Abstract
Splicing of messenger RNAs is regulated by site-specific binding of members of the serine-arginine-rich (SR) protein family, and SR protein kinases (SRPK) 1 and 2 regulate overall activity of the SR proteins by phosphorylation of their RS domains. We have reported that specifically designed SRPK inhibitors suppressed effectively several DNA and RNA viruses in vitro and in vivo. Here, we show that an SRPK inhibitor, SRPIN340, suppressed in a dose-dependent fashion expression of a hepatitis C virus (HCV) subgenomic replicon and replication of the HCV-JFH1 clone in vitro. The inhibitory effects were not associated with antiproliferative or nonspecific cytotoxic effects on the host cells. Overexpression of SRPK1 or SRPK2 resulted in augmentation of HCV replication, while small interfering RNA (siRNA) knockdown of the SRPKs suppressed HCV replication significantly. Immunocytochemistry showed that SRPKs and the HCV core and NS5A proteins colocalized to some extent in the perinuclear area. Our results demonstrate that SRPKs are host factors essential for HCV replication and that functional inhibitors of these kinases may constitute a new class of antiviral agents against HCV infection.
Hepatitis C virus (HCV) infects up to 170 million people worldwide, and these infections frequently are characterized by chronic liver inflammation, leading to decompensated liver cirrhosis and hepatocellular cancers (1). Alpha and beta interferons are the mainstay of HCV therapeutics. However, the most effective pegylated interferon plus ribavirin combination therapies can eliminate HCV from around half of the patients only (6). These difficulties in eradicating HCV are compounded by the limited treatment options. For this reason, the development of safe and effective therapeutic agents against HCV has been a strong motivation in academia and industry (23).
Serine-arginine-rich (SR) proteins are a family of non-small nuclear ribonucleoprotein particle (non-snRNP) splicing factors that are highly conserved throughout the eukaryotes. They harbor one or two RNA recognition motifs and an RS domain at the amino and carboxyl termini, respectively (29). RS domains consist of multiple consecutive Arg-Ser/Ser-Arg dipeptide repeats, in which the Ser residues are extensively phosphorylated by several kinases, including SR protein kinases (SRPKs). SRPK1 was the first SR protein kinase to be cloned, on the basis of its ability to phosphorylate SR proteins in vitro (8, 9), and two other structurally related kinases, SRPK2 and SRPK3, also have been shown to phosphorylate SR proteins (16, 31). Although the precise physiological role of this phosphorylation remains unknown, it is expected that phosphorylation of SR proteins affects their protein-protein and protein-RNA interactions, intracellular localization and trafficking, and alternative splicing of pre-mRNA (21).
As SRPK-dependent herpes simplex virus (HSV) splicing and SRPK-mediated phosphorylation of hepatitis B virus (HBV) core protein have been reported (4, 25, 33), it is reasonable to expect that SR proteins and SRPK might be suitable targets for therapeutic modulation of various viral infections. Actually, we found that increased activity of SRPK2 upregulated human immunodeficiency virus (HIV) expression and that an isonicotinamide compound, SRPIN340, which preferentially inhibited SRPK1 and SRPK2, suppressed propagation of Sindbis virus, HIV, and cytomegalovirus (7). In this study, we investigated the effects of SRPIN340 on HCV replication using the HCV subgenomic replicon system (27, 32) and HCV-JFH1 virus cell culture (30, 34). Here, we demonstrate that cellular SRPK is required for HCV replication and suggest that the inhibitor of SRPK could be used therapeutically.
MATERIALS AND METHODS
SRPK inhibitor.
SRPIN340, N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]isonicotinamide, inhibits SRPK1 and SRPK2 kinase activities potently (7). SRPIN340 does not inhibit other classes of SRPKs significantly, including Clk1 and Clk and other classes of SR kinases. SRPIN614, N-methyl-N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]isonicotinamide, is a negative-control compound that has no suppressive effects on SRPK1 or SRPK2. SRPIN340 and SRPIN614 were synthesized in-house (7).
In vitro kinase assay.
Kinase activities of SRPKs were assayed as described previously (18). Briefly, His6-tagged recombinant SRPK1 or SRPK2 was expressed in Escherichia coli and purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography. The purified SRPK1 or SRPK2 was incubated in the presence of ATP, [γ-32P]ATP, and a synthetic peptide of the SF2/ASF RS domain (NH2-RSPSYGRSRSRSRSRSRSRSRSNSRSRSY-OH) at pH 7.5 and 30°C for 10 min. The reaction mixtures were spotted onto phosphocellulose membranes (Whatman, Kent, United Kingdom) and washed with 5% phosphoric acid solution, and the radioactivity was measured using a liquid scintillation counter. The net radioactivity was deduced by subtracting the background count from the reaction mixture without kinase, and the data are expressed as the percentage of the control sample containing the solvent.
Cells and cell culture.
Huh7 and Huh7.5.1 cell lines (34) were maintained in Dulbecco's modified minimal essential medium (Sigma, St. Louis, MO) supplemented with 10% fetal calf serum at 37°C under 5% CO2. To maintain cell lines carrying the HCV replicon (Huh7/Rep-Feo cells), G418 (Nacalai Tesque, Kyoto, Japan) was added to the culture medium to a final concentration of 500 μg/ml.
HCV replicon constructs and transfection.
The HCV replicon plasmids, which contain Rep-Feo, were derived from the HCV-N strain (pHC1bneo/delS [Rep-Feo-1b]) and the HCV-JFH1 strain (pSGR-JFH1 [Rep-Feo-2a]) (10, 14). These constructs express a chimeric reporter protein of firefly luciferase (Fluc) and neomycin phosphotransferase. RNA synthesis and transfection of the replicon have been described (Huh7/Rep-Feo-1b, Huh7/Rep-Feo-2a) (27, 32).
