Abstract
A novel composite transposon (Tn6072) resembling staphylococcal cassette chromosome mercury (SCCHg) was identified in a collection of sequence type (ST) 239 methicillin (meticillin)-resistant Staphylococcus aureus (MRSA) isolates from Romanian hospitals. Tn6072 is homologous to the 5′ region of SCCHg found in staphylococcal cassette chromosome mec (SCCmec) type III prototype strain 85/2082 but lacks the characteristic mer operon. SCCHg has previously been reported to integrate downstream of orfX, at the same chromosomal location as SCCmec. Tn6072, by contrast, is demarcated by two IS431 elements, flanked by 8-bp direct repeats, and inserted upstream of the origin of replication, within an open reading frame homologous to SAR2700 of S. aureus strain MRSA252. Analysis of a geographically and temporally diverse collection of 111 strains from the ST239 clonal group uncovered 11 additional strains harboring Tn6072, demonstrating a lineage-specific insertion pattern. Complete sequence analysis of the SCCmec regions of two representative Romanian strains (BK16704, BK16691) revealed two additional novel structures derived from a type III SCCmec background. BK16704 possesses an SCCmec 3A.1.4 structure, with an IS256 insertion downstream of the right chromosomal junction. In contrast, the SCCmec element of BK16691 is truncated downstream of the mec gene complex, with a 24-kb deletion encompassing the right chromosomal junction and an inverted downstream IS256 element. This structure, tentatively named “ψSCCmec16691,” confers methicillin resistance but lacks most of the J1/J2 region, including the ccr gene complex. Taken together, these findings provide evidence for the continuing evolution of SCC elements, as well as the ST239 clonal group.
Methicillin (meticillin) resistance in staphylococci is associated with a heterogeneous mobile element known as staphylococcal cassette chromosome mec (SCCmec) (19, 22). SCCmec elements are characterized by several common features, including (i) integration at a specific chromosomal site; (ii) flanking repeat sequences; and (iii) presence of two essential genetic components, namely, a mec gene complex and a cassette chromosome recombinase (ccr) gene complex (17). Integration takes place at a unique bacterial chromosomal attachment site (attBSCC) located in the orfX gene, approximately 34 kb downstream of the origin of replication, and is achieved by recognition of a specific nucleotide sequence designated the integration site sequence of SCC (ISS) (17, 21). SCCmec elements are classified by binary combinations of different ccr and mec gene complex allotypes and can be further differentiated by variation within three “joining” (J) regions: J1, right chromosomal junction to ccr gene complex; J2, ccr gene complex to mec gene complex; and J3, mec gene complex to left chromosomal junction. Eight types of SCCmec elements (I to VIII) have been described for methicillin-resistant S. aureus (MRSA) (17), along with various J-region subtypes, with ongoing diversification driven by both recombination and de novo gene acquisition from exogenous sources (39, 40). Other SCC elements which do not harbor mecA have also been described for staphylococci (18, 23, 26, 29), integrated in the same chromosomal location as SCCmec.
Acquisition of SCCmec by methicillin-susceptible S. aureus (MSSA) is the defining characteristic of MRSA, and major “epidemic waves” in the evolution of MRSA have been linked to the emergence of SCCmec types I to IV (7, 18, 20, 38). SCCmec type III, for example, is uniquely associated with a widespread group of nosocomial clones (Brazilian, Hungarian, AUS-2 and -3, CMRSA-3, and EMRSA-1, -4, -7, -9, and -11) assigned by multilocus sequence typing (MLST) (9) to the sequence type (ST) 239 subgroup of clonal complex (CC) 8. In many geographic regions, including Brazil, Eastern Europe, the Middle East, India, East Asia, and Eastern Australia, ST239-III is the predominant nosocomial MRSA genotype, accounting for as many as 70% of hospital-acquired MRSA isolates worldwide (3, 10). The ST239 lineage is believed to have arisen via a large-scale chromosomal recombination event between genotypically disparate strains, with the majority of the chromosome (80%) derived from a CC8 background, whereas a 635-kb region surrounding the origin of replication appears to be derived from CC30 (16, 33) (Fig. 1 A). This region includes both SCCmec and the spa locus, consistent with the presence in ST239 of spa repeat motifs (33) characteristic of CC30; SCCmec type III in CC30, however, has never been reported (16).
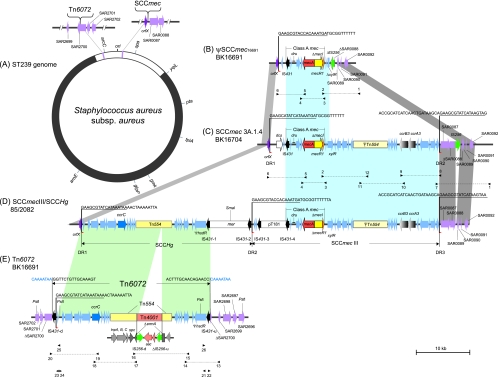
FIG. 1.
