Abstract
Small peptides that inhibit the hepatitis C virus (HCV) at the stage of viral entry have the potential to serve as attractive antiviral drugs. Ribosome display is a cell-free system for in vitro selection of peptides from large random peptide libraries. Thus, we utilized a ribosome display library technique for affinity selection of HCV envelope protein E2-binding peptide ligands. Through 13 rounds of selection, the ribosome display system generated high-affinity 12-mer peptides, and the selected peptide PE2D (MARHRNWPLVMV) demonstrated the highest specificity and affinity to the HCV E2 protein. Furthermore, amino acids 489 to 508 (YPPRPCGIVPAKSVCGPVYC) of E2 were identified as crucial for binding to PE2D. The selected peptides, especially PE2D, not only dramatically blocked E2 protein binding to hepatocytes but also dramatically inhibited HCV cell culture (HCVcc) entry into hepatocytes. HCVcc and HCV particles from HCV patient serum samples could also be specifically captured using PE2D. Our study demonstrates that the newly selected peptide ligand PE2D holds great promise for developing a new molecular probe, a therapeutic drug specifically for HCV, or an early-diagnostic reagent for HCV surface envelope antigen E2.
Hepatitis C virus (HCV) is a major cause of chronic liver disease worldwide, and infection with this virus frequently results in cirrhosis and hepatocellular carcinoma (6, 9). Humans and chimpanzees are the only species that are susceptible to HCV infection. The lack of a small-animal model and difficulties in propagating HCV in cell culture have hampered HCV research. However, the production of infectious HCV in tissue culture was recently established from a cloned JFH-1 viral genome and has since been widely applied in many labs around the world (35, 51, 60). This new technique has played an important role in HCV-related research and drug discovery.
HCV is a member of the Flaviviridae family of viruses and encodes two envelope glycoproteins, E1 and E2, which form the major antigenic components of the virion (34). E1 and E2 may play a crucial role in the initiation of infection by mediating the interaction between the virus and the host cell membrane (17, 34). While E2 is thought to initiate viral attachment, E1 may be involved in virus-cell membrane fusion (17, 47). E2 has been proposed to be responsible for recognition and binding with cellular receptors (10, 18, 20, 21, 22, 54). HCV infection is dependent on at least three coreceptors: CD81 (3, 20, 38), scavenger receptor BI (SR-BI) (3, 21, 22), and claudin-1 (CLDN1) (15, 25, 48). Recently, occludin (OCLN) was reported as a new HCV entry factor required for infection of cells (44). In addition, CD81 has been identified as a critical coreceptor for HCV particle entry (25, 31, 38, 41, 54).
To date, the only available therapy for HCV infection is alpha interferon (IFN-α), either alone or in combination with ribavirin (37, 45). Such treatments are expensive, show low response rates, and carry the risk of significant side effects. Consequently, the development of effective drugs against HCV infection remains a high priority.
Early detection of HCV infection in HCV- or HIV-infected patients has significant implications for patient management. However, the currently recommended serological screening strategy for identifying anti-HCV antibodies is unable to detect plasma donations that are anti-HCV negative and HCV RNA positive during the preseroconversion window period (PWP) (30). Another method for detecting HCV RNA is reverse transcription-PCR (RT-PCR), which is not feasible due to its high cost (12); it is not used routinely in diagnostic laboratories, especially in developing countries. No current method is available to measure HCV surface antigens in serum. More sensitive and less expensive assays for the early diagnosis of HCV antigens are needed.
In recent years, a number of different methods have been developed to screen peptide or protein libraries for specific target molecules (39). The majority of these methods, such as phage display (14, 50), cell surface display (5), plasmid display (11), and the yeast two-hybrid system (16), are termed in vivo systems because living cells are involved in these processes of library generation or screening. Therefore, the complexities of libraries are limited by transformation efficiency to about 109 sequences. This limitation could be resolved for in vitro display technologies such as ribosome displays (19, 23, 24, 26, 27, 28, 29, 32) by introducing a cell-free translation system. Since this system does not require transformation, large libraries can be prepared and utilized for selection. This technique allows a much larger sampling of random sequences (1013) than has been previously possible. Ribosome display is an in vitro cell-free system of coupling individual nascent proteins (phenotypes) to their corresponding mRNAs (genotypes) via the formation of stable protein-ribosome-mRNA complexes. This technique permits the selection of a functional nascent protein by iterative cycles of panning and RT-PCR amplification in vitro. Furthermore, diversifications can be convenient. Random peptide libraries displayed on the ribosome are increasingly being used for the in vitro selection of biologically relevant macromolecules, including epitopes, antagonists, enzymes, and cell surface receptors (1, 13, 36, 52, 55, 58). Therefore, we applied a ribosome display system to select peptides that could bind to the HCV E2 envelope protein.
Small peptides that inhibit the virus at the stage of viral entry, for example, by blocking the interactions between the viral envelope glycoprotein and the cellular receptor or coreceptor, are attractive antiviral drugs. In the present study, we have identified peptide ligands that specifically bind to the HCV E2 envelope protein by using this ribosome display technology and examined the binding affinities and functions of the selected peptides.
MATERIALS AND METHODS
Materials.
Escherichia coli DH5α and E. coli BL21(DE3) were used in this study according to procedures previously described (53). Human hepatocellular liver carcinoma (Huh-7.5.1) cells were cultured in RPMI 1640 medium with 10% (vol/vol) fetal bovine serum (HyClone). HCV cell culture (HCVcc)-JFH-1 (an E2 genotype 2a isolate) and Huh-7.5.1 cells were derived from the Huh-7.5 cell line, which was kindly provided by Wen-Zhe Ho from the University of Pennsylvania School of Medicine (56). Cloned E1E2 genes of HCV genotypes 1 through 6, in pcDNA3 expression vectors, were kindly provided by Jonathan K. Ball from the University of Nottingham (41). The CD81 recombinant protein was prepared according to a previously published paper (7). Recombinant E2-glutathione S-transferase (E2-GST) and GST proteins and anti-E2 polyclonal antibody were prepared as previously described (33). CD81 (GenBank accession number BC093047), SR-B1 (GenBank accession number BC112037), CLDN1 (GenBank accession number BC012471), and OCLN (GenBank accession number BC029886) were cloned into a pLenti 6 (Invitrogen) expression vector. Anti-human SR-B1 monoclonal antibody (MAb) (BD Pharmingen), anti-human CD81 MAb 5A6 (Santa Cruz Biotechnology), rabbit anti-human CLDN1 (Cell Signaling), and anti-human OCLN (Invitrogen) were used in this study.
Serum samples.
HCV patient serum samples or healthy donor serum samples were collected from 2007 to 2009 at the Zhongnan Hospital of Wuhan University, Wuhan, China. All patients were negative for antibodies against HIV, hepatitis B virus (HBV), and hepatitis D virus. The study was approved by the local ethics committee, and all patients and controls in the study gave informed consent before blood donation.
Construction of the ribosome display library.
A ribosome display library was constructed according to previously described methods (53). The random oligonucleotide template was synthesized as a single-stranded 90-mer with the following sequence: 5′-ACCGGATCCATGATGATGATGATGATGGCCGGA(NNM)12GCTAGCCATGGATATATGTCC-3′, where the central (NNM)12 represents the random oligonucleotides: M is either A or C, and N is either A, T, G, or C. The italics represent the BamHI site, the bold represents the start codon, and the underlined text represents the Shine-Dalgarno (SD) sequence. The template for transcription/translation was synthesized by two rounds of PCR.