HCV cell culture system.
A plasmid, pJFH1-full (30, 34), which encodes the full-length HCV-JFH1 sequence, was linearized and used as the template for synthesis of HCV RNA using the RiboMax large-scale RNA production system (Promega, Madison, WI) (26). After DNase I (RQ-1, RNase-free DNase, Promega) treatment, the transcribed HCV RNA was purified using ISOGEN (Nippon Gene, Tokyo, Japan). For the RNA transfection, Huh7.5.1 cells were washed twice, and 5 × 106 cells were suspended in Opti-MEM I (Invitrogen, Carlsbad, CA) containing 10 μg of HCV RNA, transferred into a 4-mm electroporation cuvette, and subjected to an electric pulse (1,050 μF and 270 V) using the Easy Ject system (EquiBio, Middlesex, United Kingdom). After electroporation, the cell suspension was left for 5 min at room temperature and then incubated under normal culture conditions in a 10-cm-diameter cell culture dish. The transfected cells were split every 3 to 5 days. The culture supernatants were subsequently transferred onto uninfected Huh7 cells.
RT-PCR.
SRPK mRNA was detected by reverse transcription-PCR (RT-PCR) as described previously (12). The primers used were SRPK1-S (5′-GCG AAT GCA GGA AAT TGA GG-3′) and SRPK1-AS (5′-CAT AAG CGT TTG ATC CTG GC-3′) and SRPK2-S (5′-CCC TGC GGA CTA CTG CAA AGG-3′) and SRPK2-AS (5′-CAT TGC AAC AAA TCT TTT CCC-3′).
Luciferase assays.
Luciferase activity was measured with a Lumat LM9501 luminometer (Promega) using a Bright-Glo luciferase assay system (Promega) or a Dual-Luciferase reporter assay system (Promega), as described previously (22).
MTS assays.
To evaluate cell viability, dimethylthiazol carboxymethoxy-phenyl sulfophenyl tetrazolium (MTS) assays were performed using a CellTiter 96 aqueous one-solution cell proliferation assay kit (Promega), as described previously (24).
Quantification of HCV core antigen in culture media.
Culture media from JFH1-RNA-transfected Huh7 cells were collected, passed through a 0.45-μm filter (MILLEX-HA; Millipore, Bedford, MA), and stored at −80°C. The concentrations of core antigen in the culture supernatants were measured using a chemiluminescence enzyme immunoassay (CLEIA) according to the manufacturer's protocol (Lumipulse Ortho HCV antigen; Ortho-Clinical Diagnostics, Tokyo, Japan).
Real-time RT-PCR analysis.
The real-time RT-PCR was done as previously described (11). Briefly, total cellular RNA was isolated using ISOGEN (Nippon Gene), reverse transcribed, and subjected to real-time PCR analyses. Expression of mRNA was quantified using the TaqMan universal PCR master mix and the ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA).
Western blot analysis.
Western blotting was performed as described previously (11). Briefly, 10 μg of total cell lysate was separated by SDS-PAGE and blotted onto a polyvinylidene fluoride (PVDF) membrane. The membrane was incubated with the primary antibodies, followed by a peroxidase-labeled anti-IgG antibody, and visualized by chemiluminescence using the ECL Western blotting analysis system (Amersham Biosciences, Buckinghamshire, United Kingdom). The antibodies used were mouse monoclonal anti-HCV-core antibody (Abcam, Cambridge, MA), mouse monoclonal anti-HCV-NS5A antibody (Biodesign), and mouse anti-beta-actin antibody (Sigma).
Indirect immunofluorescence assay.
Cells seeded onto tissue culture chamber slides were fixed with cold acetone. The cells were incubated with anti-hemagglutinin (HA) and anti-core or anti-NS5A antibodies and subsequently with Alexa 488- or Alexa 568-labeled secondary antibodies. Cells were mounted with VECTA SHIELD mounting medium and DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories) and visualized by fluorescence microscopy (BZ-8000; Keyence) and confocal laser microscopy (FLUOVIEW FV10i; Olympus, Tokyo, Japan).
Synthetic siRNA.
The small interfering RNAs (siRNAs) were designed to target SRPK1 and SRPK2. Sequences of SRPK1-directed siRNAs were as follows: no. 1, 5′-UUA AUG ACU UCA AUC ACU CCA UUG C-3′; no. 2, 5′-UAA GAA AUC UGU GAA GCC AGC UGC C-3′. Sequences of SRPK2-directed siRNAs were as follows: no. 3, 5′-AAU ACU GCC UAG CAG CUC UAU GAU G-3′; no. 4, 5′-UCA GCU UGG UGA UGU GUC GCA GUU C-3′. The control siRNA has been described previously (32).
Plasmid constructs.
Plasmid pEMCV/IRES/Rluc, which is a renilla luciferase expression plasmid that is driven by an encephalomyocarditis virus internal ribosome entry site (EMCV-IRES), has been described (19). Eukaryote expression plasmids for SRPK1 and SRPK2, pME-HA-SRPK1 and pME-HA-SRPK2, have been described (16).
Calculation of EC50.
The 50% effective concentration (EC50) was calculated as the concentration of an inhibitor required for 50% reduction in replicon-based luciferase activity. We used probit regression analysis to obtain values.
Statistical analyses.
Statistical analyses were performed using Student's t test; P values of less than 0.05 were considered statistically significant.
RESULTS
Immunofluorescence microscopy of SRPK and HCV proteins.
