. The fold differences of gene expression were compared to the wild-type strain 84, for which the expression is considered 100% and is represented by 1 as the baseline for all.
DISCUSSION
In this report, the DNA microarray was used as a tool for analyzing the potential drug resistance mechanisms in C. glabrata. A genome-wide transcriptional profiling showed the upregulated expression of the pleiotropic drug resistance genes, CgPDR1, CgCDR1, PDH1, CgSNQ2, and CgYOR1. No altered expression of the azole target CgERG11 was observed. The involvement of a PDR-mediated drug resistance mechanism in the clinical resistant isolates was further confirmed by molecular genetic analysis. The PDR-mediated drug resistance was concluded to be the predominant mechanism of clinical azole resistance among the C. glabrata oropharyngeal isolates we analyzed from hematopoietic stem cell transplant recipients who received fluconazole prophylaxis.
Microarray analysis was used by Vermitsky et al. to determine the effect of a GOF mutant (F15) selected by exposure of an azole-susceptible laboratory strain of C. glabrata (29). The 78 genes upregulated in that study included 16 of the 18 genes in our microarray study, those 16 genes selected by their upregulation in a majority of seven clinical resistant isolates compared to their paired sensitive clinical isolates. The two exceptions found in our microarray analysis were CgERG4 and CgSNQ2. CgSNQ2 is a known CgPDR1 target that was not upregulated in F15 (29) and not uniformly upregulated in published studies of other azole-resistant isolates (8). Seventeen out of the 18 annotated upregulated genes in our study had the putative PDRE(s) in their promoter regions, with the exception of CgERG4 (Table 5). Similar but nonidentical PDRE sequences have been identified in S. cerevisiae (6) and C. glabrata (8, 29). These sequences were confirmed here by MEME, which identifies common upstream motifs. Similar to what Vermitsky et al. identified in the laboratory strain F15, we discovered that the major upregulated gene groups are involved in transport, transcription, and metabolism of lipids, sterols, or fatty acids (29).
It is striking that all seven resistant isolates in our matched pairs had a single point mutation in the coding region of CgPDR1. Ferrari et al. found single amino acid substitutions in CgPDR1 of 57 among 77 azole-resistant isolates (8). The mutations that accounted for the azole resistance were confirmed by gene replacement in nine strains. In our study, gene replacement using CgPDR1 from five strains also demonstrated that the azole resistance was due to the CgPDR1 GOF mutations (Fig. 2 and 3). We found that the mutations did not lead to a coordinated regulation of three CgPDR1 target genes, CgCDR1, PDH1, and CgSNQ2 (Fig. 3). In addition, there was no relationship between the target effect and protein domain of the amino acid substitution. We did find that the CgPDR1 GOF mutations increased at least 2-fold the expression of CgPDR1 itself in all five replacements from our resistant isolates, whereas that degree of apparent autoupregulation was found in only 2 of 21 matched pairs studied by Ferrari et al. (8). The lack of coordinated upregulation between all CgPDR1 targets, including variable autoupregulation, indicates that factors other than increased transcriptional activity of CgPDR1 are also regulating transcriptional activity. CgPDR1 GOF mutations likely affect interactions with promoter elements, transcriptional factors, and subunits of the mediator complex.
The ability of GOF mutations to arise from such a broad area of this 126-kDa protein requires explanation. It has been shown that azoles can bind to a discrete domain of CgPdr1p, facilitating binding to the mediator complex, resulting in recruitment of RNA polymerase II to the promoter complex (25). However, it is clear that GOF mutations of CgPDR1 in C. glabrata and PDR1 or PDR3 in S. cerevisiae do not require azole to exert their regulatory effect (6). Whatever the mechanism by which transcriptional activity is increased, the effect on at least four of the targets we studied varied between mutations. The similarity of this transcriptional regulatory process between C. glabrata, S. cerevisiae, and C. albicans suggests a more general phenomenon that is certainly worthy of study.
