Abstract
A qnrVC-like gene, qnrVC4, was found in a novel complex class 1 integron gene cassette array following the ISCR1 element and blaPER-1 in a multidrug-resistant strain of the aquatic bacterium Aeromonas punctata. The deduced QnrVC4 protein sequence shares 45% to 81% amino acid identity with quinolone resistance determinants QnrB6, QnrA1, QnrS1, QnrC, QnrVC1, and QnrVC3. A Ser-83 to Ile amino acid substitution in gyrase A may be mainly responsible for ciprofloxacin resistance in this strain.
Aeromonas spp. have various class 1 integrons and have been proposed as clinical and environmental reservoirs of antibiotic determinants (2, 9). Resistance to quinolones depends on mutations in the gyrase and/or topoisomerase IV genes (6, 15), and plasmid-mediated resistance determinants (qnrS2) have been found in Aeromonas spp. (2, 12, 18). Since qnr was first detected (11), 6 qnrA variants, 20 qnrB variants, and 4 qnrS variants have been described (7). Two homologs, qnrVC1 and qnrVC2, have been found in integrons (5). Recently, qnrC and qnrD were isolated in China (3, 20). ISCR1 elements located downstream of class 1 integrons and transposed by a rolling-circle (RC) transposition mechanism are widespread (14, 17).
In 2008, we investigated the molecular diversity of class 1 integrons among bacteria from wastewater samples near Thousand-Buddha Hill Hospital, Jinan, Shandong province, China. Aeromonas punctata 159, identified by its 16S rRNA gene, was selected from kanamycin-containing MacConkey agar plates and characterized as resistant to ampicillin, kanamycin, chloramphenicol, trimethoprim, sulfisoxazole, ceftazidime, and nalidixic acid on Mueller-Hinton agar plates by the disk diffusion method (4). According to the Etest strip manufacturer's data (AB Biodisk, Solna, Sweden), the MICs of ciprofloxacin, gatifloxacin, and nalidixic acid are 0.38, 0.19, and 96 μg/ml, respectively.
Integrase (23) and ISCR1 (14) were investigated by PCR amplification of genomic DNA extracted using bacterial genome extraction kits (Bioteke, Beijing, China). Amplicons of 3.2 kb and 2.0 kb were obtained using the primers hep58 and hep59 (22) and sequenced. The 2.0-kb amplicon with dfrA12-orfF-aadA2 is widely distributed (17). The 3.2-kb band contained two aacA4 cassettes bounding a 218-amino-acid-encoding open reading frame (ORF) following a catB3 cassette (Fig. 1 A). The deduced protein sequence of the ORF included a pentapeptide repeat motif. The new gene showed 99% identity with the nonfunctional qnrVC2, which has three nucleotide insertions and one deletion compared to functional qnr genes (5, 20), and showed 75%, 75%, 68%, 62%, 60%, 48%, and 34% similarities to qnrVC1, qnrVC3, qnrC, qnrS1, qnrA1, qnrB6, and qnrD, respectively, with generally commensurate degrees of protein similarity. The attC site of the qnr-like gene was analyzed with qnrVC1 and qnrVC2 (Fig. 1C), which have been reported to form a superintegron (SI) cassette in Vibrio parahaemolyticus and Vibrio cholerae (5). The gene was named qnrVC4, and its protein was designated QnrVC4 based on the qnr nomenclature (7).
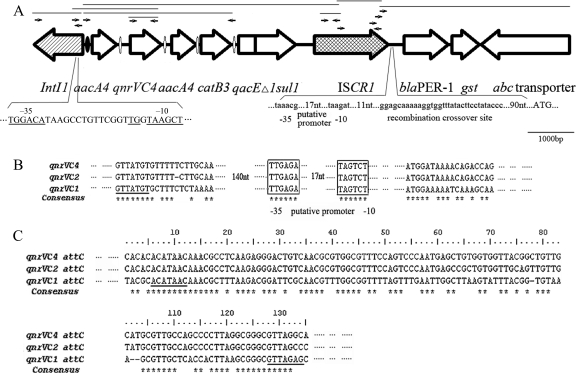
FIG. 1.