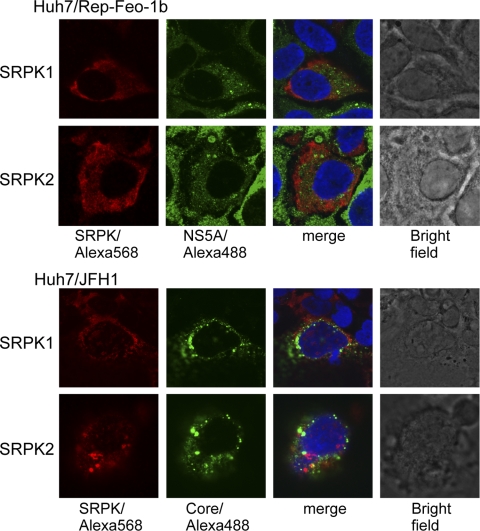
We first studied the subcellular localization of SRPK1 and SRPK2 and their association with HCV proteins. Expression plasmids for SRPK1 or SRPK2 were transfected into HCV replicon-expressing or HCV-JFH1-infected cells. Immunofluorescence analysis was performed 48 h after transfection (Fig. 1). SRPK1 and SRPK2 were distributed diffusely in the cytoplasm, and HCV core and NS5A proteins were localized at the perinuclear rim and also in the cytoplasm. Although most portions of SRPKs and the viral NS5A and core proteins were localized in different cellular compartments, SRPKs and the HCV core and NS5A proteins colocalized to some extent in the perinuclear area.
FIG. 1.
Immunofluorescence microscopy. Expression plasmids for SRPK1 and SRPK2 were transfected into Huh7/Rep-Feo-1b cells or into HCV-JFH1-infected Huh7.5.1 cells. Forty-eight hours after transfection, cells were fixed and incubated with mouse anti-NS5A or anti-core antibodies and rabbit anti-HA antibody, followed by Alexa Fluor 488-labeled anti-mouse IgG and Alexa Fluor 568-labeled anti-rabbit IgG secondary antibodies. Nuclei were stained with DAPI. Representative immunofluorescence images derived from a number of experiments are shown as three images of a single focal plane of Huh7 cells, showing NS5A and core proteins (green), SRPK1 and SRPK2 (red), DAPI staining (blue), and the superimposed images (merge).
SRPIN340 inhibits kinase activities of SRPK1 and SRPK2.
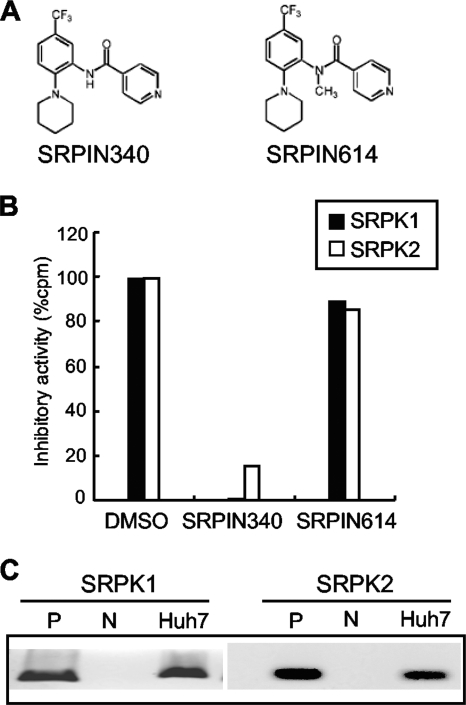
The in vitro kinase assays showed that SRPIN340 (Fig. 2 A) inhibited the kinase activities of SRPK1 and SRPK2. Ten μM SRPIN340 inhibited SRPK1- and SRPK2-mediated phosphorylation of synthetic RS-repeat peptide substrate by 99.2% and 85%, respectively (Fig. 2B), which was consistent with the results of our previous study (7). The Ki value for inhibition of SRPK1 kinase activity was 0.89 μM. SRPIN614, which lacked SRPK inhibitory action, did not inhibit SRPK1 or SRPK2 activity significantly.
FIG. 2.
Chemical structures and activities of SRPIN340 and SRPIN614. (A) Chemical structures of the SRPK inhibitor, SRPIN340, and activity-lacking control, SRPIN614. (B) Relative kinase activities of SRPK1 (black columns) and SRPK2 (white columns) in vitro, in the presence of the reagents indicated, SRPIN340, SRPIN614, and dimethyl sulfoxide (DMSO). (C) Expression of SRPK1 and SRPK2 mRNA by RT-PCR. P denotes positive controls, which are 1 ng of the respective SRPK expression plasmids. N denotes the template-lacking negative control.
SRPK inhibitor effectively suppresses HCV subgenomic replication.