Clearly, not all azole resistance arising in clinical isolates of C. glabrata can be explained by amino acid substitutions in CgPDR1 (8). The role of other mutations in causing azole resistance in clinical strains of C. glabrata remains undefined. Mutations in ERG11, which codes for the azole target, ergosterol C14-α-demethylase, have caused resistance in Aspergillus fumigatus (2) and some strains of C. albicans (22). Neither the study by Sanguinetti et al. (23) of four matched pairs nor our study of seven matched pairs (data not shown) found any increase in ERG11 expression in the azole-resistant isolates or nonsynonymous mutations in the ERG11 ORF of the resistant isolates. The finding of ERG11 (CYP51) gene duplication in an azole-resistant C. glabrata isolate by Marichal et al. (13) has not been reported by others. Mitochondrial deficiency, that may be related to upregulation of ABC transport genes, was found in four azole-resistant isolates by Ferrari et al. (8), but none of our seven matched pairs failed to grow on nonfermentable carbon sources. The infrequent observation of mutations other than those in CgPDR1 may be due to the reduced ability to conserve or improve fitness as seen by the increased mouse virulence of GOF mutants, which conferred azole resistance (8). The requirements of fitness may vary between body sites. It may be significant that sterol-dependent strains with increased azole resistance have been isolated only from urine (9). Defects in the sterol biosynthesis pathway of six such isolates have been identified (3). Because these isolates must be tested in the presence of sterols and the fact that sterols increase azole resistance in susceptible isolates, the degree of resistance is more difficult to assess.
Expression analysis by microarray confirmed that the majority of upregulated genes were explicable as CgPdr1p targets. Downregulated genes varied considerably in pattern, apparently independent of the domain in which the deduced amino acid mutation occurred. The striking difference in the downregulated genes shown in the microarray of Cg16R (Fig. 1A, panel I) was not seen when the R772I GOF mutation found in Cg16R was introduced into a different genetic background (Fig. 1B). Until the downstream effects of CgPdr1p are better understood, the relevance of downregulation to azole resistance remains unclear. The mutations of CgPDR1 had various degrees of impact on the expression of its PDR gene targets. Slightly different PDRE motifs and various numbers of the motifs were observed in the promoter regions of these target genes, which might influence the interaction between the promoter and CgPdr1p. Future analysis of the interaction between the different putative PDRE motifs and different mutated CgPdr1ps along with the wild-type CgPdr1p will be helpful in elucidating the DNA-protein interaction as well as the regulatory mechanisms underlying the azole resistance mechanism in of C. glabrata.
Acknowledgments
We thank Kathleen Meyer for her assistance in array data submission to GEO and Quentin Li and Jason Noble for their critical reading of the manuscript.
This work was supported by the Intramural Research Program from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print on 14 June 2010.
REFERENCES
- 1.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 2.Balashov, S. V., R. Gardiner, S. Park, and D. S. Perlin. 2005. Rapid, high-throughput, multiplex, real-time PCR for identification of mutations in the cyp51A gene of Aspergillus fumigatus that confer resistance to itraconazole. J. Clin. Microbiol. 43:214-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bard, M., A. M. Sturm, C. A. Pierson, S. Brown, K. M. Rogers, S. Nabinger, J. Eckstein, R. Barbuch, N. D. Lees, S. A. Howell, and K. C. Hazen. 2005. Sterol uptake in Candida glabrata: rescue of sterol auxotrophic strains. Diagn. Microbiol. Infect. Dis. 52:285-293. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, J. E., K. Izumikawa, and K. A. Marr. 2004. Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrob. Agents Chemother. 48:1773-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delahodde, A., R. Pandjaitan, M. Corral-Debrinski, and C. Jacq. 2001. Pse1/Kap121-dependent nuclear localization of the major yeast multidrug resistance (MDR) transcription factor Pdr1. Mol. Microbiol. 39:304-312. [DOI] [PubMed] [Google Scholar]