In vitro-coupled transcription/translation and affinity selection.
In vitro-coupled transcription/translation in an E. coli S30 system was performed as previously described (53). The random DNA template, amino acid mixture, S30 premix, and S30 extract were incubated at 30°C for 2.5 h. The synthetic DNA library encoding random peptide sequences was used to generate ribosome complexes.
Peptides that bound to the HCV E2 protein were selected via the ribosome display library. The coupled transcription/translation product was incubated with E2-GST-Sepharose 4B by gently shaking at 4°C for 1 h. The trapped polysomes were disrupted with EDTA to release bound mRNA, which was then converted into cDNA by RT-PCR. After being washed three times with ice-cold buffer (50 mM Tris acetate [pH 7.5], 150 mM NaCl, 1% [vol/vol] Tween 20, and 5 mM magnesium acetate), the retained ribosomal complexes were dissociated with ice-cold elution buffer (50 mM Tris acetate [pH 7.5], 150 mM NaCl, and 10 mM EDTA) for 10 min at 4°C by gentle shaking. The released mRNA was recovered using an RNaid kit. Purified RNA was subsequently used for RT-PCR. From round three to round six, RNA pools were first bound to GST-Sepharose 4B to remove nonspecifically bound RNA, and then the unbound RNA was bound to E2-GST-Sepharose 4B compounds.
Cloning, sequencing, and synthesis of selected peptides.
After selection, RT-PCR products were digested with PstI and BamHI and then subcloned into pUC19 vectors (PstI and BamHI digestion), which were then transformed into E. coli DH5α cells. Plasmid DNA was isolated from individual clones, purified, and analyzed by sequencing. Individual peptide sequences were obtained and then synthesized.
SPR affinity assay.
Surface plasmon resonance (SPR)—a label-free, biosensor-based system—has been widely used to study biomolecular interactions in real time by providing high-quality kinetic and affinity data as well as rapid active concentration determination. We analyzed the interactions between the E2 protein and selected peptides by the SPR technique using a Biacore 1000 biosensor, according to the previously published method (40). The values of the association rate (Kon) and the dissociation rate (Koff) were calculated from the forward and reverse reactions of the optimally fitted binding curves, respectively.
ELISA.
For analysis of the binding affinities of different concentrations of PE2D with the E2-GST or GST proteins, an indirect enzyme-linked immunosorbent assay (ELISA) method was used. ELISA (96-well) plates were coated with 0.3 nM E2-GST or GST protein at 4°C for 7 h. The plates were then blocked with 2% bovine serum albumin (BSA) at 37°C for 2 h. After the plates were washed with phosphate-buffered saline (PBS) containing 0.1% Tween 20 three times, different concentrations of biotin-labeled PE2D peptides were added to each well. After incubation at 37°C for 30 min, 100 μl of horseradish peroxidase (HRP)-streptavidin (1:1,000) was added and incubated at 37°C for 1 h. Finally, the samples were developed with a substrate solution containing o-phenylenediamine. The absorbance of each sample was measured at 450 nm. All data were analyzed for statistical significance.
For analysis of the PE2D binding site on the E2 protein, 15 E2 fragments (P1 through P15) that spanned different regions of E2 were synthesized by HD Biosciences Co. Ltd. The ELISA plates were coated with 0.3 nM each E2 fragment (P1 to P15, 100 μl per well) at 4°C for 7 h. After the plates were blocked with 2% BSA and washed with PBS, 250 nM biotin-labeled PE2D was added to each well. The same steps as those described above were then followed.
For the virus capture assays, both indirect and sandwich ELISA methods were used. In the indirect ELISA, some 96-well plates were coated with HCV infectious particles (3 × 105 virus particles in 180 μl per well) in carbonate buffer, pH 9.6, for 7 h at 4°C and blocked with 1% BSA at 4°C overnight. After incubation, the plates were washed, and different concentrations of biotin-labeled PE2D were added to each well. Each concentration was tested in triplicate. The same steps as those described above were then followed.
In the sandwich ELISA, 96-well plates were coated with 0.3 nM PE2D (100 μl per well) and blocked with 2% BSA at 4°C overnight. A total of 100 μl of HCV patient serum samples (n = 50, both HCV antibody and HCV RNA positive) or healthy donor serum samples (n = 55, both HCV antibody and HCV RNA negative) were added and incubated at 37°C for 30 min. The plates were then extensively washed to remove unbound viral particles, rabbit anti-E2 polyclonal antibody (1:150) was added to each well, and the plates were incubated at 37°C for 1 h. The plates were then washed again, and HRP-labeled streptavidin was added. The same steps as those described above were then followed.
Flow cytometry.
To analyze the blocking effects of the selected peptides (PE2A, PE2B, PE2C, and PE2D) on the HCV E2 binding ability to Huh-7.5.1 cells, 250 nM peptides, 50 IU heparin, 120 nM CD81, or their mixture was incubated with 0.25 μg E2 protein in a final volume of 100 μl PBS buffer at 37°C for 1 h. They were then mixed with 1 × 106 Huh-7.5.1 cells and further incubated at 37°C for 30 min. The cells were then washed in PBS and stained in 100 μl of PBS containing 1% BSA and anti-E2 polyclonal antibody for 30 min. The cells were pelleted, washed, and resuspended in PBS containing 1% BSA and phycoerythrin (PE)-labeled goat anti-rabbit IgG antibody. After incubation, the cells were washed, and cell-bound E2 was analyzed using flow cytometry.
The binding affinities of E2 to target cells by flow cytometry were obtained by monitoring the mean fluorescence intensity of target cells bound to the fluorescein isothiocyanate (FITC)-labeled anti-E2 IgG as previously described (8, 49). The equation Y = BmaxX/(Kd + X) (Origin Pro 7.5) was used to calculate the target antigen E2 binding equilibrium dissociation constant Kd, where Bmax is the maximum percentage of fluorescence, Y is the mean percentage of fluorescence, and X is the ligand E2 protein concentration expressed in moles.
Fluorescence microscopy analysis of the blocking effects of peptides on HCVcc infection.
Huh-7.5.1 cells were cultured in 6-well plates at a concentration of 4.5 × 105 cells/well with 200 μl (7 × 107 copies) of JFH-1 HCVcc and 250 nM PE2D, PE2D scramble mutant (PE2D-mut), or 10,000 IU/well IFN-α. The mixtures were incubated at 37°C overnight. Cells were washed with diethyl pyrocarbonate (DEPC)-treated PBS to remove HCVcc in the supernatant and then fixed with 4% paraformaldehyde. The cells were then washed with PBS containing 0.2% BSA. FITC-labeled anti-rabbit IgG and rabbit anti-E2 polyclonal antibodies were added to the Huh-7.5.1 cells, incubated for 4 h, and washed with PBS three times. Cells were then permeabilized with buffers containing 0.5% Triton X-100, 0.1% SDS, and 50 mmol/liter Tris (pH 8.0) and stained with 100 μg/ml of the DNA-binding dye propidium iodide (PI) (Sigma) at room temperature for 5 min. The cells were washed three times with PBS. Imaging of the cells on the cover slide was performed with a confocal fluorescence microscope (Leica DM RXA) under 488-nm excitation light (green for FITC and red for PI).