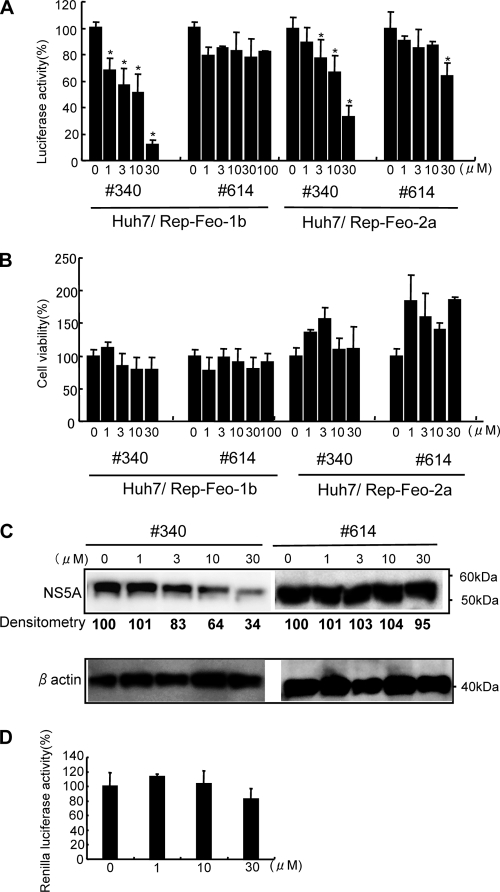
Next, we detected expression of SRPK1 and SRPK2 mRNAs in Huh7 cell lines using RT-PCR. As shown in Fig. 2C, both SRPK1 and SRPK2 mRNAs were detectable in Huh7 cells. Next, we assessed the effects of SRPIN340 on replication of the HCV genotype 1b and 2a replicons. SRPIN340 was added to HCV replicon-expressing cells, Huh7/Rep-Feo-1b and Huh7/Rep-Feo-2a. After 48 h of incubation, expression levels of the HCV replicons were measured by luciferase assay. SRPIN340 suppressed HCV 1b and 2a replication in a dose-dependent manner (Fig. 3 A). The 50% effective concentrations (EC50) for the HCV 1b and 2a replicons were 4.7 μM and 15.8 μM, respectively. In contrast, SRPIN614, which did not possess SRPK inhibitory activity, did not suppress expression of the replicon even at a concentration of 100 μM. MTS-mediated cell viability assays showed no significant effects of SRPIN340 or SRPIN614 (Fig. 3B). Similarly, we assessed the effect by Western blotting. SRPIN340 suppressed cellular HCV NS5A protein expression levels in a dose-dependent manner (Fig. 3C). SRPIN340 showed no effect on EMCV-IRES-mediated protein expression (Fig. 3D). These results indicated that the SRPK inhibitor had specific suppressive effects on HCV subgenomic replication and that these effects are not due to cytotoxicity.
FIG. 3.
Effects of SRPIN340 and SRPIN614 on expression of HCV subgenomic replicons. Huh7/Rep-Feo-1b or Huh7/Rep-Feo-2a cells were cultured in the presence of SRPIN340 (no. 340) or SRPIN614 (no. 614) at the concentrations indicated. After 48 h of culture, a luciferase assay (A), a cell viability assay (B), and Western blotting (C) were performed. (A) Effect of SRPIN340 and SRPIN614 on levels of HCV replication represented by replicon-dependent internal luciferase activities. Bars indicate luciferase activities relative to that of the drug-negative control. (B) Effect of SRPIN340 and SRPIN614 on cell viability. MTS assays were performed after culture in the presence of the drugs indicated. Bars indicate values relative to that of the drug-negative control. Asterisks indicate P values of less than 0.05. (C) Western blotting analyses. The expression levels of NS5A and beta-actin were detected by using anti-NS5A and anti-beta-actin antibodies. Densitometry of NS5A protein was performed, and results are indicated as percentages of the drug-negative control. The assay was repeated three times, and a representative result is shown. (D) Effect of SRPIN340 on EMCV-IRES-driven protein expression. Plasmid pECMV/IRES-Rluc was transfected into Huh7 cells. Twenty-four hours after transfection, the cells were incubated in indicated concentrations of SRPIN340. The renilla luciferase assay was performed at 48 h after incubation. In panels A, B, and D, assays were done in quadruplicate, and error bars indicate standard deviations.
SRPIN340 suppresses HCV-JFH1 in cell culture.
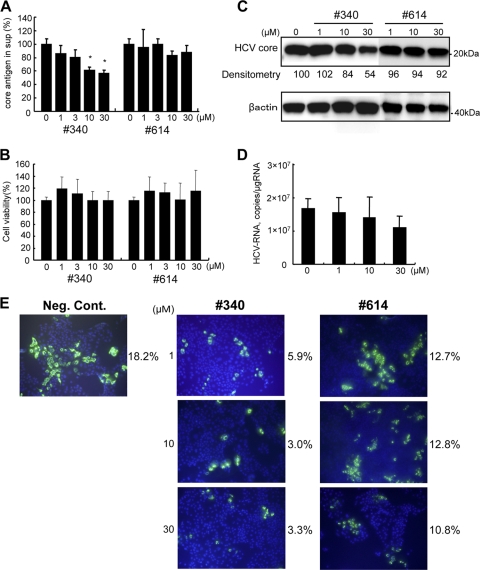
Next, we assessed the effects of the SRPK inhibitor on HCV-JFH1 in cell culture. Various concentrations of SRPIN340 were added to HCV-JFH1-infected Huh7 cells, and core antigen was quantified in the medium after 48 h of incubation. As shown in Fig. 4 A, SRPIN340 significantly suppressed HCV core antigen secretion in a dose-dependent manner. An MTS-based cell viability assay did not show significant cytotoxicity from these inhibitors (Fig. 4B). In Western blotting, SRPIN340 suppressed expression of intracellular core protein by HCV-JFH1-infected cells in a dose-dependent manner; incubation with 30 μM SRPIN340 suppressed core protein expression by 54% of the drug-negative control, while SRPIN614 did not suppress core protein expression substantially (Fig. 4C). The effects of SRPIN340 on cellular HCV RNA were confirmed by real-time RT-PCR analyses (Fig. 4D). Similarly, in immunofluorescence microscopy, treatment with SRPIN340 resulted in a dose-dependent decrease in the number of HCV core-positive cells, but no effect was detected following treatment with SRPIN614 (Fig. 4E). These data indicate that SRPK inhibitors have antiviral effects on HCV infection and replication in vitro.
FIG. 4.
