Abstract
The in vitro activity of azithromycin against 1,237 nontyphoidal Salmonella enterica isolates collected from Finnish patients between 2003 and 2008 was investigated. Only 24 (1.9%) of the isolates tested and 15 (5.1%) of the 294 isolates with reduced fluoroquinolone susceptibility had azithromycin MICs of ≥32 μg/ml. These data show that azithromycin has good in vitro activity against nontyphoidal S. enterica, and thus, it may be a good candidate for clinical treatment studies of salmonellosis.
Salmonella is one of the most common causes of food-borne illnesses and a major cause of human infections all over the world (23). Salmonella infections are usually treated with fluoroquinolones or extended-spectrum cephalosporins. Unfortunately, excessive use of fluoroquinolones both in human and in veterinary medicine has led to increasing numbers of resistant isolates, including nontyphoidal strains of Salmonella enterica (18, 19, 28). In addition, the nonclassical quinolone resistance phenotype (the Qnr phenotype), showing reduced susceptibility to ciprofloxacin (MIC of ≥0.125 μg/ml) but susceptibility or only low-level resistance to nalidixic acid (MIC of ≤32 μg/ml), has become more common (6, 14, 19, 20, 22). Extended-spectrum β-lactamase (ESBL) producers have emerged in Enterobacteriaceae and in Salmonella, and there are reports of the coappearance of ESBL and qnr genes in the same transferable genetic elements (5, 10, 21, 27). These resistance problems may jeopardize the treatment of severe Salmonella infections. Thus, alternative antibiotics for the treatment of Salmonella infections are needed.
Salmonella isolates are intrinsically resistant to erythromycin via active efflux (2) but naturally susceptible to azithromycin (29), which is a 15-membered erythromycin derivative. Resistance to macrolides is usually conferred by mutations in nucleotides A2058 and A2059 of the 23S rRNA, according to the Escherichia coli numbering (26). Also, the alteration of the 50S ribosomal subunit proteins L4 (rlpD) and L22 (rlpV) may lead to macrolide resistance (4).
The purpose of the present study was to determine the in vitro activity of azithromycin against nontyphoidal Salmonella isolates collected between 2003 and 2008 from Finnish patients. Special attention was paid to isolates with reduced fluoroquinolone susceptibility or showing the Qnr phenotype. In addition, mutations in the 23S rRNA and in the L4 and L22 ribosomal proteins were investigated.
(This work was presented in part at the 19th European Congress of Clinical Microbiology and Infectious Diseases [ECCMID], Helsinki, Finland, 2009 [P1043].)
A total of 1,237 nontyphoidal Salmonella isolates (638 domestic and 599 foreign) collected from Finnish patients between 2003 and 2008 were included in this study. Starting in January each year, we collected the first 100 domestic and the first 100 foreign, i.e., collected from Finnish travelers returning from abroad, Salmonella isolates. The strains were serotyped at the National Salmonella Reference Centre in Finland.
The MICs of antimicrobial agents for the isolates were determined by the agar dilution method according to the CLSI guidelines (8). Mueller-Hinton II agar (Becton Dickinson, Cockeysville, MD) was used as the culture medium. All 1,237 Salmonella isolates were tested for susceptibility to ciprofloxacin, nalidixic acid, and azithromycin (all from Sigma, Steinheim, Germany). We tested 809 selected isolates for erythromycin (Sigma) and 635 for telithromycin (Sanofi Aventis, Paris, France) susceptibility. Control strains for susceptibility testing were as described previously (14). On the basis of earlier publications (1, 16, 17), the MIC breakpoint chosen for reduced ciprofloxacin susceptibility was ≥0.125 μg/ml. For nalidixic acid, CLSI breakpoints were used (9). Based on the EUCAST recommendation for S. enterica serovar Typhi (www.eucast.org) and a previous publication (3), the epidemiological cutoff value chosen for azithromycin was ≥32 μg/ml. The susceptibility data were analyzed by using the WHONET 5.4 computer program.