Real-time quantitative RT-PCR analysis of the inhibitory effects of peptides on HCVcc infection.
Huh-7.5.1 cells were cultured in 6-well plates at a concentration of 4.5 × 105 cells/well with different final concentrations of PE2D or PE2D-mut or with 10,000 IU/well IFN-α, along with 200 μl (7 × 107 copies) of JFH-1 HCVcc. The mixtures were incubated at 37°C overnight. Cells were washed with DEPC-treated PBS to remove any HCVcc in the supernatant, and the total RNA was extracted using Trizol reagent (Invitrogen Life Technologies) and reverse transcribed using the first-strand cDNA synthesis kit (Fergment). The HCV primers were designed based on the HCV sequence (GenBank accession number M67463) selected within the 5′ noncoding region (NCR) of the HCV genome according to a previous publication (56). RNA was quantified using RT-PCR and the QuantiTect SYBR green PCR handbook kit (Qiagen).
Statistical analysis.
Differences were considered to be statistically significant at P values of <0.05. Data were analyzed using the Student t test and the nonparametric method with SPSS software.
RESULTS
Selection of peptides that bind to HCV E2 by ribosome display.
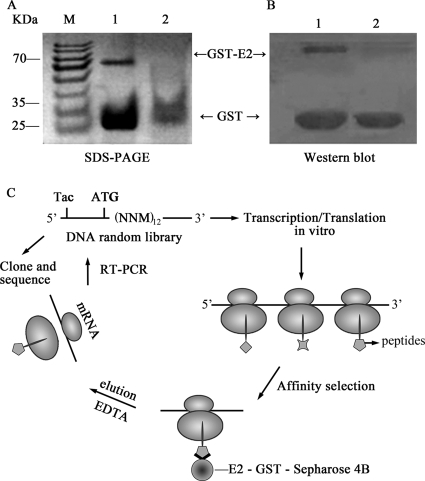
First, E2-GST and GST proteins were purified and identified by SDS-PAGE (Fig. 1 A) and Western blot analysis (Fig. 1B). To isolate peptides that specifically bound to the HCV E2 protein, we utilized E2-GST-Sepharose 4B as a selection target and GST-Sepharose 4B as a counterselection target. The ribosome display system with a distinct random 12-mer peptide library was utilized (53), and mRNA-carrying peptide products were bound to E2-GST-Sepharose 4B beads (Fig. 1C). After a 12-cycle selection, the 12th-pool clones from RNA that bound to E2-GST-Sepharose 4B beads were reverse transcribed by RT-PCR and cloned into pUC19 vectors. Thirty individual clones were selected for DNA sequencing and then translated into 12-mer amino acid sequences. The results of an NCBI BLAST analysis showed that the four selected 12-mer peptides have protein-binding potentials, and they were then further selected for chemical synthesis and named PE2A, PE2B, PE2C, and PE2D (Table 1).
FIG. 1.
High-affinity peptides for HCV-E2 were isolated by the ribosome display technique. (A and B) SDS-PAGE (A) and Western blot analysis (B) of the purified E2-GST and GST proteins. (C) The random DNA library, which contained open reading frames and lacked stop codons, was transcribed and translated in an E. coli S30 system. The ribosome was stalled at the end of the mRNA. The encoded protein was not released and folded correctly on the ribosome. The mRNA-ribosome-protein complexes were used for affinity selection of E2-GST-Sepharose 4B. After being washed, the bound ribosomal complexes were dissociated. The mRNA was purified and used for reverse PCR amplification. After 12 rounds of selection, RT-PCR products were cloned into pUC19 for subsequent DNA sequencing and then translated into amino acid sequences.
TABLE 1.
Kinetic binding parameter data of the interactions between E2 and PE2A to -D by SPR analysisa
| Peptide (amino acid sequence) | Kon (cells/s) | Koff (cells/s) | Kd (nM) |
|---|---|---|---|
| PE2A (GFGRYRRHGSPW) | 2.56 × 104 | 2.22 × 103 | 50 |
| PE2B (MAIGPYPACGSG) | 3.22 × 104 | 1.39 × 10−3 | 43 |
| PE2C (LSVLVISMFNAV) | No binding | No binding | No binding |
| PE2D (MARHRNWPLVMV) | 7.51 × 104 | 1.45 × 10−3 | 19 |
The peptide sequences are protected by patent (X.-L. Zhang, 17 June 2009, Chinese patent applications 200,710,168,887.5 and 200,710,168,890.7). Kon, association rate; Koff, dissociation rate; Kd, Koff/Kon.
Determination of the binding affinities between the selected peptides and the E2 protein.
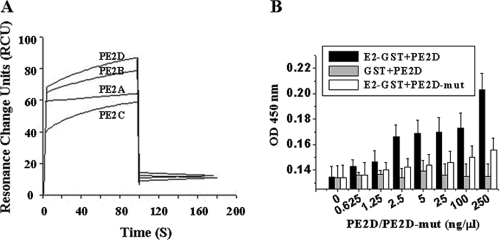
The binding affinities of the four selected peptides to the HCV E2 protein were measured by SPR (Fig. 2 and Table 1). The differences in dissociation constants indicated that PE2D bound to E2 with the highest binding affinity (with a Kd of 19 nM) in the low nanomolar range. In contrast, PE2B and PE2A had relatively low affinities (Kds of 43 nM and 50 nM, respectively) for E2, and PE2C exhibited undetectable binding with the E2 protein (Table 1). The sequence of binding affinities was as follows: PE2D (Kd, 19 nM) > PE2B (Kd, 43 nM) > PE2A (Kd, 50 nM) > PE2C (undetectable Kd). Usually, the peptide with the lowest Kd displays the highest binding affinity (40, 53). Therefore, PE2D was chosen for further characterization, as it had the highest E2 binding affinity based on dissociation constants.
FIG. 2.
Binding affinity analysis of the interaction between selected peptides and the HCV E2 protein. (A) A typical time profile of the SPR signal. The SPR analysis was performed as described in Materials and Methods. The data demonstrate the interaction between E2 and the four peptides, PE2A to -D. PE2C has the lowest binding affinity. (B) The indirect ELISA method was used to analyze the binding affinities of different doses of PE2D and PE2D-mut with the E2-GST or GST proteins. OD, optical density.
An ELISA analysis showed that PE2D bound to E2-GST in a peptide concentration-dependent manner (Fig. 2B). However, different concentrations of PE2D did not have any detectable binding affinities to the GST control protein (Fig. 2B). However, different concentrations of scrambled PE2D (PE2D-mut) had a significantly lower binding affinity to E2-GST than PE2D (Fig. 2B).
The selected peptide PE2D specifically targets HCV particles.
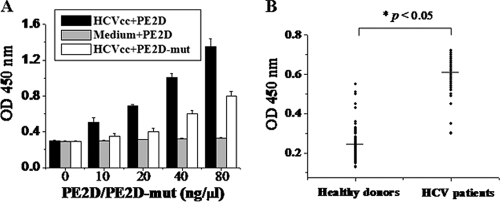
To determine whether PE2D could bind to HCV particles, virus capture assays were performed by both indirect and sandwich ELISA methods. In the indirect ELISA, virus samples from HCVcc or control cell culture medium were used to coat ELISA plates and incubated with different concentrations of biotin-labeled PE2D, followed by extensive washing to remove unbound peptide. The peptide-bound viral particles on the ELISA plate were revealed using HRP-labeled streptavidin, as described in Materials and Methods. The results showed that PE2D bound to HCVcc in a concentration-dependent manner (Fig. 3 A), while it did not bind to the control cell culture medium (Fig. 3A). These results are consistent with the binding affinities of PE2D with the different concentrations of E2 protein shown in Fig. 2B. However, the different concentrations of PE2D-mut had significantly lower binding abilities to HCVcc than PE2D (Fig. 3B).