Effect of SRPIN340 and SRPIN614 on HCV-JFH1 virus replication. HCV-JFH1-stably infected Huh7 cells of ∼14 days were cultured in the presence of SRPIN340 or SRPIN614 at the concentrations indicated. After 48 h, cellular and supernatant HCV core antigens were detected. (A) HCV core antigen assays of culture supernatant (sup). Bars indicate values relative to that of the drug-negative control. Asterisks indicate P values of less than 0.05. (B) Effect of SRPIN340 and SRPIN614 on cell viability. MTS assays were performed 48 h after culture in the presence of the drugs indicated. Bars indicate values relative to that of the drug-negative control. (C) Western blotting analyses. The expression of HCV core and beta-actin was detected using anti-core and anti-beta-actin antibodies. Densitometry of HCV core protein was performed, and results are indicated as percentages of the drug-negative control. (D) Real-time RT-PCR analyses. Cells were harvested at 48 h after SRPIN340 treatment. (E) Immunofluorescence microscopy. Naïve Huh7.5.1 cells were infected with HCV-JFH1 culture supernatant at a multiplicity of infection of 0.1. Three days after infection, SRPK340 or SRPIN614 was added. After 48 h, cells were incubated with anti-core antibodies followed by Alexa Fluor-conjugated secondary antibody (green). Nuclei were stained with DAPI (blue). The percentages of HCV core-positive cells were calculated and are indicated on the right of each view. The assay was repeated three times, and a representative result is shown. Neg. cont., negative control. In panels A, B, and D, assays were done in triplicate, and error bars indicate standard deviations.
Overexpression and knockdown of SRPKs regulated HCV subgenomic replication.
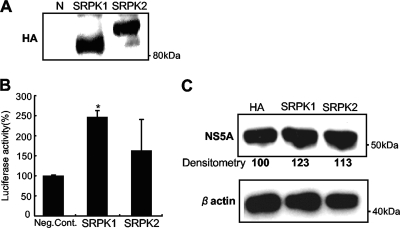
Next, we investigated the effects of the cellular expression levels of SRPK on HCV replication by overexpression and knockdown experiments. Expression plasmids for SRPK1 and SRPK2 were transfected individually into Huh7/Rep-Feo-1b cells, and internal luciferase activities were measured 72 h after transfection. The SRPK plasmid-transfected Huh7 cells expressed HA-tagged SRPK1 and SRPK2 proteins (Fig. 5 A). Transfection efficiencies were ∼20% in each experiment and were not different between expression plasmids. As shown in Fig. 5B, the luciferase activities were significantly increased in Huh7/Rep-Feo-1b cells transfected with SRPK1 or SRPK2. Western blotting showed that cellular expression of the HCV NS5A protein was increased in replicon-expressing cells with overexpression of SRPK1 or SRPK2 (Fig. 5C).
FIG. 5.
Effects of overexpression of SRPK1 and SRPK2 on HCV replication. (A) The expression of transfected HA-tagged SRPK1 and SRPK2 was detected by anti-HA antibody. (B) Huh7/Rep-Feo-1b cells seeded on 24-well plates were transfected with 0.2 μg of expression plasmids for SRPK1 or SRPK2 or empty vector. Forty-eight hours after transfection, the levels of HCV replication were measured by luciferase assay. Bars indicate values relative to that of the empty vector-transfected control. Assays were done in triplicate, and error bars indicate standard deviations. Asterisks indicate P values of less than 0.05 compared with the control. (C) Expression of HCV NS5A and beta-actin was detected using anti-NS5A and anti-beta-actin antibodies. Densitometry of HCV core protein was performed, and results are indicated as percentages of the control.
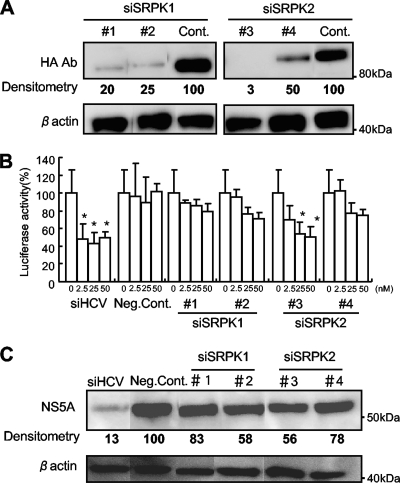
Four synthetic siRNAs were used to investigate the effects on HCV replication of suppression of expression of SRPK1 and SRPK2 proteins. These were directed against SRPK1 (siRNA 1 and siRNA 2) and SRPK2 (siRNA 3 and siRNA 4). Transgenic expression of SRPK1 and SRPK2 was specifically suppressed by transfection of the relevant siRNAs into Huh7 cells (Fig. 6 A). Next, various amounts of individual siRNA (siRNA 1, 2, 3, or 4) were transfected into Huh7/Rep-Feo-1b cells, and luciferase assays were carried out 48 h after transfection. As shown in Fig. 6B, each siRNA suppressed expression of the HCV replicon. Western blotting also showed suppression of HCV protein expression after transfection of each siRNA (Fig. 6C). These results indicated that expression of SRPK1 and SRPK2 is positively correlated with the efficiency of HCV replication.
FIG. 6.
Effects of siRNA knockdown of SRPK1 and SRPK2. (A) Huh7 cells were transfected with SRPK1 or SRPK2 expression plasmids and siRNA directed against SRPK1 (siSRPK1 no. 1 and siSRPK1 no. 2) or SRPK2 (siSRPK2 no. 3 and siSRPK2 no. 4) or control siRNA (32). Forty-eight hours after transfection, Western blotting was performed using anti-HA and anti-beta-actin antibodies. (B) Effects of siRNAs on HCV replication. The siRNAs indicated were transfected into Huh7/Rep-Feo-1b cells, and luciferase activities were measured 48 h after transfection. siHCV denotes the positive control, siRNA directed against the 5′-untranslated region of the HCV genome, and Neg. Cont. denotes a negative-control siRNA targeting an unrelated gene, which has been described previously (32). Bars indicate values relative to that of the mock-transfected control. Assays were done in triplicate, and error bars indicate standard deviations. Asterisks indicate P values of less than 0.05. (C) Western blotting analyses. Fifty micromoles of the siRNAs indicated was transfected into Huh7/Rep-Feo-1b cells. Forty-eight hours after transfection, cells were harvested and subjected to Western blotting. Expression of NS5A and beta-actin was detected with the relevant antibodies. Densitometry of NS5A protein was performed, and results are indicated as percentages of the control.