Pyrosequencing was used to detect macrolide resistance causing point mutations in the ribosomal target sites A2058 and A2059 (E. coli numbering) of the 23S rRNA in 22 isolates with azithromycin MICs of ≥32 μg/ml, 44 isolates belonging to the Qnr phenotype, and 73 isolates showing erythromycin MICs of ≥32 μg/ml. Pyrosequencing was performed with previously described primers and protocols (15) using a PSQ 96MA pyrosequencer.
The 50S ribosomal proteins L4 and L22 were amplified, and mutations were screened in 24 isolates having azithromycin MICs of ≥32 μg/ml, of which 6 were of the Qnr phenotype. In addition, 13 isolates having only erythromycin MICs of ≥32 μg/ml and showing the Qnr phenotype were tested. DNA was prepared as described above. The specific primers used for amplification of the complete L4 and L22 genes rlpD and rlpV were Salm_L4_f (5′-TGAAGGCGTAAGGGGATAGCA-3′) and Salm_L4_r (5′-TCAGCAGA CGTTCTTCACGAA-3′) and Salm_L22_f (5′-GAAATAAGGTAG GAGGAAGAG-3′) and Salm_L22_r (5′-CCATTGCTAGTCTCCAGAGTC-3′). The PCR conditions were as follows: 94°C for 10 min, 94°C for 30 s, 56°C for 30 s, and 72°C for 60 s for 33 cycles. Any L4- or L22-positive results were confirmed by direct sequencing of both strands of amplicons using specific PCR primers as previously described (24). Amino acid sequences were then compared with the known rlpD and rlpV genes by a BLAST search through the European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/blast/).
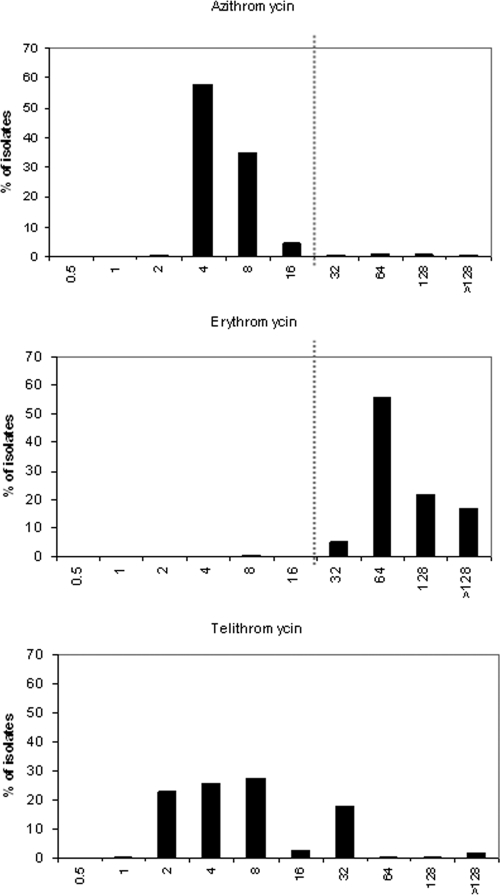
Two different populations of S. enterica were detected regarding the azithromycin MIC distribution. The majority of the isolates had azithromycin MICs of 4 to 8 μg/ml, i.e., representing the wild-type population, whereas a minority of the isolates had MICs of 32 to ≥128 μg/ml, i.e., over the epidemiological cutoff value. Between 2003 and 2008, 24 (1.9%) of the 1,237 isolates had azithromycin MICs of ≥32 μg/ml (Fig. 1; Table 1). Nine (1.4%) of the 638 domestic isolates and 15 (2.1%) of the 599 foreign isolates had azithromycin MICs over the epidemiological cutoff value.
FIG. 1.
Histograms of azithromycin (n = 1,237), erythromycin (n = 809), and telithromycin (n = 635) MICs for S. enterica isolates collected from Finnish travelers between 2003 and 2008. The vertical line represents the resistance breakpoint.
TABLE 1.