FIG. 3.
Virus capture assays. (A) Binding affinity analysis of the interaction between different doses of PE2D and HCVcc (3 × 105 virus particles in 180 μl per well) in 96-well plates by indirect ELISA. The data shown are the means ± standard errors of the mean (SEM) from six separate experiments. (B) Serum samples from both HCV patients and healthy donors were analyzed by the sandwich ELISA. HCV particles from HCV patient serum samples could be specifically captured using peptide PE2D. Peptide PE2D-bound viral particles in the ELISA plate were revealed by addition of anti-E2 polyclonal antibody and HRP-labeled IgG. The absorbance of each sample was measured at 450 nm. Data were analyzed with the nonparametric statistical analysis method.
In the sandwich ELISA, PE2D was adsorbed to the ELISA plate, and serum samples from 50 HCV patients and 55 healthy donors were added and incubated. The PE2D-bound viral particles were revealed following the addition of anti-E2 polyclonal antibody and HRP-labeled IgG. As shown in Fig. 3B, biotin-labeled PE2D could bind to HCV particles in HCV patient samples with much higher affinities than it could to healthy donor samples (P < 0.05). These data suggest that the selected peptide PE2D specifically targets HCV particles and could be used to diagnose early HCV infection by detecting HCV surface antigen in sera that is present during the early stage or before seroconversion.
Identification of the PE2D peptide-binding site on the E2 protein.
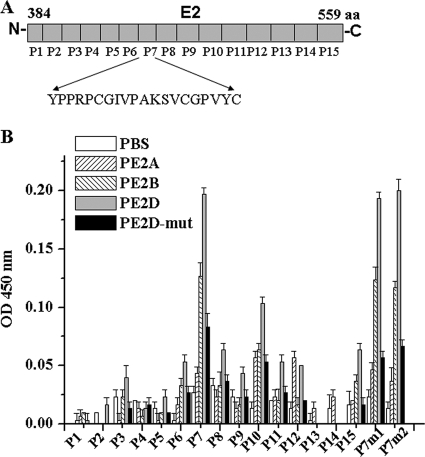
To further identify the PE2D peptide-binding site on E2, 15 peptide fragments (P1 to P15), which were based on the genotype 1a E2 amino acid sequence and secondary structure, were synthesized (Fig. 4 A). In the indirect ELISAs, each E2 fragment (P1 to P15) was used to coat ELISA plates and incubated with biotin-labeled PE2D. The biotin-PE2D-bound E2 fragments in the ELISA plate were revealed by the HRP-labeled streptavidin, as described in Materials and Methods. Of all the E2 fragments tested, the results showed that PE2D bound most strongly to the E2-P7 fragment sequence (YPPRPCGIVPAKSVCGPVYC) (Fig. 4B). The binding affinity between PE2D and P7 was 2- to 20-fold higher than those between PE2D and the other 14 peptides (Fig. 4A). While PE2A exhibited much lower binding affinities to each E2 peptide (P1 to P15) of E2, it still exhibited binding affinities that were higher than those of the PBS control, while PE2D-mut showed significantly lower binding affinities to each E2 peptide (P1 to P15) than PE2D (Fig. 4B).
FIG. 4.
Identification of the PE2D peptide-binding site of the E2 protein. (A) Diagram of the 15 E2 peptide fragments (P1 to P15) that spanned different regions of E2.
Since PE2D bound most strongly to the E2-P7 fragment sequence, we compared the recognized E2 sequences (the P7 fragment sequences) in different genotypes of HCV E2 (Table 2) and found several conserved amino acids in the E2-P7 fragment. Two regions of conserved amino acids of the P7 fragment were mutated to create P7m1 (P491, C494G495 to A491, A494A495) and P7m2 (C503G504 to A503A504), and then we detected their binding abilities to PE2D. Results (Fig. 4B) showed that P7m1 and P7m2 had no reduced blocking effects on E2-receptor interaction, which indicates that the mutated amino acids of P7m1 (P491 and C494G495) and P7m2 (C503G504) are not the critical sites for PE2D binding to E2. Some other conserved amino acids (for example, P505VYC508 or P498A499, shown in Table 2) might be the important sites for PE2D binding to E2.
TABLE 2.
Comparison of E2-P7 fragment sequences in several HCV E2 genotypes
| HCV genotype | Isolated strain | GenBank accession no. | HCV E2-P7 sequencea |
|---|---|---|---|
| 1a | H77 | AAB67038.1 | Y489PPRPCGIVPAKSVCGPVYC508 |
| 1b | HC-J4/91 | AAC15725 | H488YAPRPCGVPASQVCGPVYC508 |
| 1b | CON1 | Q9WMX2.3 | Y489APRPCGIVPAAGVCGPVYC508 |
| 2a | JFH-1 | BAB32872.1 | Y489PPKPCG-WPARSVCGPVYC508 |
| 3a | K3a/650 | BAA06044 | Y489APRPCGIVPALNVCGPVYC508 |
| 4a | ED43 | CAA72338 | Y489APRPCGIVPASSVCGPVYC508 |
| 5a | EUH1480 | CAA73640 | Y489PPRPCGVVPARDVCGPVYC508 |
| 6a | 6a33 | AAW56714 | Y489APRPCDVVPASTVCGPVYC508 |
Underlined residues were mutated from P491 to A491 and C494G495 to A494A495 in P7m1 or from C503G504 to A503A504 in P7m2. The bold letters represent conserved amino acids among six genotypes of HCV E2.
PE2D and CD81 may share a partial binding site on the E2 protein.
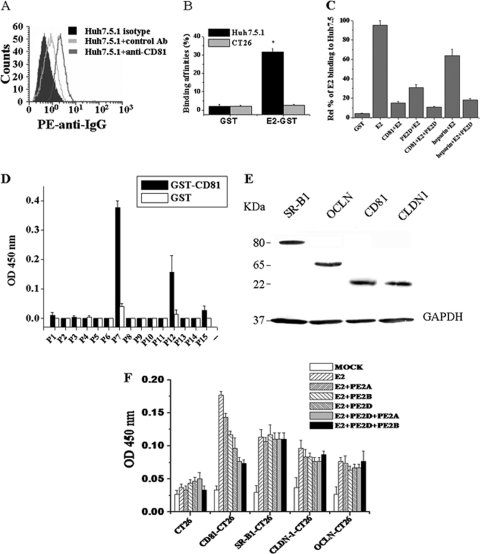
Human CD81, which is expressed on the surface of the human hepatocyte cell line Huh-7.5.1 (47), has been described as a main receptor for HCV (10, 31, 59) and may represent a molecular target for clinical intervention in HCV infection. We used a PE-labeled anti-CD81 antibody to detect whether Huh-7.5.1 cells expressed CD81. Flow cytometric analysis showed that PE-labeled anti-CD81 antibody bound to Huh-7.5.1 cells with a much higher binding affinity than that of the PE-labeled control antibody (Ab) (Fig. 5 A), which indicates that CD81 is expressed on the surfaces of Huh-7.5.1 cells. Flow cytometric analysis showed that the E2-GST protein bound to Huh-7.5.1 cells with a significantly higher affinity than that of the GST protein (Fig. 5B). The E2 protein specifically bound to Huh7.5.1 cells but not to CT26 cells (Fig. 5B). All of the above data suggest that Huh-7.5.1 cellular surfaces have HCV E2-binding CD81 receptor molecules.