Schematic map of the complex class 1 integron in A. punctata 159. (A) Open arrows indicate open reading frames, white ovals indicate attC recombination sites, a black oval indicates the attI1 recombination site, the arrow with diagonal lines represents the class 1 integrase, and the cross-hatched arrow represents a putative recombinase encoded by ISCR1 element. The −35 and −10 motifs of the putative promoter in the class 1 integrase are shown in uppercase. The −35 and −10 motifs of the putative promoter of blaPER-1, the ISCR1 recombination crossover sites, and right-hand boundary of the CR1 element are shown in lowercase. The ATG start codon of the blaPER-1 gene is shown in uppercase. The locations of the primers are indicated by small arrows, and PCR products are indicated by lines above the structure. nt, nucleotides; *, conserved residues. (B) Analysis of the putative promoter of the qnrVC4 gene cassette. (C) qnrVC4 attC recombination site analysis showing 91% and 69% identities with qnrVC2 and qnrVC1, respectively.
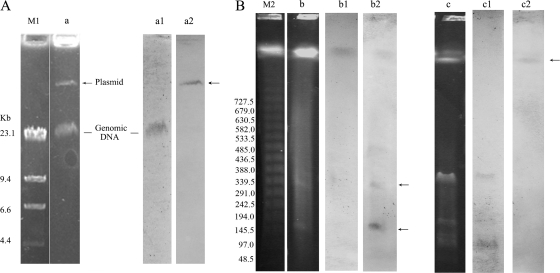
Although the qnrVC4 gene of A. punctata was not transferable by transformation of plasmids or conjugation (19), the location of the gene was studied by DNA-DNA hybridizations with probes for the qnrVC4 gene and for the 23S rRNA gene. Both hybridizations were carried out using plasmid isolation by alkaline lysis (16), followed by agarose gel electrophoresis and total DNA digestion with S1 nuclease and I-CeuI, followed by pulsed-field gel electrophoresis (PFGE) (1, 10). The results indicated that qnrVC4 was located on a large plasmid (Fig. 2).
FIG. 2.
qnrVC4 location identified by agarose gel electrophoresis (A), genomic mapping with S1 nuclease and I-CeuI by pulsed-field gel electrophoresis (PFGE) (B), and relative hybridization with the 23S rRNA gene and the qnrVC4 probe. Lane a shows large-plasmid analysis of A. punctata 159; lanes b and c show genomic mapping with S1 nuclease and I-CeuI digestion by PFGE. Lanes a1, b1, and c1 show hybridization results with the 23S rRNA gene. Lanes a2, b2, and c2 show hybridization with the qnrVC4 probe. M1, HindIII-digested λ DNA marker; M2, λ concatemer marker for PFGE. Arrows indicate plasmid locations.
Since ISCR1 was found, primers hep58 and orf341B were used to determine the adjacent region. A 5.3-kb amplicon was obtained and sequenced, and this procedure demonstrated that aacA4-qnrVC4-aacA4-catB3 is followed by qacEΔ1, sul1, and ISCR1 (Fig. 1A). The primer aadA2F was used with orf341B to investigate the relationship between dfrA12-orfF-aadA2 and ISCR1, but no amplicon was obtained.
Self-formed adaptor PCR (SEFA-PCR) (21) was used to amplify the region downstream of ISCR1, initially using 513Sp1 and 513Sp3, and then 513Sp2 (Table 1) was used for nested PCR. A 3.9-kb region was obtained, and sequencing revealed three ORFs. A 927-bp ORF encoding extended-spectrum β-lactamase PER-1 and a putative promoter were observed upstream of blaPER-1 (Fig. 1A). The only other reported blaPER-1 associated with ISCR1 is located downstream of ant(3′)-Ij-aac(6′)Ib-nit1-nit2-catB3 in A. punctata integron In39 (AY740681). Most blaPER-1 genes are associated with the Tn1213 transposon backbone and IS4 family (13). Another 657-bp ORF appears to encode a glutathione S-transferase, and the 1.8-kb ORF may encode an ABC transporter.
TABLE 1.