Twenty-four S. enterica isolates showing azithromycin MICs of ≥32 μg/ml
| Isolate no. | Yr | Strain | Origin | S. enterica serovar | AZMa MIC (μg/ml) | qnr geneb |
|---|---|---|---|---|---|---|
| 1 | 2003 | s2099 | Thailand | Newport | 32 | − |
| 2 | 2003 | s2018 | Thailand | Stanley | 64 | + |
| 3 | 2003 | s2021 | Thailand | Rissen | >128 | − |
| 4 | 2003 | s2085 | Thailand | Stanley | 64 | + |
| 5 | 2003 | s2086 | Thailand | Stanley | 64 | + |
| 6 | 2003 | s2181 | Finland | Poona | 32 | − |
| 7 | 2003 | s2137 | Finland | Typhimurium | 128 | − |
| 8 | 2003 | s2195 | Finland | Virchow | >128 | − |
| 9 | 2004 | s2236 | Thailand | Stanley | 32 | + |
| 10 | 2004 | s2265 | Thailand | Stanley | 128 | + |
| 11 | 2004 | s2280 | Thailand | Stanley | 64 | + |
| 12 | 2004 | s2389 | Finland | Typhimurium | 128 | − |
| 13 | 2004 | s2391 | Finland | Typhimurium | 128 | − |
| 14 | 2005 | s2439 | Egypt | Blockley | 64 | − |
| 15 | 2005 | s2477 | Egypt | Bredeney | >128 | − |
| 16 | 2005 | s2608 | Finland | Blockley | 64 | − |
| 17 | 2006 | s2635 | Thailand | Emek | 64 | − |
| 18 | 2006 | s2768 | Finland | Blockley | 128 | − |
| 19 | 2006 | s2829 | Finland | Saintpaul | 64 | − |
| 20 | 2007 | s2868 | Malaysia | Stanley | 128 | − |
| 21 | 2007 | s2948 | Finland | Hvittingfoss | 128 | − |
| 22 | 2008 | s3141 | Thailand | Rissen | 64 | − |
| 23 | 2008 | s3082 | Thailand | Typhimurium | 128 | − |
| 24 | 2008 | s3139 | Thailand | Enteritidis | 128 | − |
AZM, azithromycin.
+, present; −, absent.
Two hundred ninety-four (23.8%) S. enterica isolates showed reduced fluoroquinolone susceptibility, and 53 (18.0%) of them showed the Qnr phenotype. Among the isolates with reduced fluoroquinolone susceptibility, 4 (5.2%) domestic and 11 (5.1%) foreign isolates had azithromycin MICs of ≥32 μg/ml. Among the Qnr phenotype isolates, six (11.3%) had azithromycin MICs of ≥32 μg/ml (Table 1). Azithromycin MICs of ≥32 μg/ml were detected among 12 different serovars, S. enterica serovar Stanley being the most common one, and all isolates with a Qnr phenotype belonged to S. enterica serovar Stanley (Table 1).
Of the S. enterica isolates tested, 99.6% (806/809) had erythromycin MICs of ≥32 μg/ml. The erythromycin MICs varied between 8 and ≥128 μg/ml, and the vast majority of the isolates had erythromycin MICs of ≥64 μg/ml (Fig. 1).
No mutations in A2058 or A2059 of the 23S rRNA gene were detected among any of the isolates tested. Sequencing of the 50S ribosomal proteins L4 and L22 revealed G235A and C379T mutations in the rlpD gene and G25A in the rlpV gene, respectively. A Glu79-Lys substitution in the rlpD gene was found in six isolates belonging to three different serovars (Table 2). An Arg127-Trp substitution in the rlpD gene was found in three S. enterica serovar Montevideo isolates, which also had an Asp9-Asn substitution outside the coding region (Table 2).
TABLE 2.