FIG. 5.
PE2D can partially page HCV E2 binding with the CD81 receptor. (A) CD81 molecules are expressed on the cellular surface of Huh-7.5.1 cells, as demonstrated by flow cytometric analysis with PE-labeled anti-CD81 antibody. (B) HCV E2 had a much higher binding affinity for Huh-7.5.1 cells than for CT26 cells by flow cytometric analysis. E2-GST or GST proteins (0.25 μg) were preincubated with 1 × 106 Huh-7.5.1 or CT26 cells, respectively. Anti-GST antibody and FITC-anti-IgG were added and analyzed by flow cytometric analysis. (C) PE2D and CD81 may share a partial binding site on the E2 protein. Huh-7.5.1 cells mixed with PE2D, CD81, heparin, CD81 plus PE2D, or heparin plus PE2D were incubated with E2 protein at 37°C for 1 h, as described in Materials and Methods, and then cell-bound E2 was detected by flow cytometric analysis with rabbit anti-E2 polyclonal antibody stained with PE-conjugated rabbit IgG. The data are shown as the means ± SEM from three separate experiments. (D) The binding affinities of GST-CD81 for the different E2 fragments (P1 to P15) were measured using ELISA. The data are shown as means ± SEM from six separate experiments. (E) Determination of CD81, SR-B1, CLDN1, or OCLN expression in the CD81-, SR-B1-, CLDN1-, or OCLN-transfected CT26 cells by Western blot analysis, using anti-CD81 MAb 5A6, anti-SR-B1 MAb, anti-CLDN1, or anti-OCLN, respectively. (F) Comparison of the blocking effects of PE2D, PE2B, PE2A, or PE2D-mut on the E2 receptor interactions. The E2 protein (1 μg/100 μl) was premixed with the selected peptide (100 pmol/100 μl) and incubated with CT26 cells (1 × 105) stably expressing CD81, SR-B1, CLDN1, or OCLN in 96-well plates. The binding of E2 to receptor-expressing CT26 cells was detected using 50 nM biotin-anti-E2 aptamer ZE2, followed by the addition of HRP-streptavidin. The absorbance of each sample was measured at 450 nm.
Some other reports have demonstrated that cellular binding of the HCV E2 envelope protein requires cell surface heparan sulfate (2). Next, we examined whether PE2D and CD81 or heparan sulfate shared similar binding sites on the HCV E2 protein. Huh-7.5.1 cells were incubated with the HCV E2 protein in the presence or absence of the CD81 protein or heparan sulfate. Anti-E2 antibody was used to detect E2 binding with Huh-7.5.1 cells by flow cytometric analysis. As shown in Fig. 5C, CD81 and PE2D blocked approximately 82% and 70% of HCV E2 protein binding to Huh-7.5.1 cells, respectively (Fig. 5C). CD81 and PE2D may occupy overlapping binding sites on the E2 protein, since we observed that the binding percentages were reduced only by a small degree after coincubation, from 18% (CD81 plus E2) to 15% (CD81 plus E2 plus PE2D) (Fig. 5C). The addition of heparan sulfate blocked approximately 34% of HCV E2 protein binding to Huh-7.5.1 cells. However, the addition of both PE2D and heparan sulfate dramatically blocked approximately 82% of HCV E2 protein binding to Huh-7.5.1 cells (Fig. 5C), indicating that these may not have overlapping binding sites on the E2 protein.
We further examined the interactions between CD81 and different regions (P1 to P15) of E2. CD81 bound most tightly to P7 and second most tightly to P12 (Fig. 5D). These data further demonstrate that PE2D and CD81 may share similar binding sites on the P7 fragment of E2.
It is believed that HCV infection is dependent on four major coreceptors (CD81, SR-B1, CLDN1, and OCLN) (43). We further tested which of these coreceptor interactions is the target for PE2D. The E2 protein was premixed with the selected peptide and incubated with CT26 cells stably expressing CD81, SR-B1, CLDN1, or OCLN. The binding of E2 to receptor-expressing CT26 cells was detected using biotin-anti-E2 aptamer ZE2 (ZE2 is similar to anti-E2 MAb) (8). We detected the blocking effects of PE2D on the binding of E2 to CT26 cells expressing each receptor (Fig. 5E). As shown in Fig. 5F, PE2D dramatically blocked E2 binding to CD81-expressing CT26 cells, while it did not affect the binding of E2 to SR-B1-, CLDN1-, or OCLN-expressing CT26 cells. PE2D had the best blocking effects among the selected peptides. Furthermore, PE2D plus PE2A and PE2D plus PE2B have stronger blocking effects than PE2D alone (Fig. 5F), which indicates that there is some functional synergy among these peptides.
Selected peptides block HCV E2 binding to Huh-7.5.1 target cells.
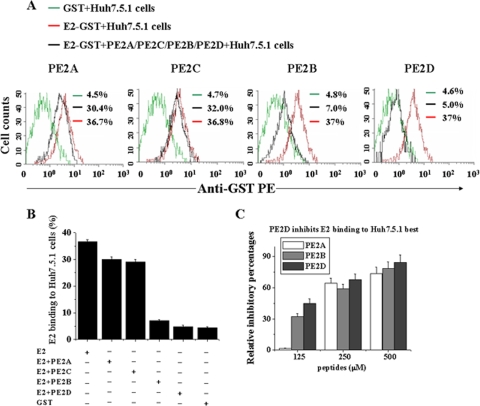
To determine whether the selected peptides were able to block HCV entry into target cells, increasing amounts of the selected peptides were added together with the E2 protein to Huh-7.5.1 target cells. Flow cytometric analysis (Fig. 6 A and B) demonstrated that PE2D had the strongest effect, reducing E2-binding affinities from 37% to 5%. PE2B had a slightly smaller effect (with reduction from 37% to 7%), and PE2C and PE2A exhibited the weakest inhibitory effects (with reductions from 37% to 30% and 37% to 32%, respectively) among the four peptides (Fig. 6A to C). PE2D and PE2B significantly inhibited E2 protein binding to Huh-7.5.1 cells in a concentration-dependent manner (Fig. 6C).
FIG. 6.
Inhibitory effects of the selected peptides on E2 protein binding to Huh-7.5.1 cells. (A) Huh-7.5.1 cells (1 × 106) were incubated with 0.25 μg E2-GST, GST alone, or E2-GST plus the 250 nM peptides (PE2A to D), followed by staining with anti-GST antibody. Cell-bound GST-E2 was detected with rabbit polyclonal anti-GST antibody stained with anti-rabbit FITC-IgG. Binding was analyzed by flow cytometric analysis. The data shown are representative of five experiments. (B) Statistical analysis of the E2-GST or GST binding percentages of Huh-7.5.1 cells as determined by flow cytometry. The data shown are the means ± SEM from four separate experiments. (C) Cell-bound E2 was examined using flow cytometry following the addition of different concentrations of PE2D, PE2A, or PE2B. The inhibitory percentages of selected peptides on the Huh-7.5.1 cell-bound E2 were calculated. All data shown are means ± SEM from four separate experiments.