Absence of viral or cellular resistance to SRPIN340.
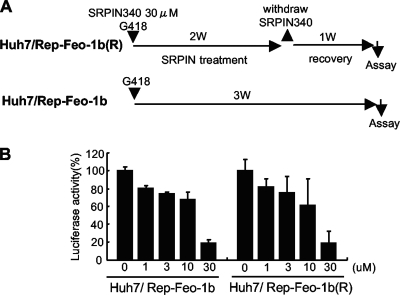
In order to assess whether long-term exposure to the antiviral molecule could select a resistant replicon, we compared sensitivity to SRPIN340 between HCV replicon cells after continuous treatment of the drug and their control cells (Fig. 7 A). Huh7/Rep-Feo-1b cells were treated with or without 30 μM SRPIN340 for 2 weeks. After 1 week of recovery culture without SRPIN340, a cell line, designated Huh7/Rep-Feo-1b(R), was established. As shown in Fig. 7B, the suppressive effect of SRPIN340 was not significantly different between Huh/Rep-Feo-1b(R) and its control cell line. These results suggest that SRPIN340 treatment under these conditions may not see the emergence of drug-resistant HCV replicons or cellular hyporesponsiveness to the drug.
FIG. 7.
Drug resistance assay of HCV replicon cells. (A) Schema for the establishment of SRPIN340-resistant cells and the control cells. Huh7/Rep-Feo-1b cells were treated with or without 30 μM SRPIN340 for 2 weeks in the presence of 500 μg/ml of G418. After 1 week of recovery culture without SRPIN340, a cell line, Huh7/Rep-Feo-1b(R), was established. (B) Huh/Rep-Feo-1b and Huh7/Rep-Feo-1b(R) cell lines were cultured in the presence of indicated concentrations of SRPIN340. Forty-eight hours after culture, internal luciferase assays were performed. Bars indicate luciferase activities relative to that of the drug-negative control. Assays were done in quadruplicate, and error bars indicate standard deviations.
DISCUSSION
These results demonstrate that small molecule inhibitors of cellular SRPK1 and SRPK2 (Fig. 2A) efficiently and specifically suppress intracellular replication of HCV subgenomic replicons and HCV-JFH1 viruses in cell culture, in a dose-dependent manner (Fig. 3 and 4). Real-time RT-PCR and Western blot analyses revealed that both RNA synthesis and its translation were reduced by SRPIN340. This inhibition was not associated with antiproliferative or nonspecific cytotoxic effects on the host cells (Fig. 3B and 4B). Transgenic overexpression of SRPK1 or SRPK2 resulted in augmentation of HCV replication and infection (Fig. 5). On the other hand, siRNA-mediated knockdown of these SRPKs suppressed HCV replication significantly (Fig. 6). These results demonstrate the dependence of the virus on the host RNA processing machinery that consists of SR proteins and their regulator, SRPK, and indicate that the inhibition of host SRPKs by small molecules may constitute a novel antiviral treatment against HCV.
SRPK1 and SRPK2 belong to the serine/threonine protein kinases. The two SRPKs efficiently phosphorylate SR proteins, such as the splicing factors ASF/SF2 and SC35, at their RS domains (3, 31). Overexpression of either SRPK1 or SRPK2 induces the phosphorylation-dependent shift of SR proteins from nuclear speckles to the nucleoplasm (8). Because SR proteins regulate splice site selection and spliceosome assembly, SRPK-mediated phosphorylation and cellular redistribution of SR proteins have been implicated in the control of mRNA maturation and alternative RNA splicing (31).
It remains to be clarified how the SRPK and SR proteins are involved in HCV replication and how the SRPIN340-directed suppression of such proteins leads to suppression of replication. There are several possibilities: that SRPIN340 may suppress processing of mRNAs that encode essential host proteins for HCV replication, that it suppresses alternative processing of the viral genomic RNA, and that certain viral proteins are substrates of host SRPK. Li et al. screened host factors required for HCV propagation through genome-wide siRNA targeting (17). They did not identify SRPKs as essential host proteins for HCV infection. Because our SRPIN340 inhibits both SRPK1 and SRPK2 and may target other family members of SRPK that possess the same target domain, it is still possible that the maintenance of overall SRPK activity may be essential for cellular HCV replication.
Several lines of evidence suggest that the viral life cycle may be partly governed by the regulation of SR protein phosphorylation as part of the RNA-processing machinery. It has been reported that virus infection induces dephosphorylation and functional inactivation of SR proteins. As a possible mechanism, Kanj et al. (13) have reported that adenoviral infection caused cellular accumulation of ceramide, which induces dephosphorylation of SR proteins by activation of the host protein phosphatase (PP)1 and consequently suppresses viral replication (2). At an early stage of adenoviral infection, the viral E4-ORF4 protein binds to the host PP2A and SR proteins, resulting in dephosphorylation of the SR proteins and consequent activation of IIIa splicing of the viral precursor mRNA that is the dominant transcript of the late phase of infection (5). In HIV infection, the role of SR proteins in the splicing of the proviral RNA has been demonstrated by a report that overexpression of SRp40, SRp55, or SRp75 caused overproduction of HIV (7). HIV Tat controls subcellular localization of SR proteins and establishes efficient HIV replication. These findings suggest that the levels of SR protein phosphorylation are positively correlated with early viral replication in host cells and that SRPIN340 treatment suppresses viral replication at an early stage.