Structure of Tn6072 and SCC elements in strains BK16691, BK16704, and 85/2082. (A) Schematic representation of S. aureus ST239 chromosome showing the integration sites of Tn6072 and SCCmec. A 635-kb segment corresponding to the ST30-derived chromosomal replacement is depicted in white. The locations of the seven MLST and spa loci are also indicated. (B) ψSCCmec16691 element from strain BK16691; (C) SCCmec 3A.1.4 element from strain BK16704; (D) composite SCCmec III/SCCHg element from prototype strain 85/2082; (E) Tn6072 element from strain BK16691, shown in the opposite orientation relative to representation shown in Fig. 1A, with PstI restriction sites used for inverse PCR. Blue shading denotes regions of homology shared by SCCmec elements in strains BK16691, BK16704, and 85/2082; green shading denotes homology between SCCHg and Tn6072; gray shading indicates homology shared by flanking chromosomal junctions. ORFs are portrayed by colored arrows, with chromosomal ORFs indicated in purple, IS431 elements in black, and IS256 elements in green. Dotted lines to the left of IS256 elements in Fig. 1B and 1E denote the truncated regions of ΔIS256 and ΔIS256-u, respectively, in BK16691. Tn554, Tn4001, pT181, and mer elements are depicted as rectangles. ISS sites are depicted by small red arrowheads, with characteristic ISS sequences underlined. Integration site sequences and direct repeats in Tn6072 are shown in blue (direct repeats in Tn6072 are shown in reverse complement, since the orientation of Tn6072 is inverted relative to SCCHg). Small black arrowheads represent the locations of primers used for long-range, inverse, and outward-directed PCR (see Table S1 in supplemental material).
The 67-kb structure originally reported for SCCmec III prototype strain 85/2082 (18) was subsequently shown to consist of an SCCmec III element (class A mec with ccrAB3) in tandem arrangement with another SCC element, staphylococcal cassette chromosome mercury (SCCHg) (5). The latter element harbors a mercury resistance operon (mer), a Tn554 element, and a site-specific recombinase gene complex (ccrC) and integrates into the S. aureus genome at the aforementioned insertion sequence site within orfX (Fig. 1D). Precise chromosomal excision of SCCHg has been previously demonstrated (5, 18), presumably mediated by ccrC, but SCCHg in any other chromosomal location has not been reported.
In this study, a novel transposon (Tn6072) closely resembling SCCHg was identified in multiple isolates of two ST239 MRSA clones circulating in Romanian hospitals and in 11 additional strains from 6 other countries. We demonstrate that Tn6072 is located upstream of the origin of replication, within an open reading frame (ORF) corresponding to SAR2700 in CC30 strain MRSA252 (15). In addition, one of the two Romanian clones is shown to possess a 24-kb deletion encompassing the entire region from the mec gene complex to the right chromosomal junction, thereby lacking a ccr gene complex and flanking repeat sequences. This structure, while conferring resistance to methicillin, does not satisfy current criteria for SCCmec elements (17) and has tentatively been designated “ψSCCmec16691.”
MATERIALS AND METHODS
Bacterial strains.
Thirty-four MRSA strains were selected from a recent study involving approximately 150 S. aureus isolates, collected between 2004 and 2005 from three affiliated hospitals of the Clinic County Hospital in Braçsov, Romania (R. Ionescu, J. R. Mediavilla, L. Chen, D. O. Grigorescu, M. Idomir, B. N. Kreiswirth, and R. B. Roberts, unpublished data). All 34 isolates chosen for this study belonged to the ST239 clonal group, as determined by multilocus sequence typing (ST239/ST1312) and spa typing (spa type 351/WGKAQQ/t030; spa type 3/WGKAOMQ/t037; or spa type 1020/WFGKAOMQ/t074). SCCmec III prototype strain 85/2082 (18) was kindly provided by T. Ito (Juntendo University, Tokyo, Japan), while SCCmec IIIB prototype strain HDG2 (32) was a gift from H. de Lencastre (Rockefeller University, New York, NY). A previously described collection of 111 diverse strains from the ST239 clonal group, collected over 34 years from 29 countries (36), was used to assess the distribution of Tn6072. Strains MRSA252 (NRS071) and MSSA476 (NRS072) were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) (http://www.narsa.net).
MIC testing.
MIC values for oxacillin (Sigma-Aldrich, St. Louis, MO) were determined for all 34 Romanian isolates using the broth dilution method, according to CLSI guidelines (6). Bacterial suspensions were incubated for 24 h at 37°C, and optical density at 600 nm (OD600) values were obtained using a VersaMax tunable microplate reader (Molecular Devices, Sunnyvale, CA). S. aureus strains MRSA252 and MSSA476 were used as positive and negative controls (15), respectively.
DNA isolation.