Mutations found in the 50S ribosomal proteins L4 and L22 and corresponding MICs
| S. enterica serovar | Origin | Yr | Mutation in L4 and L22 |
MICa |
|||||
|---|---|---|---|---|---|---|---|---|---|
| rlpD | rlpV | AZM | ERY | TEL | CIP | NAL | |||
| Blockley | Egypt | 2005 | G235A | 64 | >128 | >128 | 0.5 | >512 | |
| Blockley | Finland | 2005 | G235A | 64 | >128 | >128 | 0.25 | >512 | |
| Blockley | Finland | 2006 | G235A | 128 | >128 | >128 | 0.25 | >512 | |
| Saintpaul | Finland | 2006 | G235A | 64 | >128 | >128 | 0.03 | 16 | |
| Typhimurium | Thailand | 2008 | G235A | 128 | >128 | >128 | 0.06 | 16 | |
| Typhimurium | Finland | 2004 | G235A | 128 | 64 | NDb | 0.25 | 512 | |
| Montevideo | Thailand | 2004 | C379T | G25A | 4 | 64 | 32 | 0.5 | 32 |
| Montevideo | Thailand | 2007 | C379T | G25A | 8 | >128 | 32 | 0.5 | 16 |
| Montevideo | Thailand | 2008 | C379T | G25A | 8 | >128 | 4 | 0.5 | 16 |
AZM, azithromycin; ERY, erythromycin; TEL, telithromycin; CIP, ciprofloxacin; NAL, nalidixic acid.
ND, not determined.
Salmonella isolates are intrinsically resistant to erythromycin via the AcrAB efflux pump (2) but naturally susceptible to azithromycin (29). Azithromycin has shown good efficacy in the treatment of patients suffering from typhoid fever (7, 12, 13, 25, 30), and there has been speculation that azithromycin could be used for empirical therapy of traveler's diarrhea (11, 31). The present study was performed to determine the in vitro activity of azithromycin toward nontyphoidal Salmonella isolates collected from Finnish patients between 2003 and 2008. Our results show that while nearly all (99.6%) of our S. enterica isolates had erythromycin MICs of ≥32 μg/ml, only 24 isolates had azithromycin MICs of ≥32 μg/ml. Azithromycin showed good in vitro efficacy also against S. enterica isolates with reduced fluoroquinolone susceptibility, although 11.3% of the Qnr phenotype isolates showed azithromycin MICs of ≥32 μg/ml.
These data show that azithromycin has good in vitro activity against nontyphoidal S. enterica isolates. Although highly azithromycin-resistant isolates did occur, azithromycin was effective even against the isolates with reduced fluoroquinolone susceptibility, including those showing the Qnr phenotype. Based on these results, azithromycin is a good candidate for clinical treatment studies of salmonellosis.
Acknowledgments
We thank Minna Lamppu, Toni Huovinen, and Erkki Nieminen for their skillful technical assistance.
This work has been financially supported by the EVO Research Fund of the Turku University Central Hospital and the Turku University Foundation (to M.G.).
We have no conflicts of interest to declare.
Footnotes
Published ahead of print on 24 May 2010.
REFERENCES
- 1.Aarestrup, F. M., C. Wiuff, K. Molbak, and E. J. Threlfall. 2003. Is it time to change fluoroquinolone breakpoints for Salmonella spp.? Antimicrob. Agents Chemother. 47:827-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braoudaki, M., and A. C. Hilton. 2005. Mechanisms of resistance in Salmonella enterica adapted to erythromycin, benzalkonium chloride and triclosan. Int. J. Antimicrob. Agents 25:31-37. [DOI] [PubMed] [Google Scholar]
- 3.Butler, T. 2001. Effect of increased inoculum of Salmonella typhi on MIC of azithromycin and resultant growth characteristics. J. Antimicrob. Chemother. 48:903-906. [DOI] [PubMed] [Google Scholar]
- 4.Caldwell, D. B., Y. Wang, and J. Lin. 2008. Development, stability, and molecular mechanisms of macrolide resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 52:3947-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattoir, V., L. Poirel, V. Rotimi, C. J. Soussy, and P. Nordmann. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60:394-397. [DOI] [PubMed] [Google Scholar]