It has been reported that HBV core protein is a substrate of SRPK1- and SRPK2-mediated phosphorylation (4). Phosphorylation of RS domains in HBV core prevents nonspecific RNA binding, which facilitates specific interaction of HBV core with the pregenomic RNA and formation of immature capsids. A functional similarity between HBV core protein and SR proteins has been reported. Our preliminary results showed that SRPIN340 suppressed expression of the viral proteins and secretion of HBe and HBs antigens. While we have not demonstrated SRPK-mediated phosphorylation of HCV proteins, our immunofluorescence microscopy has demonstrated partial colocalization of SRPKs and HCV NS5A and core proteins. These findings may suggest a possible direct interaction between SRPKs and HCV proteins, and those interactions may be the targets of SRPIN340.
Given the current situation of limited therapeutic options against HCV, searching for more potent and less toxic antiviral drugs is needed to improve clinical anti-HCV chemotherapeutics. Several direct antiviral agents against HCV are currently undergoing clinical trials; these include NS3 protease inhibitors and NS5B polymerase inhibitors (28). However, the frequent emergence of drug-resistant mutant viruses is a major weakness of such agents (15). Because our compound, SRPIN340, targets host proteins, it may be effective against multiple HCV genotypes and it is less likely that drug-resistant viruses will emerge (20). Furthermore, the toxicity data available for SRPIN340 are promising (7). No adverse effects were observed when SRPIN340 was administered orally to rats, even at the highest dose (2,000 mg/kg of body weight) for 2 weeks (data not shown). These data support the feasibility of long-term in vivo use of this compound to suppress HCV replication. On the other hand, the fact that this inhibitor acts through cellular components still raises concerns regarding its safety in the case of human use. We should not be reassured by the cytotoxicity data and the small-animal data, and further preclinical studies should be planned to address this issue. Overall, our results indicate that SRPIN340, which suppresses a wide range of DNA and RNA viruses, also is effective at suppressing HCV infection and replication. Future studies with SRPIN340, its derivatives, and other chemicals that target SRPKs could be directed toward developing a new class of antiviral treatment regimens and drugs.
Acknowledgments
We thank Frank Chisari for providing Huh7.5.1 cells.
This study was supported by grants from Ministry of Education, Culture, Sports, Science and Technology—Japan, the Japan Society for the Promotion of Science, Research on Hepatitis and BSE, and Intractable Diseases, Ministry of Health, Labor and Welfare-Japan, Japan Health Sciences Foundation, and the National Institute of Biomedical Innovation.
We declare that we have nothing to disclose regarding funding from industries or conflicts of interest with respect to the study.
Footnotes
Published ahead of print on 24 May 2010.
REFERENCES
- 1.Alter, M. J.1997. Epidemiology of hepatitis C. Hepatology 26:62S-65S. [DOI] [PubMed] [Google Scholar]
- 2.Chalfant, C. E., B. Ogretmen, S. Galadari, B. J. Kroesen, B. J. Pettus, and Y. A. Hannun. 2001. FAS activation induces dephosphorylation of SR proteins; dependence on the de novo generation of ceramide and activation of protein phosphatase 1. J. Biol. Chem. 276:44848-44855. [DOI] [PubMed] [Google Scholar]
- 3.Colwill, K., L. L. Feng, J. M. Yeakley, G. D. Gish, J. F. Caceres, T. Pawson, and X. D. Fu. 1996. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J. Biol. Chem. 271:24569-24575. [DOI] [PubMed] [Google Scholar]
- 4.Daub, H., S. Blencke, P. Habenberger, A. Kurtenbach, J. Dennenmoser, J. Wissing, A. Ullrich, and M. Cotten. 2002. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J. Virol. 76:8124-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estmer Nilsson, C., S. Petersen-Mahrt, C. Durot, R. Shtrichman, A. R. Krainer, T. Kleinberger, and G. Akusjarvi. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20:864-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marionos, F. L. Goncales, D. Häussinger, M. Diago, G. Garosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 7.Fukuhara, T., T. Hosoya, S. Shimizu, K. Sumi, T. Oshiro, Y. Yoshinaka, M. Suzuki, N. Yamamoto, L. A. Herzenberg, and M. Hagiwara. 2006. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. U. S. A. 103:11329-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gui, J. F., W. S. Lane, and X. D. Fu. 1994. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369:678-682. [DOI] [PubMed] [Google Scholar]
- 9.Gui, J. F., H. Tronchere, S. D. Chandler, and X. D. Fu. 1994. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl. Acad. Sci. U. S. A. 91:10824-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itsui, Y., N. Sakamoto, S. Kakinuma, M. Nakagawa, Y. Sekine-Osajima, M. Tasaka-Fujita, Y. Nishimura-Sakurai, G. Suda, Y. Karakama, M. Yamamoto, T. Watanabe, M. Ueyama, Y. Funaoka, S. Azuma, and M. Watanabe. Antiviral effects of the interferon-induced protein GBP-1 and its interaction with the hepatitis C virus NS5B protein. Hepatology, in press. [DOI] [PubMed]
- 12.Itsui, Y., N. Sakamoto, M. Kurosaki, N. Kanazawa, Y. Tanabe, T. Koyama, Y. Takeda, M. Nakagawa, S. Kakinuma, Y. Sekine, S. Maekawa, N. Enomoto, and M. Watanabe. 2006. Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J. Viral Hepat. 13:690-700. [DOI] [PubMed] [Google Scholar]