Primers used in this study
| Primer | Target gene or DNA fragment | Sequence (5′-3′) | PCR product size | Reference |
|---|---|---|---|---|
| IntI1F | intI1 | GTTCGGTCAAGGTTCTGG | 890 bp | 23 |
| IntI1R | CGTAGAGACGTCGGAATG | |||
| K90 | 16S rRNA gene | GAGAGTTTGATCCTGGCTCAG | 1.4 kb | 23 |
| K94 | CGGCTACCTTGTTACGACTTC | |||
| orf341A | orf513 | CGCCCACTCAAACAAACG | 452 bp | 14 |
| orf341B | GAGGCTTTGGTGTAACCG | |||
| hep58 | Class 1 gene cassette array | TCATGGCTTGTTATGACTGT | Variable | 22 |
| hep59 | GTAGGGCTTATTATGCACGC | |||
| aadA2F | GCTAAGCAAGCTTATCTGGGAC | This study | ||
| 513Sp1 | The region downstream of ISCR1 | TCGGCCATTCCGACGTCTCTACGA | Variable | This study |
| 513Sp3 | CGCTCACCGCTTGATNNNNNNNNNCCCCTC | |||
| 513Sp2 | CATGTGCTGAAAGTTGGCGGTGCC | |||
| qnrVC4XF | qnrVC4 | CCCTCGAGCATGGATAAAACAGACCAGTTATA | 657 bp | This study |
| qnrVC4BR | CGGGATCCTTAGTCAGGAACTACTATTAAACCT | |||
| Pant-XF (with qnrVC4BR) | Pant, aacA4, qnrVC4 | CCCTCGAGCGAAACGGATGAAGGCAC | 1.8 kb | This study |
| gyrAF | gyrA | TCCTATCTTGATTACGCCATG | 482 bp | This study |
| gyrAR | CATGCCATRCCYACCGCRAW | |||
| gyrBF | gyrB | GGGGTCTACTGCTTCACCAA | 704 bp | This study |
| gyrBR | GCATCTGTCATGATGATGATG | |||
| parCF | parC | GTKCAGCGSCGCATCATCTAC | 243 bp | This study |
| parCR | CGGTRTAACGCATKGCSGC |
The qnrVC4 coding sequence (CDS) was amplified using primers qnrVC4XF and qnrVC4BR, and a 1.8-kb fragment containing the native Pant promoter of aacA4 and qnrVC4 was obtained by PCR using the primers Pant-XF and qnrVC4BR, digested with XhoI and BamHI, ligated into pBCKS(+), and transformed into Escherichia coli Top10. Recombinants were selected and verified by sequencing. The MICs of ciprofloxacin, gatifloxacin, and nalidixic acid for clones carrying qnrVC4 were 0.032, 0.047, and 4 μg/ml, respectively, and the corresponding MICs for an alternate insert with a Pant recombinant were 0.008, 0.006, and 2 μg/ml, respectively. As a negative control, E. coli Top10 showed MICs of 0.002 μg/ml for ciprofloxacin and gatifloxacin and 0.5 μg/ml for nalidixic acid. Pant of A. punctata 159 belonged to PcWTGN-10 containing the weak promoter (PcW: −35 TGGACA and −10 TAAGCT) with a TG motif at positions −15 and −14 before the −10 region (Fig. 1A). The PcWTGN-10 was 1.7-fold less active than the strong promoter (PcS: −35 TTGACA and −10 TAAACT) and 15-fold more active than PcW (8). Another putative promoter like qnrVC1 was found in the qnrVC4 cassette (Fig. 1B). The MIC levels of qnrVC4 in the integron are probably determined by the promoters and the environment downstream of the aacA4 cassette.
Therefore, critical regions of gyrA, gyrB, and parC, including quinolone resistance-determining regions (QRDR), were sequenced (with primers listed in Table 1). In the 482-bp gyrA amplicon, “X” mutation (random mutation) leads to replacement of a conserved Ser-83 with Ile, which is associated with reduced sensitivity to quinolones in A. punctata (6, 15). GyrB QRDR peptides are identical to those of other Aeromonas spp. Analysis of ParC QRDR fragments showed that they were the same as those of the sensitive strain Aeromonas caviae CIP 7616 (6). The gyrase A subunit containing Ser-83 may form a turn in the secondary structure, but the Leu-83 mutant would form an α-helix (24), which may affect binding of quinolones to the target GyrA.
Quinolone resistance is caused by both topoisomerase mutations and plasmid-mediated determinants in Aeromonas spp. The qnrVC4 cassette shows strong nucleotide similarity to qnrVC2 (99%), and QnrVC4 shows 81% and 77% identities with QnrVC3 and QnrVC1, respectively, indicating that qnrVC4 would have originated from Vibrionaceae (5). The qnrVC4 gene, with its novel gene cassette array and blaPER-1, is the first qnrVC gene associated with blaPER-1 and a complex class 1 integron in Aeromonas spp. These results suggest that there may be further variants of qnr genes or new complex structures.