Single colonies of S. aureus were isolated on BBL CHROMagar Staph aureus (Becton-Dickinson, Franklin Lakes, NJ), restreaked onto Luria-Bertani (LB) agar, and grown overnight at 37°C. DNA was isolated using a Wizard genomic DNA purification kit (Promega, Madison, WI), following treatment with 20 μg/ml lysostaphin (Sigma-Aldrich, St. Louis, MO) for 30 min at 37°C. Genomic DNA was stored at 4°C and used as template for all PCRs.
Genotypic analysis.
S. aureus isolates were spa typed using eGenomics software, as described previously (27, 35). Ridom spa types were subsequently assigned using the spa typing website (http://www.spaserver.ridom.de) developed by Ridom GmbH and curated by SeqNet.org (http://www.SeqNet.org) (13). MLST was also performed as described previously (9), using the database maintained at http://saureus.mlst.net. Haplotype analysis based on sequences at 32 loci (36) was performed on two representative Romanian strains (BK16691 and BK16704) and on reference strain 85/2082 in order to place them within the global population structure of the ST239 clonal group. SCCmec typing was performed using a recent multiplex real-time PCR method (4), which targets mecA and the two essential gene complexes (ccr and mec) found in all SCCmec elements. Other SCCmec or SCCHg targets, such as the mer operon, pT181, dcs, and the SCCHg J region (CZ055 to CZ066 in strain 85/2082) were detected using conventional PCR methods described elsewhere (24, 31). Lastly, the variable-number tandem repeats (VNTR) adjacent to IS431mec, referred to as dru (direct repeat unit) or HVR (hypervariable region), have demonstrated utility in molecular analyses of MRSA (11, 30, 34, 36). dru typing was therefore performed as described previously (11) using DruID software, and new dru types were submitted to http://www.dru-typing.org. DNA sequencing for spa typing, dru typing, and MLST was performed commercially.
Long-range PCR.
Several long-range PCR assays were employed to amplify the entire SCCmec regions (orfX to right chromosomal junction) and Tn6072 elements from two representative Romanian strains (BK16691 and BK16704). Primers used for long-range PCR are listed in Table S1 in the supplemental material and depicted in Fig. 1. Hot-Start Taq DNA polymerase Mastermix (Denville Scientific, Metuchen, NJ), 400 nmol of each primer, and 100 ng of template DNA were combined into 50-μl reaction mixtures. Cycling conditions were as follows: initial denaturation at 94°C for 5 min; 10 cycles of 94°C/10 s followed by 65°C/10 min; and an additional 20 cycles of 94°C/10 s and 65°C/10 min, increased by 20 s per cycle at 65°C.
Inverse PCR.
Inverse PCR was used in order to sequence the chromosomal junctions surrounding Tn6072. Briefly, 5 μg of S. aureus genomic DNA was digested overnight using 20 U of PstI (New England BioLabs, Ipswich, MA) and, subsequently, 0.5 μg of PstI-digested fragments was ligated for 3 h at 16°C using T4 DNA ligase (New England BioLabs). Ligation products were purified using equal volumes of phenol-chloroform, and the aqueous phase was precipitated with ethanol and resuspended in double distilled water (ddH2O). Approximately 100 ng of circularized DNA was used as template to amplify both Tn6072 flanking regions, using the long-range PCR conditions described above.
Outward-directed PCR.
To detect the presence of circularized extrachromosomal Tn6072 intermediates (minicircles), two outward-directed primers (3F7 and 3R8) (see Table S1 in the supplemental material) matching the terminal sequences of Tn6072 were designed. Outward-directed PCR was then performed as described elsewhere (37), using genomic DNA as a template. An alternative primer set (3F7 and SCCHg-R) was used to detect the extrachromosomal form of SCCHg in strains 85/2082 and BK16684 (SCCmec III with SCCHg).
Excision of SCCmec.
PCR was also employed to identify the precise excision of SCCmec, using methods described elsewhere (5, 19). Briefly, primer pair cR2/cLt2 was used to detect attB within the type 3A.1.4 SCCmec strains found in this study, while primer pairs LF10/cR2 and LF12/orfR1 were used to detect potential excision of the truncated SCCmec element in ψSCCmec16691-bearing strains.
Other PCR methods.
Additional PCR assays were employed to characterize the structures and integration sites of SCCmec 3A.1.4, ψSCCmec16691, and Tn6072. Primer pairs LF10/mecI-R and LF10/IS256-P3 were used to identify ψSCCmec16691 and the associated downstream IS256 insertion, with ψSCCmec16691-bearing strains yielding 2.5- and 1.1-kb fragments (see Table S1 in the supplemental material). Similarly, primer set LF10/IS256-F1 was used to confirm the downstream IS256 insertion in all type 3A.1.4 SCCmec-bearing strains, yielding a single 0.9-kb amplicon. Carriage of Tn6072 was determined using primers 3F2 and IS431-r-1, while integration within the SAR2700 homolog was confirmed by amplifying the upstream and downstream junctions using primer pairs SAR2699-F3/typeIII-F and 3F2/SAR2700-R4, respectively (see Table S1). Primer set SAR2699-F3/SAR2700-R4 was used as a negative control, yielding a 0.5-kb amplicon for all strains not harboring Tn6072 (e.g., 85/2082). Lastly, insertion of Tn4001 within the Tn554 element of Tn6072 was identified using primer pairs 3F3/aac2-R1 and ermA-F/aac-R1 (see Table S1).