- 6.Cavaco, L. M., R. S. Hendriksen, and F. M. Aarestrup. 2007. Plasmid-mediated quinolone resistance determinant qnrS1 detected in Salmonella enterica serovar Corvallis strains isolated in Denmark and Thailand. J. Antimicrob. Chemother. 60:704-706. [DOI] [PubMed] [Google Scholar]
- 7.Chinh, N. T., C. M. Parry, N. T. Ly, H. D. Ha, M. X. Thong, T. S. Diep, J. Wain, N. J. White, and J. J. Farrar. 2000. A randomized controlled comparison of azithromycin and ofloxacin for treatment of multidrug-resistant or nalidixic acid-resistant enteric fever. Antimicrob. Agents Chemother. 44:1855-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2007. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—7th edition, M07-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing: 17th informational supplement, M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Doublet, B., S. A. Granier, F. Robin, R. Bonnet, L. Fabre, A. Brisabois, A. Cloeckaert, and F. X. Weill. 2009. Novel plasmid-encoded ceftazidime-hydrolyzing CTX-M-53 extended-spectrum beta-lactamase from Salmonella enterica serotypes Westhampton and Senftenberg. Antimicrob. Agents Chemother. 53:1944-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuPont, H. L. 2009. Clinical practice. Bacterial diarrhea. N. Engl. J. Med. 361:1560-1569. [DOI] [PubMed] [Google Scholar]
- 12.Frenck, R. W., Jr., A. Mansour, I. Nakhla, Y. Sultan, S. Putnam, T. Wierzba, M. Morsy, and C. Knirsch. 2004. Short-course azithromycin for the treatment of uncomplicated typhoid fever in children and adolescents. Clin. Infect. Dis. 38:951-957. [DOI] [PubMed] [Google Scholar]
- 13.Girgis, N. I., T. Butler, R. W. Frenck, Y. Sultan, F. M. Brown, D. Tribble, and R. Khakhria. 1999. Azithromycin versus ciprofloxacin for treatment of uncomplicated typhoid fever in a randomized trial in Egypt that included patients with multidrug resistance. Antimicrob. Agents Chemother. 43:1441-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunell, M., M. A. Webber, P. Kotilainen, A. J. Lilly, J. M. Caddick, J. Jalava, P. Huovinen, A. Siitonen, A. J. Hakanen, and L. J. Piddock. 2009. Mechanisms of resistance in nontyphoidal Salmonella enterica strains exhibiting a nonclassical quinolone resistance phenotype. Antimicrob. Agents Chemother. 53:3832-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haanperä, M., P. Huovinen, and J. Jalava. 2005. Detection and quantification of macrolide resistance mutations at positions 2058 and 2059 of the 23S rRNA gene by pyrosequencing. Antimicrob. Agents Chemother. 49:457-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hakanen, A., P. Kotilainen, P. Huovinen, H. Helenius, and A. Siitonen. 2001. Reduced fluoroquinolone susceptibility in Salmonella enterica serotypes in travelers returning from Southeast Asia. Emerg. Infect. Dis. 7:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakanen, A., P. Kotilainen, J. Jalava, A. Siitonen, and P. Huovinen. 1999. Detection of decreased fluoroquinolone susceptibility in salmonellas and validation of nalidixic acid screening test. J. Clin. Microbiol. 37:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakanen, A. J., P. Kotilainen, S. Pitkanen, S. Huikko, A. Siitonen, and P. Huovinen. 2006. Reduction in fluoroquinolone susceptibility among non-typhoidal strains of Salmonella enterica isolated from Finnish patients. J. Antimicrob. Chemother. 57:569-572. [DOI] [PubMed] [Google Scholar]