Nucleotide sequence accession numbers.
The complex class 1 integron sequences and gyrA, gyrB, and parC nucleotide sequences in A. punctata 159 have been deposited into the GenBank database with the accession numbers GQ891757, GQ891754, GQ891755, and GQ891756, respectively.
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China (no. 30870084) and State Key Laboratory of Microbial Technology, Shandong University.
We thank George A. Jacoby for providing E. coli J53 (azide resistant) and Vivian Miao from University of British Columbia for helpful comments on manuscript preparation.
Footnotes
Published ahead of print on 1 June 2010.
REFERENCES
- 1.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 2.Cattoir, V., L. Poirel, C. Aubert, C. J. Soussy, and P. Nordmann. 2008. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg. Infect. Dis. 14:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavaco, L. M., H. Hasman, S. Xia, and F. M. Aarestrup. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 53:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Fonseca, É. L., F. Dos Santos Freitas, V. V. Vieira, and A. C. P. Vicente. 2008. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg. Infect. Dis. 14:1129-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goñi-Urriza, M., C. Arpin, M. Capdepuy, V. Dubois, P. Caumette, and C. Quentin. 2002. Type II topoisomerase quinolone resistance-determining regions of Aeromonas caviae, A. hydrophila, and A. sobria complexes and mutations associated with quinolone resistance. Antimicrob. Agents Chemother. 46:350-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby, G., V. Cattoir, D. Hooper, L. Martínez-Martínez, P. Nordmann, A. Pascual, L. Poirel, and M. Wang. 2008. qnr gene nomenclature. Antimicrob. Agents Chemother. 52:2297-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jové, T., S. Da Re, F. Denis, D. Mazel, and M. C. Ploy. 2010. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet. 6:e1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, M. F., C. F. Peng, Y. H. Lin, S. R. Lin, and Y. H. Chen. 2008. Molecular diversity of class 1 integrons in human isolates of Aeromonas spp. from southern Taiwan. Jpn. J. Infect. Dis. 61:343-349. [PubMed] [Google Scholar]
- 10.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martínez-Martínez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 12.Picão, R. C., L. Poirel, A. Demarta, C. S. Silva, A. R. Corvaglia, O. Petrini, and P. Nordmann. 2008. Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J. Antimicrob. Chemother. 62:948-950. [DOI] [PubMed] [Google Scholar]
- 13.Poirel, L., L. Cabanne, H. Vahaboglu, and P. Nordmann. 2005. Genetic environment and expression of the extended-spectrum beta-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob. Agents Chemother. 49:1708-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabaté, M., F. Navarro, E. Miró, S. Campoy, B. Mirelis, J. Barbé, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying bla(CTX-M-9). Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha, S., S. Chattopadhyay, S. K. Bhattacharya, G. B. Nair, and T. Ramamurthy. 2004. An unusually high level of quinolone resistance associated with type II topoisomerase mutations in quinolone resistance-determining regions of Aeromonas caviae isolated from diarrhoeal patients. Res. Microbiol. 155:827-829. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi, S., and Y. Nagano. 1984. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J. Clin. Microbiol. 20:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verner-Jeffreys, D. W., T. J. Welch, T. Schwarz, M. J. Pond, M. J. Woodward, S. J. Haig, G. S. Rimmer, E. Roberts, V. Morrison, and C. Baker-Austin. 2009. High prevalence of multidrug-tolerant bacteria and associated antimicrobial resistance genes isolated from ornamental fish and their carriage water. PLoS One 4:e8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, M., Q. Guo, X. Xu, X. Wang, X. Ye, S. Wu, D. C. Hooper, and M. Wang. 2009. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 53:1892-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, S., J. He, Z. Cui, and S. Li. 2007. Self-formed adaptor PCR: a simple and efficient method for chromosome walking. Appl. Environ. Microbiol. 73:5048-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White, P. A., C. J. McIver, Y. Deng, and W. D. Rawlinson. 2000. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182:265-269. [DOI] [PubMed] [Google Scholar]
- 23.Xu, H., J. Davies, and V. Miao. 2007. Molecular characterization of class 3 integrons from Delftia spp. J. Bacteriol. 189:6276-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]