Sequence analysis.
Complete nucleotide sequences of the SCCmec and Tn6072 elements from two representative Romanian strains (BK16691 and BK16704) were obtained using long-range PCR, inverse PCR, and primer walking. Amplicons generated by long-range and inverse PCR were verified by agarose gel electrophoresis and sequenced commercially. ORFs were identified with GLIMMER V3.02 software (8) using default parameters. ClustalW2 (25) was used to align and compare multiple sequences, and annotation was performed using BLASTN and BLASTX (http://blast.ncbi.nlm.nih.gov/).
Nomenclature assignment for Tn6072.
Transposon number 6072 was assigned using the Tn Number Registry maintained by the Eastman Dental Institute at University College London (http://www.ucl.ac.uk/eastman/tn).
Nucleotide sequence accession number.
The complete nucleotide sequences of the following elements have been deposited in GenBank under the following accession numbers indicated: (i) the ψSCCmec16691 element from strain BK16691, GU235983; (ii) the SCCmec 3A.1.4 element from strain BK16704, GU235984; and (iii) the Tn6072 element from strain BK16691, GU235985.
RESULTS AND DISCUSSION
Characteristics of Romanian strains.
Thirty-four clinical MRSA strains used in this study were obtained between March 2004 and June 2005 from the Clinic County Hospital in Braçsov, Romania, as part of another study (Ionescu et al., unpublished data). In that study, approximately 150 S. aureus isolates were analyzed by spa typing and SCCmec typing, among other methods. Of these, 32 MRSA strains possessed the same spa repeat pattern (spa type 351/WGKAQQ/Ridom t030), while another two strains (BK16684 and BK16672) displayed closely related patterns (3/WGKAOMQ/t037 and 1020/WFGKAOMQ/t074, respectively) (Table 1 ). Subsequent MLST typing indicated that 32/34 strains belonged to ST239 (2-3-1-1-4-4-3), while 2 spa type 351 strains (BK16658 and BK16666) were classified as ST1312 (2-3-176-1-4-4-3), a single-locus variant of ST239. However, SCCmec typing suggested the presence of two different variants among the 32 spa type 351 strains. The majority (20/32) exhibited a signature (class A mec, ccrAB3, ccrC) consistent with SCCmec III prototype strain 85/2082, which harbors the SCCHg element (Fig. 1). The remaining 12/32 spa type 351 strains, however, were positive only for class A mec and ccrC and were therefore classified as nontypeable. Sequencing of the entire SCCmec region from one of the latter strains (BK16691) revealed the presence of a truncated SCCmec element, subsequently dubbed ψSCCmec16691 (see below). Of the remaining two MRSA strains, one possessed an SCCmec III signature (BK16684), while the other strain (BK16672) possessed a signature consistent with SCCmec IIIB (class A mec and ccrAB3, without ccrC). Conventional PCR analysis of other SCC targets (Table 1) showed that all of the spa type 351 strains possessed the SCCHg J region (CZ055 to CZ056 in strain 85/2082) but did not harbor the mer operon or pT181. Interestingly, the 20 spa type 351 strains with the SCCmec III signature also possessed the dcs locus, previously reported only for the J3 regions of SCCmec types I, II, and IV (31) but recently described in an ST239 strain from Saudi Arabia (JCSC1716, SCCmec type 3A.1.4) (5). These 20 strains were accordingly classified SCCmec type 3A.1.4. In contrast, the dcs locus was not present with the 12 spa type 351 strains with the nontypeable ψSCCmec16691 element. The latter group of strains all possessed the same dru type (dt9x), whereas three different dru types (dt10a, dt10g, and dt7f) were found among the 20 SCCmec 3A.1.4 strains (Table 1). Lastly, MIC testing indicated that all 34 strains were highly resistant to oxacillin (≥256 μg/ml), with the exception of BK16672 (8 μg/ml) (Table 1).
TABLE 1.