PE2D blocks HCVcc infection of hepatocytes.
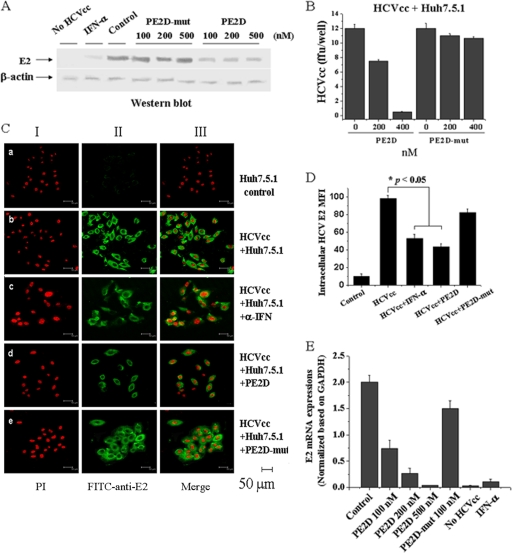
Finally, we examined the inhibitory effects of PE2D on HCV infection of human hepatocyte Huh-7.5.1 cells. Increasing concentrations of PE2D were added to HCVcc and target Huh-7.5.1 cell culture, and the inhibitory effects were determined. Both Western blot analysis and immunofluorescence staining for HCV E2 indicated that HCV E2 expression and the mean fluorescence densities of E2-positive cells in the HCVcc-Huh-7.5.1 cells decreased in a PE2D concentration-dependent manner (Fig. 7 A and B). Immunofluorescence microscopy demonstrated that HCV was detected in the cytoplasm of these cells (Fig. 7C) and the fluorescence densities were decreased upon addition of PE2D or IFN-α (Fig. 7C), while addition of the PE2D mutant had no or much less effect (Fig. 7A to D). Results similar to these were obtained with real-time quantitative RT-PCR analysis (Fig. 7E). In addition, both Western blot and real-time RT-PCR analyses showed that the inhibitory effects of 350 nM PE2D were virtually equivalent to those of 10,000 IU of IFN-α (Fig. 7A and E). These data illustrate that PE2D specifically inhibits HCV infection of human hepatocytes.
FIG. 7.
PE2D dramatically inhibits HCVcc infection of Huh-7.5.1 cells. (A) Western blot analysis of HCVcc incubated with different concentrations of PE2D or PE2D-mut in Huh-7.5.1 cells with anti-E2 antibody. The β-actin gene, a housekeeping gene with constant expression, was used as an internal control. (B) The differences in fluorescence of HCVcc incubated with different concentrations of PE2D or PE2D-mut in Huh-7.5.1 cells are shown as the means ± SEM from three separate experiments. ffu, focus-forming units. (C) Confocal fluorescence microscopic analysis of immunofluorescent detection of E2 in HCVcc-labeled Huh-7.5.1 cells. Cell nuclei were stained with the red fluorescent dye PI, and HCV was stained with green FITC-conjugated anti-E2 antibody. The illustrated data are representative of three separate experiments. (D) Quantitation (means ± SEM from three separate experiments) of the intracellular HCV E2 mean fluorescence intensities (MFI) shown in panel C. (E) Levels of intracellular HCV-E2 RNA were measured by real-time RT-PCR analysis. HCV-E2 RNA levels were normalized based on GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA levels assessed by real-time RT-PCR. The data shown are the means ± SEM from three independent experiments.
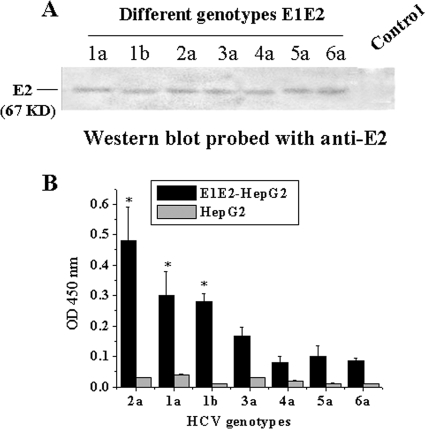
HCV has six major genotypes and numerous subtypes based on its positive-stranded RNA genome (20). HepG2 cells stably expressing the E1E2 proteins encoded by the E1E2 genes (from genotypes 1 to 6) were established by G418 selection, and E2 expressions from these cells were determined by Western blot analysis (Fig. 8 A). Different genotypes of HepG2 cells stably expressing the E1E2 gene were used to coat ELISA plates. We utilized ELISA to determine which genotypes possessed E1E2 that could be bound by PE2D. The results showed that the sequence of binding affinities of PE2D to E2 of each genotype was as follows: genotype 2a > 1a, 1b > 3a > 5a > 6a > 4a, and the binding affinities of PE2D with genotypes 1a, 1b, and 2a were 2- to 3-fold higher than those of genotypes 3a, 4a, 5a, and 6a (Fig. 8B). This result suggests that HCV genotypes 1 and 2 could be most effectively blocked by PE2D.
FIG. 8.
Different genotypes of E2 detected by PE2D. (A) Determination of E2 protein expressions of different genotypes of HepG2 cells stably expressing the E1E2 gene with anti-E2 antibody by Western blot analysis. HepG2 cells were used as the control. (B) Different genotypes of E2 were detected by PE2D. Data shown are the means ± SEM from three independent experiments.
DISCUSSION
Ribosome display has been used for the selection of peptide ligands (19, 32, 52) and proteins in vitro (4, 13, 24). One of the key advantages of ribosome display is that the diversity of the library is not limited by the transformation efficiency of bacterial cells (23). Additionally, ribosome display has been used to improve the affinity (4, 32) and the stability (28) of existing protein ligands. Using ribosome display, this study successfully generated 12-mer peptides that bound specifically to the HCV E2 protein. To our knowledge, this is the first time this finding has been demonstrated. We applied the SPR sensor system (Table 1 and Fig. 2A), ELISA (Fig. 2B and 3), and flow cytometric analysis methods (Fig. 5 and 6) to investigate the kinetic parameters and binding affinities of the selected peptides to the HCV E2 protein and HCV particles. Notably, we identified the strongest binding and interaction between the selected peptide PE2D and the E2 protein. The results of an NCBI BLAST analysis further demonstrated that the PE2D sequence was 75% identical (8/12 amino acid residues) to laminin (GenBank accession number EAW 75372) amino acids 3357 to 3368 (LARHRNWPSLSM [underlined letters represent identical residues]). These data illustrate that PE2D most likely has the binding domain of the laminin-alpha5 chain domain, which functions normally for protein binding (57). Because the molecular weight of laminin is much larger than that of PE2D, whether laminin can bind to E2 and block E2 binding to its receptors needs to be further investigated.
Recently, several reports showed that HCV cell entry is a multistep process (43). It is believed that the initial capture of HCV particles by glycosaminoglycans and/or lipoprotein receptors is followed by coordinated interactions with the SR-BI, the CD81 tetraspanin, the tight junction protein CLDN1, and/or OCLN. The different regions, structures, and conformations of E2 might be involved in binding to different coreceptors.