- 13.Kanj, S. S., N. Dandashi, A. El-Hed, H. Harik, M. Maalouf, L. Kozhaya, T. Mousallem, A. E. Tollefson, W. S. Wold, C. E. Chalfant, and G. S. Dbaibo. 2006. Ceramide regulates SR protein phosphorylation during adenoviral infection. Virology 345:280-289. [DOI] [PubMed] [Google Scholar]
- 14.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 15.Kuntzen, T., J. Timm, A. Berical, N. Lennon, A. M. Berlin, S. K. Young, B. Lee, D. Heckerman, J. Carlson, L. L. Reyor, M. Kleyman, C. M. McMahon, C. Birch, J. Schulze Zur Wiesch, T. Ledlie, M. Koehrsen, C. Kodira, A. D. Roberts, G. M. Lauer, H. R. Rosen, F. Bihl, A. Cerny, U. Spengler, Z. Liu, A. Y. Kim, Y. Xing, A. Schneidewind, M. A. Madey, J. F. Fleckenstein, V. M. Park, J. E. Galagan, C. Nusbaum, B. D. Walker, G. V. Lake-Bakaar, E. S. Daar, I. M. Jacobson, E. D. Gomperts, B. R. Edlin, S. M. Donfield, R. T. Chung, A. H. Talal, T. Marion, B. W. Birren, M. R. Henn, and T. M. Allen. 2008. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology 48:1769-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroyanagi, N., H. Onogi, T. Wakabayashi, and M. Hagiwara. 1998. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem. Biophys. Res. Commun. 242:357-364. [DOI] [PubMed] [Google Scholar]
- 17.Li, Q., A. L. Brass, A. Ng, Z. Hu, R. J. Xavier, T. J. Liang, and S. J. Elledge. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl. Acad. Sci. U. S. A. 106:16410-16415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muraki, M., B. Ohkawara, T. Hosoya, H. Onogi, J. Koizumi, T. Koizumi, K. Sumi, J. Yomoda, M. V. Murray, H. Kimura, K. Furuichi, H. Shibuya, A. R. Krainer, M. Suzuki, and M. Hagiwara. 2004. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J. Biol. Chem. 279:24246-24254. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa, M., N. Sakamoto, N. Enomoto, Y. Tanabe, N. Kanazawa, T. Koyama, M. Kurosaki, S. Maekawa, T. Yamashiro, C. H. Chen, Y. Itsui, S. Kakinuma, and M. Watanabe. 2004. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem. Biophys. Res. Commun. 313:42-47. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa, M., N. Sakamoto, Y. Tanabe, T. Koyama, Y. Itsui, Y. Takeda, C. H. Chen, S. Kakinuma, S. Oooka, S. Maekawa, N. Enomoto, and M. Watanabe. 2005. Suppression of hepatitis C virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology 129:1031-1041. [DOI] [PubMed] [Google Scholar]
- 21.Ngo, J. C., S. Chakrabarti, J. H. Ding, A. Velazquez-Dones, B. Nolen, B. E. Aubol, J. A. Adams, X. D. Fu, and G. Ghosh. 2005. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol. Cell 20:77-89. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura-Sakurai, Y., N. Sakamoto, K. Mogushi, S. Nagaie, M. Nakagawa, Y. Itsui, Y. Sekine-Osajima, M. Tasaka-Fujita, Y. Onuki-Karakama, G. Suda, K. Mishima, M. Yamamoto, M. Ueyama, Y. Funaoka, T. Watanabe, S. Azuma, S. Kakinuma, K. Tsuchiya, H. Tanaka, N. Enomoto, and M. Watanabe. 2010. Comparison of HCV-associated gene expression and cell signaling pathways in cells with or without HCV replicon and in replicon-cured cells. J. Gastroenterol. 45:523-536. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto, N., and M. Watanabe. 2009. New therapeutic approaches to hepatitis C virus. J. Gastroenterol. 44:643-649. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto, N., M. Yoshimura, T. Kimura, K. Toyama, Y. Sekine-Osajima, M. Watanabe, and M. Muramatsu. 2007. Bone morphogenetic protein-7 and interferon-alpha synergistically suppress hepatitis C virus replicon. Biochem. Biophys. Res. Commun. 357:467-473. [DOI] [PubMed] [Google Scholar]
- 25.Sciabica, K. S., Q. J. Dai, and R. M. Sandri-Goldin. 2003. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 22:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekine-Osajima, Y., N. Sakamoto, K. Mishima, M. Nakagawa, Y. Itsui, M. Tasaka, Y. Nishimura-Sakurai, C. H. Chen, T. Kanai, K. Tsuchiya, T. Wakita, N. Enomoto, and M. Watanabe. 2008. Development of plaque assays for hepatitis C virus-JFH1 strain and isolation of mutants with enhanced cytopathogenicity and replication capacity. Virology 371:71-85. [DOI] [PubMed] [Google Scholar]
- 27.Tanabe, Y., N. Sakamoto, N. Enomoto, M. Kurosaki, E. Ueda, S. Maekawa, T. Yamashiro, M. Nakagawa, C. H. Chen, N. Kanazawa, and M. Watanabe. 2004. Synergistic inhibition of intracellular hepatitis C virus replication by combination of ribavirin and interferon-alpha. J. Infect. Dis. 189:1129-1139. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, A. J., and J. G. McHutchison. 2009. Antiviral resistance and specifically targeted therapy for HCV (STAT-C). J. Viral Hepat. 16:377-387. [DOI] [PubMed] [Google Scholar]
- 29.Valcarcel, J., and M. R. Green. 1996. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 21:296-301. [PubMed] [Google Scholar]
- 30.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, H. Y., W. Lin, J. A. Dyck, J. M. Yeakley, Z. Songyang, L. C. Cantley, and X. D. Fu. 1998. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol. 140:737-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokota, T., N. Sakamoto, N. Enomoto, Y. Tanabe, M. Miyagishi, S. Maekawa, L. Yi, M. Kurosaki, K. Taira, M. Watanabe, and H. Mizusawa. 2003. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 4:602-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, Y., X. D. Fu, and J. H. Ou. 2005. Suppression of hepatitis B virus replication by SRPK1 and SRPK2 via a pathway independent of the phosphorylation of the viral core protein. Virology 342:150-158. [DOI] [PubMed] [Google Scholar]
- 34.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]