- 6.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 7.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari, S., F. Ischer, D. Calabrese, B. Posteraro, M. Sanguinetti, G. Fadda, B. Rohde, C. Bauser, O. Bader, and D. Sanglard. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 5:e1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazen, K. C., J. Stei, C. Darracott, A. Breathnach, J. May, and S. A. Howell. 2005. Isolation of cholesterol-dependent Candida glabrata from clinical specimens. Diagn. Microbiol. Infect. Dis. 52:35-37. [DOI] [PubMed] [Google Scholar]
- 10.Izumikawa, K., H. Kakeya, H. F. Tsai, B. Grimberg, and J. E. Bennett. 2003. Function of Candida glabrata ABC transporter gene, PDH1. Yeast 20:249-261. [DOI] [PubMed] [Google Scholar]
- 11.Kitada, K., E. Yamaguchi, and M. Arisawa. 1996. Isolation of a Candida glabrata centromere and its use in construction of plasmid vectors. Gene 175:105-108. [DOI] [PubMed] [Google Scholar]
- 12.Koszul, R., A. Malpertuy, L. Frangeul, C. Bouchier, P. Wincker, A. Thierry, S. Duthoy, S. Ferris, C. Hennequin, and B. Dujon. 2003. The complete mitochondrial genome sequence of the pathogenic yeast Candida (Torulopsis) glabrata. FEBS Lett. 534:39-48. [DOI] [PubMed] [Google Scholar]
- 13.Marichal, P., H. Vanden Bossche, F. C. Odds, G. Nobels, D. W. Warnock, V. Timmerman, C. Van Broeckhoven, S. Fay, and P. Mose-Larsen. 1997. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 41:2229-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morschhäuser, J. 2010. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 47:94-106. [DOI] [PubMed] [Google Scholar]
- 15.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaller, M. A., and D. J. Diekema. 2004. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin. Microbiol. Infect. 10(Suppl. 1):11-23. [DOI] [PubMed] [Google Scholar]
- 17.Pfaller, M. A., R. N. Jones, G. V. Doern, H. S. Sader, S. A. Messer, A. Houston, S. Coffman, R. J. Hollis, and the Sentry Participant Group. 2000. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997-1998. Antimicrob. Agents Chemother. 44:747-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaller, M. A., S. A. Messer, L. Boyken, R. J. Hollis, C. Rice, S. Tendolkar, and D. J. Diekema. 2004. In vitro activities of voriconazole, posaconazole, and fluconazole against 4,169 clinical isolates of Candida spp. and Cryptococcus neoformans collected during 2001 and 2002 in the ARTEMIS global antifungal surveillance program. Diagn. Microbiol. Infect. Dis. 48:201-205. [DOI] [PubMed] [Google Scholar]
- 19.Pfaller, M. A., S. A. Messer, L. Boyken, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2003. Variation in susceptibility of bloodstream isolates of Candida glabrata to fluconazole according to patient age and geographic location. J. Clin. Microbiol. 41:2176-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., and W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanguinetti, M., B. Posteraro, B. Fiori, S. Ranno, R. Torelli, and G. Fadda. 2005. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob. Agents Chemother. 49:668-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakur, J. K., H. Arthanari, F. Yang, S. J. Pan, X. Fan, J. Breger, D. P. Frueh, K. Gulshan, D. K. Li, E. Mylonakis, K. Struhl, W. S. Moye-Rowley, B. P. Cormack, G. Wagner, and A. M. Naar. 2008. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452:604-609. [DOI] [PubMed] [Google Scholar]
- 26.Torelli, R., B. Posteraro, S. Ferrari, M. La Sorda, G. Fadda, D. Sanglard, and M. Sanguinetti. 2008. The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol. Microbiol. 68:186-201. [DOI] [PubMed] [Google Scholar]
- 27.Trick, W. E., S. K. Fridkin, J. R. Edwards, R. A. Hajjeh, and R. P. Gaynes. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35:627-630. [DOI] [PubMed] [Google Scholar]
- 28.Tsai, H. F., A. A. Krol, K. E. Sarti, and J. E. Bennett. 2006. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50:1384-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vermitsky, J. P., K. D. Earhart, W. L. Smith, R. Homayouni, T. D. Edlind, and P. D. Rogers. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol. Microbiol. 61:704-722. [DOI] [PubMed] [Google Scholar]
- 30.Vermitsky, J. P., and T. D. Edlind. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 48:3773-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]