Several conserved residues in HCV E2 for CD81 binding were identified as W420, Y527, W529, G530, and D535 (41). Three putative CD81 interaction sites on HCV E2 have also been previously identified: region 1, amino acids 474 to 492 (YANGSGLDERPYCWHYPPR) (47); region 2, amino acids 522 to 551 (SGAPTYSWGANDTDVFVLNNTRPPLGNWFG) (47); and region 3, amino acids 612 to 619 (PYRLWHYPC) (47). We found that PE2D binds to amino acids 489 to 508 of E2 (Fig. 4A and B), and CD81 binds most tightly with the P7 fragment (amino acids 489 to 508) of E2 as well (Fig. 5D). These residues partly overlap with the previously identified CD81-binding region 1 (amino acids 474 to 492) of E2 (47) and are close to the CD81-binding conserved residues W420 and Y527 and region 2 (amino acids 522 to 551) of E2 (41, 47). These data demonstrate that PE2D and CD81 share an overlapping binding site on the P7 fragment of E2 and that PE2D can partially block HCV E2 binding with the CD81 receptor and subsequent viral entry by binding to these residues of E2.
Finally, using Western blot, real-time RT-PCR, and immunofluorescence staining analyses, we found that the selected peptides, especially PE2D, bound to E2 and HCV particles in a concentration-dependent manner (Fig. 3A) and that PE2D, but not its mutant, significantly inhibits HCVcc binding and entry into human hepatocyte Huh-7.5.1 cells (Fig. 7). These data suggest that PE2D may also hold potential for use as a drug against HCV infection; this potential of PE2D is strengthened by its small molecular weight (12-mer), its low immunogenicity, and the ease of its chemical synthesis in large quantities at a relatively low cost. The newly selected peptides, especially PE2D, are worthy of further clinical and basic research.
The limitations of the methods of detection of anti-HCV antibodies or HCV RNA in serum that are currently used for the diagnosis of HCV infection have enhanced efforts to find a rapid, simple, sensitive, and specific alternative diagnostic approach to detect viral antigens. Similar to the detection of an HBV surface antigen for the diagnosis of HBV infection, the selected E2-binding peptides may be helpful for the diagnosis of early HCV infection through a simple ELISA method to detect HCV E2 envelope surface antigen. Although HCV E2 contains approximately 1 or 2 amino acid sequence hypervariable regions (HVR), most of the E2 residues and the chemicophysical properties and conformation of HVR are highly conserved (42, 46). Our experimental results showed that PE2D specifically targets HCV particles and could also be used to diagnose early HCV infection by detecting HCV surface antigen in sera (Fig. 3A and B), especially for HCV genotypes 1 and 2 (Fig. 8B). Therefore, PE2D holds great potential for the detection of HCV surface antigen in both clinical applications and basic research.
In summary, our data demonstrate that the selected peptides, especially PE2D, hold great promise for developing new molecular probes, as therapeutic drugs, or as early diagnostic reagents targeting HCV. The selected peptides can also serve as tools for analyzing HCV-host cell interactions both in vitro and in vivo.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (20532020, 30670098, and 30870122), the 973 Program of China (2006CB504300 and 2009CB522507), the National Special Fund of China for the Important Infectious Diseases (2008ZX10003-005 and 2008ZX10002-013), and the Hubei Province Natural Science Foundation (2006ABD007).
The authors declare that there are patent applications for this work and for peptide sequences (X.-L. Zhang, 17 June 2009, Chinese patent applications 200,710,168,887.5 and 200,710,168,890.7).
Footnotes
Published ahead of print on 17 May 2010.
REFERENCES
- 1.Amstutz, P., P. Forrer, C. Zahnd, and A. Pluckthun. 2001. In vitro display technologies: novel developments and applications. Curr. Opin. Biotechnol. 12:400-405. [DOI] [PubMed] [Google Scholar]
- 2.Barth, H., C. Schäfer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. K. Toyoda, T. Toida, T. H. Kuppevelt, E. Depla, F. von Weizsäcker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 4.Binz, H. K., P. Amstutz, A. Kohl, M. T. Stumpp, C. Briand, P. Forrer, M. G. Grütter, and A. Plückthun. 2004. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 22:575-582. [DOI] [PubMed] [Google Scholar]
- 5.Boder, E. T., and K. D. Wittrup. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553-557. [DOI] [PubMed] [Google Scholar]
- 6.Boyer, N., and P. Marcellin. 2000. Pathogenesis, diagnosis and management of hepatitis C. J. Hepatol. 32:98-112. [DOI] [PubMed] [Google Scholar]
- 7.Cao, J., P. Zhao, X. H. Miao, L. J. Zhao, L. J. Xue, and Z. Z. Qi. 2003. Phage display selection on whole cells yields a small peptide specific for HCV receptor human CD81. Cell Res. 13:473-479. [DOI] [PubMed] [Google Scholar]
- 8.Chen, F., Y. Hu, D. Li, H. Chen, and X. L. Zhang. 2009. CS-SELEX generates high-affinity ssDNA aptamers as molecular probes for hepatitis C virus envelope glycoprotein E2. PLoS One 4:e8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 10.Cocquerel, L., C. Voisset, and J. Dubuisson. 2006. Hepatitis C virus entry: potential receptors and their biological functions. J. Gen. Virol. 87:1075-1084. [DOI] [PubMed] [Google Scholar]
- 11.Cull, M. G., J. F. Miller, and P. J. Schatz. 1992. Screening for receptor ligands using large libraries of peptides linked to the C terminus of the lac repressor. Proc. Natl. Acad. Sci. U. S. A. 89:1865-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Cock, L., V. Hutse, and R. Vranckx. 2005. Correlation between detection of antibodies against hepatitis C virus in oral fluid and hepatitis C virus RNA in serum. Eur. J. Clin. Microbiol. Infect. Dis. 24:566-568. [DOI] [PubMed] [Google Scholar]
- 13.Dower, W. J., and L. C. Mattheakis. 2002. In vitro selection as a powerful tool for the applied evolution of proteins and peptides. Curr. Opin. Chem. Biol. 6:390-398. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, I. S. 1996. Phage display of proteins. Curr. Opin. Biotechnol. 7:547-553. [DOI] [PubMed] [Google Scholar]
- 15.Evans, M. J., T. von Hahn, N. M. Tscherne, A. J. Svder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 16.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 17.Flint, M., and J. A. McKeating. 2000. The role of the hepatitis C virus glycoproteins in infection. Rev. Med. Virol. 10:101-117. [DOI] [PubMed] [Google Scholar]
- 18.Flint, M., T. von Hahn, J. Zhang, M. Farquhar, C. T. Jones, P. Balfe, C. M. Rice, and J. A. McKeating. 2006. Diverse CD81 proteins support hepatitis C virus infection. J. Virol. 80:11331-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gersuk, G. M., M. J. Corey, F. Corey, J. E. Stray, G. H. Kawasaki, and R. L. Vessella. 1997. High-affinity peptide ligands to prostate-specific antigen identified by polysome selection. Biochem. Biophys. Res. Commun. 232:578-582. [DOI] [PubMed] [Google Scholar]
- 20.Gottwein, J. M., T. K. Scheel, T. B. Jensen, J. B. Lademann, J. C. Prentoe, M. L. Knudsen, A. M. Hoegh, and J. Bukh. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364-377. [DOI] [PubMed] [Google Scholar]