- 19.Hakanen, A. J., M. Lindgren, P. Huovinen, J. Jalava, A. Siitonen, and P. Kotilainen. 2005. New quinolone resistance phenomenon in Salmonella enterica: nalidixic acid-susceptible isolates with reduced fluoroquinolone susceptibility. J. Clin. Microbiol. 43:5775-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins, K. L., M. Day, and E. J. Threlfall. 2008. Plasmid-mediated quinolone resistance in Salmonella enterica, United Kingdom. Emerg. Infect. Dis. 14:340-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavilla, S., J. J. Gonzalez-Lopez, M. Sabate, A. Garcia-Fernandez, M. N. Larrosa, R. M. Bartolome, A. Carattoli, and G. Prats. 2008. Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona, Spain. J. Antimicrob. Chemother. 61:291-295. [DOI] [PubMed] [Google Scholar]
- 22.Lindgren, M. M., P. Kotilainen, P. Huovinen, S. Hurme, S. Lukinmaa, M. A. Webber, L. J. Piddock, A. Siitonen, and A. J. Hakanen. 2009. Reduced fluoroquinolone susceptibility in Salmonella enterica isolates from travelers, Finland. Emerg. Infect. Dis. 15:809-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukinmaa, S., U. M. Nakari, A. Liimatainen, and A. Siitonen. 2006. Genomic diversity within phage types of Salmonella enterica ssp. enterica serotypes Enteritidis and Typhimurium. Foodborne Pathog. Dis. 3:97-105. [DOI] [PubMed] [Google Scholar]
- 24.Nyberg, S. D., M. Osterblad, A. J. Hakanen, P. Huovinen, and J. Jalava. 2007. Detection and molecular genetics of extended-spectrum beta-lactamases among cefuroxime-resistant Escherichia coli and Klebsiella spp. isolates from Finland, 2002-2004. Scand. J. Infect. Dis. 39:417-424. [DOI] [PubMed] [Google Scholar]
- 25.Parry, C. M., V. A. Ho, T. Phuong le, P. V. Bay, M. N. Lanh, T. Tung le, N. T. Tham, J. Wain, T. T. Hien, and J. J. Farrar. 2007. Randomized controlled comparison of ofloxacin, azithromycin, and an ofloxacin-azithromycin combination for treatment of multidrug-resistant and nalidixic acid-resistant typhoid fever. Antimicrob. Agents Chemother. 51:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfister, P., S. Jenni, J. Poehlsgaard, A. Thomas, S. Douthwaite, N. Ban, and E. C. Bottger. 2004. The structural basis of macrolide-ribosome binding assessed using mutagenesis of 23S rRNA positions 2058 and 2059. J. Mol. Biol. 342:1569-1581. [DOI] [PubMed] [Google Scholar]
- 27.Poirel, L., J. D. Pitout, L. Calvo, J. M. Rodriguez-Martinez, D. Church, and P. Nordmann. 2006. In vivo selection of fluoroquinolone-resistant Escherichia coli isolates expressing plasmid-mediated quinolone resistance and expanded-spectrum beta-lactamase. Antimicrob. Agents Chemother. 50:1525-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson, J. E., K. Gay, T. J. Barrett, F. Medalla, T. M. Chiller, and F. J. Angulo. 2007. Increase in nalidixic acid resistance among non-Typhi Salmonella enterica isolates in the United States from 1996 to 2003. Antimicrob. Agents Chemother. 51:195-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stock, I., and B. Wiedemann. 2000. Natural antibiotic susceptibility of Salmonella enterica strains. Int. J. Antimicrob. Agents 16:211-217. [DOI] [PubMed] [Google Scholar]
- 30.Threlfall, E. J., E. de Pinna, M. Day, J. Lawrence, and J. Jones. 2008. Alternatives to ciprofloxacin use for enteric fever, United kingdom. Emerg. Infect. Dis. 14:860-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tribble, D. R., J. W. Sanders, L. W. Pang, C. Mason, C. Pitarangsi, S. Baqar, A. Armstrong, P. Hshieh, A. Fox, E. A. Maley, C. Lebron, D. J. Faix, J. V. Lawler, G. Nayak, M. Lewis, L. Bodhidatta, and D. A. Scott. 2007. Traveler's diarrhea in Thailand: randomized, double-blind trial comparing single-dose and 3-day azithromycin-based regimens with a 3-day levofloxacin regimen. Clin. Infect. Dis. 44:338-346. [DOI] [PubMed] [Google Scholar]