Characteristics of Romanian MRSA strains used in this study
| Strain IDa | Oxacillin MIC (μg/ml) | MLSTb | spa typec | spa repeat patterns | SCCmec type | dru type | dru repeat patterns | SCC components |
Excision of SCCmec | Tn6072 |
IS256 (16691)e | IS256 (16704)f | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mecI | ccrC | mer | Hg-Jd | pT181 | dcs | SAR2700 insertion | Tn6072 minicircle | Tn4001 insertion | |||||||||||
| 16684 | 256 | ST239 | 3 | WGKAOMQ | III | dt8t | 5a-2d-5b-3a-4c-3b-4e-4j | + | + | + | + | + | − | + | − | g | − | − | − |
| 16672 | 8 | ST239 | 1020 | WFGKAOMQ | IIIB | dt11ac | 5a-2d-4a-0-2d-6f-2a-3o-3b-4e-3e | + | − | − | − | − | − | + | − | − | − | − | − |
| 16645 | >512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16646 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16651 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16652 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16653 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16656 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16658 | 256 | ST1312 | 351 | WGKAQQ | 3A.1.4 | dt10g | 5a-2d-4a-0-2d-5b-2a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16666 | 256 | ST1312 | 351 | WGKAQQ | 3A.1.4 | dt10g | 5a-2d-4a-0-2d-5b-2a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16670 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16671 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16676 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16677 | 256 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt7f | 5a-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16680 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16686 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16687 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16698 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16699 | 256 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16700 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16702 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16704 | 512 | ST239 | 351 | WGKAQQ | 3A.1.4 | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | + | + | − | + | − | + | + | + | + | + | − | + |
| 16674 | 256 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16678 | 256 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16679 | 512 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16683 | 256 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16688 | 512 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16689 | 512 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16691 | 256 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16692 | 256 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16695 | 512 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16696 | 512 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16703 | 256 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| 16708 | 256 | ST239 | 351 | WGKAQQ | ψSCCmec16691 | dt9x | 5a-2d-3p-5b-3a-2g-2c-4e-3e | + | + | − | + | − | − | − | + | + | + | + | − |
| HDG2 | 64 | ST239 | 121 | WGKAOM | IIIB | dt11l | 5a-2d-4a-2d-2d-5b-3a-2g-3b- 4e-3e | + | − | − | − | − | − | + | − | − | − | − | − |
| 85/2082 | 16 | ST239 | 3 | WGKAOMQ | III | dt10c | 5a-2d-4a-0-2d-1b-2g-3b-4e-3e | + | + | + | + | + | − | + | − | e | − | − | − |
Prototype strains used in this study are shown in boldface. ID, identification number.
Multilocus sequence types correspond to the following allelic profiles: ST239 (2-3-1-1-4-4-3) and ST1312 (2-3-176-1-4-4-3).
Ridom equivalents for eGenomics spa types are as follows: spa type 3, Ridom t037; spa type 121, Ridom t421; spa type 351, Ridom t030; and spa type 1020, Ridom t074.
J region of SCCHg element (corresponding to CZ055 to CZ066 in strain 85/2082), detected using primers MN21/MN22 (see Table S1 in supplemental material) (24).
Downstream ΔIS256 within ΔSAR0088 in strain BK16691, detected with primer set LF10/IS256-P3 (see Table S1 in the supplemental material) (12).
Downstream IS256 within ΔSAR0088 in strain BK16704, detected with primer set LF10/IS256-F1 (see Table S1 in the supplemental material).
Tn6072 minicircle is not present, but an SCCHg minicircle was detected using primer set 3F7/SCCHg-R (see Table S1 in the supplemental material).
SCCmec structure in BK16691 and BK16704.
Complete SCCmec sequencing of BK16704 confirmed it as a type III SCCmec element, possessing ccrAB3 in combination with the class A mec gene complex (IS431-mecA-mecR1-mecI) (Fig. 1C). Two SCCmec integration site sequences (ISS) containing direct repeats (DR) were likewise identified. A dcs locus was identified in the J3 region of this strain, in agreement with PCR results described above. The right chromosomal junction of the SCCmec element in BK16704 was also sequenced, revealing an IS256 insertion within an open reading frame corresponding to SAR0088 of MRSA252, which is located downstream of SCCmec in the genome of MRSA252 (Fig. 1C) (15).
In contrast to BK16704, complete nucleotide sequencing of the SCCmec region of BK16691 (Fig. 1B) confirmed the presence of a class A mec gene complex with no downstream ccr loci, in agreement with real-time PCR SCCmec typing results (Table 1) (4). Surprisingly, BK16691 possesses a 24-kb deletion spanning most of the region between the mec gene complex and the right chromosomal junction. The deleted region originates in the xylR gene, just downstream of mecI, and terminates within a truncated homolog of MRSA252 SAR0088 (Fig. 1B). Moreover, the J3 region of BK16691 lacks the aforementioned dcs locus, while the IS256 insertion in BK16691 is inverted relative to the one in BK16704. Only one ISS sequence was found within the attBSCC attachment site in orfX, and no DR were identified at either chromosomal junction. Since the SCC region of BK16691 does not possess ccr loci or flanking direct repeats, it cannot be designated an SCCmec element according to current nomenclature guidelines (20), despite possessing a mec gene complex and chromosomal integration site (attBSCC) identical to those of other SCCmec strains. Consequently, the entire SCC region of strain BK16691, from the ISS in orfX to the inverted copy of IS256, has tentatively been named ψSCCmec16691.