- 21.Grove, J., S. Nielsen, J. Zhong, M. F. Bassendine, H. E. Drummer, P. Balfe, and J. A. McKeating. 2008. Identification of a resident in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J. Virol. 82:12020-12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grove, J., T. Huby, Z. Stamataki, T. Vanwolleghem, P. Meuleman, M. Farquhar, A. Schwarz, M. Moreau, J. S. Owen, G. Leroux-Roels, P. Balfe, and J. A. McKeating. 2007. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J. Virol. 81:3162-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanes, J., and A. Pluckthun. 1997. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. U. S. A. 94:4937-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanes, J., C. Schatzel, A. Knappik, and A. Pluckthun. 2000. Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nat. Biotechnol. 18:1287-1292. [DOI] [PubMed] [Google Scholar]
- 25.Harris, H. J., M. J. Farquhar, C. J. Mee, C. Davis, G. M. Reynolds, A. Jennings, K. Hu, F. Yuan, H. Deng, S. G. Hubscher, J. H. Han, P. Balfe, and J. A. McKeating. 2008. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J. Virol. 82:5007-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, M., and M. J. Taussig. 2007. Rapid discovery of protein interactions by cell-free protein technologies. Biochem. Soc. Trans. 35:962-965. [DOI] [PubMed] [Google Scholar]
- 27.Irving, R. A., G. Coia, A. Roberts, S. D. Nuttall, and P. J. Hudson. 2001. Ribosome display and affinity maturation: from antibodies to single V-domains and steps towards cancer therapeutics. J. Immunol. Methods 248:31-45. [DOI] [PubMed] [Google Scholar]
- 28.Jermutus, L., A. Honegger, F. Schwesinger, J. Hanes, and A. Plückthun. 2001. Tailoring in vitro evolution for protein affinity or stability. Proc. Natl. Acad. Sci. U. S. A. 98:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keefe, A. D. 2001. Protein selecting using mRNA display. Curr. Protoc. Mol. Biol. Unit 24.5, p. 24.5.1-24.5.34. doi: 10.1002/0471142727.mb2405s53. [DOI] [PubMed]
- 30.Kita, M., M. Deguchi, M. Kagita, N. Yoshioka, E. Kobayashi, M. Watanabe, S. Asari, K. Yamanaka, and Y. Iwatani. 2009. Clinical utility and characteristics of nine anti-HCV antibody screening reagents used in Japan. Clin. Lab. 55:9-22. [PubMed] [Google Scholar]
- 31.Koutsoudakis, G., E. Herrmann, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamla, T., and V. A. Erdmann. 2003. Searching sequence space for high-affinity binding peptides using ribosome display. J. Mol. Biol. 329:381-388. [DOI] [PubMed] [Google Scholar]
- 33.Li, P., Q. Wan, Y. Feng, M. Liu, J. G. Wu, X. W. Chen, and X. L. Zhang. 2007. Engineering of N-glycosylation of hepatitis C virus envelope protein E2 enhances T cell responses for DNA immunization. Vaccine 25:1544-1551. [DOI] [PubMed] [Google Scholar]
- 34.Lindenbach, B. D., and C. M. Rice. 2003. Molecular biology of flaviviridae. Adv. Virus Res. 59:23-61. [DOI] [PubMed] [Google Scholar]
- 35.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wölk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 36.Mattheakis, L. C., R. R. Bhatt, and W. J. Dower. 1994. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc. Natl. Acad. Sci. U. S. A. 91:9022-9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alpha-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 38.Meuleman, P., J. Hesselgesser, M. Paulson, T. Vanwolleghem, I. Desombere, H. Reiser, and G. Leroux-Roles. 2008. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology 48:1761-1768. [DOI] [PubMed] [Google Scholar]
- 39.Mondon, P., O. Dubreuil, K. Bouayadi, and H. Kharrat. 2008. Human antibody libraries: a race to engineer and explore a large diversity. Front. Biosci. 13:1117-1129. [DOI] [PubMed] [Google Scholar]
- 40.Murphy, M., L. Jason-Moller, and J. Bruno. 2006. Using Biacore to measure the binding kinetics of an antibody-antigen interaction. Curr. Protoc. Protein Sci. Chapter 19, Unit 19.14. doi: 10.1002/0471142301.ps1914s45. [DOI] [PubMed]
- 41.Owsianka, A. M., J. M. Timms, A. W. Tarr, R. J. Brown, T. P. Hickling, A. Szwejk, K. Bienkowska-Szewczyk, B. J. Thomson, A. H. Patel, and J. K. Ball. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penin, F., C. Combet, G. Germanidis, P. O. Frainais, G. Deléage, and J. M. Pawlotsky. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pietschmann, T. 2009. Virology: final entry key for hepatits C virus. Nature 457(7231):797-798. [DOI] [PubMed] [Google Scholar]
- 44.Ploss, A., M. J. Evans, V. A. Gaysinskaya, M. Panis, H. You, Y. P. de Jong, and C. M. Rice. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457(7231):882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, and G. Ideo. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 46.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothwangl, K. B., B. Manicassamy, S. L. Uprichard, and L. Rong. 2008. Dissecting the role of putative CD81 binding regions of E2 in mediating HCV entry: putative CD81 binding region 1 is not involved in CD81 binding. Virol. J. 5:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwarz, A. K., J. Grove, K. Hu, C. J. Mee, P. Balfe, and J. A. McKeating. 2009. Hepatoma cell density promotes claudin-1 and scavenger receptor BI expression and hepatitis C virus internalization. J. Virol. 83:12407-12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shangguan, D. H., Y. Li, Z. W. Tang, Z. C. Cao, H. W. Chen, P. Mallikaratchy, K. Sefah, C. J. Yang, and W. Tan. 2006. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. U. S. A. 103:11838-11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, G. P., and V. A. Petrenko. 1997. Phage display. Chem. Rev. 97:391-410. [DOI] [PubMed] [Google Scholar]
- 51.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weichhart, T., M. Horky, J. Söllner, S. Gangl, T. Henics, E. Nagy, A. Meinke, A. von Gabain, C. M. Fraser, S. R. Gill, M. Hafner, and U. von Ahsen. 2003. Functional selection of vaccine candidate peptides from Staphylococcus aureus whole-genome expression libraries in vitro. Infect. Immun. 71:4633-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, H. Y., X.-L. Zhang, Q. Pan, and J. Wu. 2005. Functional selection of a type IV pili-binding peptide that specifically inhibits Salmonella typhi adhesion to/invasion of human monocytic cells. Peptide 26:2057-2063. [DOI] [PubMed] [Google Scholar]
- 54.Wünschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, L. M., J. L. Wang, L. Kang, S. Gao, Y. H. Liu, and T. M. Hu. 2008. Construction and analysis of high-complexity ribosome display random peptide libraries. PLoS One 3:e2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, J. H., J. P. Lai, S. D. Douglas, D. Metzger, X. H. Zhu, and W.-Z. Ho. 2002. Real-time RT-PCR for quantitation of hepatitis C virus RNA. J. Virol. Methods 102:119-128. [DOI] [PubMed] [Google Scholar]
- 57.Yu, H., and J. F. Talts. 2003. Beta1 integrin and alpha-dystroglycan binding sites are localized to different laminin-G-domain-like (LG) modules within the laminin alpha5 chain G domain. Biochem. J. 371(Pt. 2):289-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zahnd, C., P. Amstutz, and A. Plückthun. 2007. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods 4:269-279. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]