Structure of Tn6072.
As described above, SCCmec typing demonstrated that BK16704 and BK16691 both possess a ccrC locus, and an additional PCR confirmed the presence of an element closely resembling the J region of SCCHg, minus the characteristic mer operon. However, complete sequencing from orfX to the right chromosomal junctions of both strains did not reveal any ccrC loci, suggesting that an SCCHg-like fragment might be located elsewhere on the chromosome or within an extrachromosomal element. Inverse PCR was therefore used to determine the insertion junctions for the SCCHg homolog in both BK16691 and BK16704. A single PCR product was obtained using primer pairs typeIII-R/IS431-r-1 (8.7 kb) and 3F2/3R2 (5.3 kb), corresponding to the 5′ and 3′ junctions, respectively (data not shown). The results of inverse PCR in both BK16691 and BK16704 also indicated that the SCCHg homology is located in an open reading frame corresponding to MRSA252 SAR2700, within the 635-kb chromosomal replacement region previously described for ST239 (Fig. 1A) (33). The SAR2700 homolog is located 108 kb upstream from the origin of replication (ori), thereby confirming the insertion of an SCCHg-like element outside the SCCmec region in BK16691 and BK16704. Surprisingly, complete nucleotide sequencing of the putative SCCHg homolog from strain BK16691 revealed it to be a novel composite transposon, and it was accordingly named Tn6072.
The Tn6072 element from strain BK16691 is illustrated in Fig. 1E, with its chromosomal location depicted in Fig. 1A. Tn6072 is 29,422 bp in length, with an overall structure highly similar to the 5′ region of the SCCHg element from 85/2082 (18). However, the chromosomal orientation of Tn6072 is inverted relative to the full-length SCCHg element in 85/2082 (Fig. 1A, D, and E). In strain BK16691, Tn6072 is flanked by two IS431 elements, one of which displaces the orfX locus found in previously described SCCHg elements. Inspection of the junction sequences at both termini determined that each IS431 element is also flanked by an 8-bp direct repeat (TTATTTTG, shown as a reverse complement in Fig. 1E, CAAAATAA) as a result of target site duplication following insertion. Moreover, a characteristic ISS sequence (GAAGCGTATCATAAATAA) was identified near the downstream IS431 element (IS431-d), as depicted in Fig. 1E. The 36-bp sequence between the ISS and IS431-d is nearly identical to the one between IS431-2 and DR-2 in the SCCHg element of strain 85/2082, while the right contiguous sequence of ISS is identical to the one immediately downstream of orfX in 85/2082 (Fig. 2).
FIG. 2.
Sequence comparison of the region upstream of IS431-d in the Tn6072 element of BK16691 (shown in Fig. 1E) and the extrachromosomal circular form of SCCHg from 85/2082, with the regions downstream of orfX and IS431-2 in the SCCHg element of 85/2082 (shown in Fig. 1D). Asterisks denote nucleotide identity in two or more strains, while the consensus ISS region common to all three sequences is surrounded by a box. The ISS sequences for orfX, IS431-2, and IS431-d correspond to the underlined sequences in Fig. 1D and 1E, respectively. Flanking inverted repeat (IR) sequences for the IS256 elements are displayed in italics, and the transcriptional directions of orfX and the IS431 elements are indicated by boxed arrows.
SCCHg can be excised from the chromosome of 85/2082, as reported previously (5); in this study, we found that SCCHg can also form extrachromosomal circles in 85/2082 (Table 1), using outward-oriented PCR with primer set 3F7/SCCHg-R (see Table S1 in the supplemental material). Sequence analysis revealed that the outward PCR product of SCCHg comprises both termini, with only one intervening copy of IS431. The sequence upstream of IS431 in the SCCHg minicircle is identical to that of IS431-d in Tn6072 and includes the characteristic ISS sequence (Fig. 2). These results suggest that Tn6072 may have originated from an IS431-flanked SCCHg homolog containing the region from DR1 to DR2 in 85/2082, but without the mer operon and IS431-1 (Fig. 1D). Such an element may have been excised from the chromosomal ISS sites, forming a transpositionally active extrachromosomal circle in the same manner as the SCCHg minicircle, and then integrated into the aforementioned SAR2700 homolog.
Recently, an extrachromosomal SCCHg element was described in plasmid pTW20_1 from ST239 strain TW20, with IS431 elements flanking both the plasmid and chromosomal SCCHg homologs (16). Similar findings have been described elsewhere (1, 14), suggesting that IS431-mediated recombination may provide a hypothesis to explain the homoplasies inferred from the distribution of SCCHg and other SCC components within the ST239 clonal group (36). Outward-directed PCR experiments indicate that Tn6072 also forms extrachromosomal circles (Table 1), almost identical in structure to the SCCHg minicircles described above, with both termini connected by a single copy of IS431. Tn6072 is therefore a potentially active transposon, and integration of Tn6072 into other genomic locations, including other S. aureus clonal backgrounds and staphylococcal species, is possible. Consequently, screening for the presence of Tn6072-like elements in staphylococcal strains bearing SCCHg-associated loci is advisable, since current SCCmec typing methods cannot differentiate between Tn6072 and SCCHg (4, 24, 31, 41).
Distribution of Tn6072 and ψSCCmec16691.
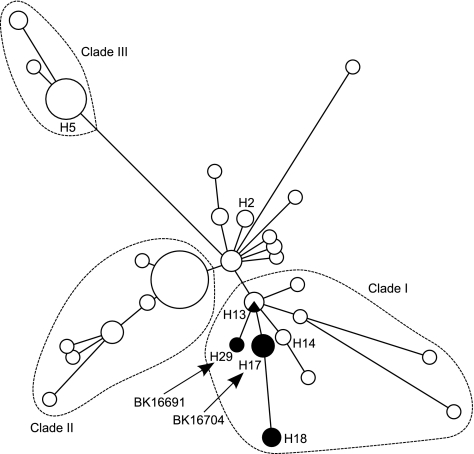
An additional 19 Romanian strains were found with an SCCmec 3A.1.4 structure like that of BK16704, whereas another 11 Romanian strains possessed a truncated ψSCCmec16691 element (Table 1). Subsequent screening of a geographically and temporally diverse collection of 111 strains from the ST239 clonal group (36) identified 11 additional strains carrying Tn6072 elements, all integrated within the same chromosomal site as the Romanian strains (see Table S2 in the supplemental material). Comparative analysis suggested that the distribution of Tn6072 is limited to a predominantly European clade of the ST239 clonal group (clade I, Fig. 3) which includes the previously described “Czech clone” (28). BK16691 comprises a novel haplotype (H29) within this lineage, whereas BK16704 bears the same haplotype (H17) consistent with Tn6072-positive strains from the Netherlands, Germany, Hungary, Russia, and Turkey (see Table S2). All but one of the non-Romanian strains possessed an SCCmec 3A.1.4 structure, whereas no additional strains with truncated ψSCCmec16691-like elements were observed. These results demonstrate that Tn6072 insertions have been present in European ST239 strains for at least 2 decades (as early as 1994), whereas ψSCCmec16691 has been observed only with the Romanian collection analyzed in this study.
FIG. 3.
Distribution of Tn6072 within the ST239 clonal group. Maximum parsimony tree depicts the global population structure of this clonal group, as described previously (36). Each circle represents a distinct haplotype, defined by sequences at 32 chromosomal loci. Circle sizes represent the relative frequencies of individual haplotypes within a sample of previously characterized strains (36). Dark shading within circles denotes the presence of Tn6072, while the locations of Tn6072-bearing strains BK16691 and BK16704 are indicated by arrows. Haplotype H18 represents the location of Czech clone prototype strain 2HK, while haplotypes H2, H5, and H14 denote SCCmec prototype strains HDG2 (Portuguese clone, SCCmec IIIB), HU25 (Brazilian clone, SCCmec IIIA), and 85/2082 (SCCmec III), respectively. (Adapted from reference 36 with permission of the publisher.)
To our knowledge, the only other description of a mec gene complex with no accompanying ccr loci involves the putative “primordial” mecIRAm element described recently for plasmid pMCCL2 of Macrococcus caseolyticus strain JCSC5402 (2). ψSCCmec16691 is therefore a novel example of a chromosomal mecA-bearing SCC element without ccr loci, which appears to have lost the capability for chromosomal excision (Table 1). All ψSCCmec16691-bearing isolates used in this study were highly resistant to oxacillin (Table 1), consistent with carriage of mecA and the associated mec gene complex. Consequently, ψSCCmec16691 may represent the smallest example to date (10.5 kb) of a genetic element conferring methicillin resistance in staphylococci. In addition, ψSCCmec16691-bearing strains were isolated from more than one hospital on multiple occasions, suggesting that they remain viable in nosocomial environments despite losing most of the SCCmec III element. Further studies are required to compare the relative fitness levels of these strains with those of other ST239-MRSA-III clones. It remains to be seen if ψSCCmec16691-bearing strains are still circulating in Romanian hospitals and whether they will spread beyond this setting. In conclusion, our findings provide additional evidence of the ongoing diversification of type III SCCmec elements within the ST239 clonal group, while further elucidating the evolutionary mechanisms of SCC elements.
Supplementary Material
Acknowledgments
This work was supported in part by the American Heart Association (D.A.R.), NIH grant GM080602 (D.A.R.), and The Cary L. Guy Foundation, New York (R.B.R.).
Footnotes
Published ahead of print on 17 May 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Arakere, G., S. Nadig, T. Ito, X. X. Ma, and K. Hiramatsu. 2009. A novel type-III staphylococcal cassette chromosome mec (SCCmec) variant among Indian isolates of methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 292:141-148. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., K. Kuwahara-Arai, I. Uchiyama, F. Takeuchi, T. Ito, and K. Hiramatsu. 2009. Complete genome sequence of Macrococcus caseolyticus strain JSCS5402, reflecting the ancestral genome of the human-pathogenic staphylococci. J. Bacteriol. 191:1180-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranovich, T., H. Zaraket, I. I. Shabana, V. Nevzorova, V. Turcutyuicov, and H. Suzuki. 2010. Molecular characterization and susceptibility of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from hospitals and the community in Vladivostok, Russia. Clin. Microbiol. Infect. 16:575-582. [DOI] [PubMed] [Google Scholar]
- 4.Chen, L., J. R. Mediavilla, D. C. Oliveira, B. M. Willey, H. de Lencastre, and B. N. Kreiswirth. 2009. Multiplex real-time PCR for rapid staphylococcal cassette chromosome mec (SCCmec) typing. J. Clin. Microbiol. 47:3692-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 50:1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CLSI. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344-1347. [DOI] [PubMed] [Google Scholar]
- 8.Delcher, A. L., K. A. Bratke, E. C. Powers, and S. L. Salzberg. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., E. K. Nickerson, N. Chantratita, V. Wuthiekanun, P. Srisomang, R. Cousins, W. Pan, G. Zhang, B. Xu, N. P. J. Day, and S. J. Peacock. 2008. Rapid detection of the pandemic methicillin-resistant Staphylococcus aureus clone ST 239, a dominant strain in Asian hospitals. J. Clin. Microbiol. 46:1520-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goering, R. V., D. Morrison, Z. Al-Doori, G. F. Edwards, and C. G. Gemmell. 2008. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin. Microbiol. Infect. 14:964-969. [DOI] [PubMed] [Google Scholar]
- 12.Gu, J., H. Li, M. Li, C. Vuong, M. Otto, Y. Wen, and Q. Gao. 2005. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J. Hosp. Infect. 61:342-348. [DOI] [PubMed] [Google Scholar]
- 13.Harmsen, D., H. Claus, W. Witte, J. Rothganger, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heusser, R., M. Ender, B. Berger-Bachi, and N. McCallum. 2007. Mosaic staphylococcal cassette chromosome mec containing two recombinase loci and a new mec complex, B2. Antimicrob. Agents Chemother. 51:390-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holden, M. T., J. A. Lindsay, C. Corton, M. A. Quail, J. D. Cockfield, S. Pathak, R. Batra, J. Parkhill, S. D. Bentley, and J. D. Edgeworth. 2009. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J. Bacteriol. 192:888-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements. 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updat. 6:41-52. [DOI] [PubMed] [Google Scholar]
- 22.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama, Y., F. Takeuchi, T. Ito, X. X. Ma, Y. Ui-Mizutani, I. Kobayashi, and K. Hiramatsu. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 185:2711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 26.Luong, T. T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 184:3623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathema, B., J. Mediavilla, and B. N. Kreiswirth. 2008. Sequence analysis of the variable number tandem repeat in Staphylococcus aureus protein A gene: spa typing. Methods Mol. Biol. 431:285-305. [DOI] [PubMed] [Google Scholar]
- 28.Melter, O., M. Aires de Sousa, P. Urbaskova, V. Jakubu, H. Zemlickova, and H. de Lencastre. 2003. Update on the major clonal types of methicillin-resistant Staphylococcus aureus in the Czech Republic. J. Clin. Microbiol. 41:4998-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mongkolrattanothai, K., S. Boyle, T. V. Murphy, and R. S. Daum. 2004. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1823-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nahvi, M. D., J. E. Fitzgibbon, J. F. John, and D. T. Dubin. 2001. Sequence analysis of dru regions from methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococcal isolates. Microb. Drug Resist. 7:1-12. [DOI] [PubMed] [Google Scholar]
- 31.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 33.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryffel, C., R. Bucher, F. H. Kayser, and B. Berger-Bachi. 1991. The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to the Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J. Bacteriol. 173:7416-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth, D. S., L. K. McDougal, F. W. Gran, A. Manoharan, M. C. Enright, J.-H. Song, H. de Lencastre, and D. A. Robinson. 2010. Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS One 5:e8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth, D. S., and D. A. Robinson. 2009. Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. J. Bacteriol. 191:5964-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wielders, C. L., M. R. Vriens, S. Brisse, L. A. de Graaf-Miltenburg, A. Troelstra, A. Fleer, F. J. Schmitz, J. Verhoef, and A. C. Fluit. 2001. In-vivo transfer of mecA DNA to Staphylococcus aureus. Lancet 357:1674-1675. [DOI] [PubMed] [Google Scholar]
- 40.Wu, S. W., H. de Lencastre, and A. Tomasz. 2001. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J. Bacteriol. 183:2417-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.