Abstract
Simple sequence repeats (SSRs) and their role in phase variation have been extensively studied in Gram-negative organisms, where they have been associated with antigenic variation and other adaptation strategies. In this study, we apply comparative genomics in order to find evidence of slipped-strand mispairing in the human Gram-positive pathogen Streptococcus agalactiae. In two consecutive screenings, 2,233 (650 + 1,583) SSRs were identified in our reference genome 2603V/R, and these loci were examined in seven other S. agalactiae genomes. A total of 56 SSR loci were found to exhibit variation, where gain or loss of repeat units was observed in at least one other genome, resulting in aberrant genotypes. Homopolymeric adenine tracts predominated among the repeats that varied. Positional analysis revealed that long polyadenine tracts were overrepresented in the 5′ ends of open reading frames (ORFs) and underrepresented in the 3′ ends. Repeat clustering in ORFs was also examined, and the highest degree of clustering was observed for a capsule biosynthesis gene and a pilus sortase. A statistical analysis of observed over expected ratios suggested a selective pressure against long homopolymeric tracts. Altered phenotypes were verified for three genes encoding surface-attached proteins, in which frameshifts or fusions led to truncation of proteins and/or affected surface localization through loss or gain of the cell wall sorting signal. The data suggest that SSRs contributes to genome plasticity in S. agalactiae but that the bet-hedging strategy is different from Gram-negative organisms.
Pathogenic organisms use a variety of strategies to evade the human immune system and to efficiently adapt to rapidly changing microenvironmental conditions. In the last decade, a number of reports have emphasized the importance of phase variation as a means to increase fitness (19, 42). Several important Gram-negative pathogens have been systematically evaluated for the presence of putative or confirmed phase-varying genes (1, 11, 13, 18, 20, 30, 34, 44). From those studies, it has been understood that DNA simple sequence repeats (SSRs) are instrumental for phase variation in several Gram-negative bacteria. SSRs in the chromosome have the capacity to form transient mispaired regions, and during replication the DNA polymerase may undergo slippage at SSR locations, which results in either expansion or contraction of tandem repeat units (16). When the SSR is located in coding sequences, and the repeat unit is not a multiple of three, this results in a frameshift which may either truncate an open reading frame (ORF), if resulting in a premature stop codon, or fuse with a second ORF by avoiding an impending stop. In addition to such ON-OFF events, slipped-strand mispairing in promoter regions may modulate transcription levels by changing the distance between the −10 and −35 elements in promoters or by changing the binding of transcription factors (22, 43). It has thus been argued that SSRs represent so-called contingency loci, enabling a series of alternative geno-/phenotypes during replication in a bacterial population, rather than clonal expansion (19). Such a mechanism also implies antigenic variation that could be used to escape the selective pressure of specific antibodies. The availability of complete genome sequences has made it possible to use bioinformatic analysis to identify potentially phase-variable genes, through probabilistic analysis of repeat regions, comparative genomics, and/or sequencing of putative phase-variable loci in multiple strains. Such approaches have been successfully applied to Haemophilus influenzae (11), Helicobacter pylori (1, 27, 30, 40), Campylobacter jejuni (20, 44), and pathogenic Neisseria species (18, 29, 34, 39). In contrast, very little is known about the role of SSRs in Gram-positive bacteria. There are selected examples of genes undergoing phase variation through slipped-strand mispairing in Streptococcus spp. (21, 22, 24), and Streptococcus pneumoniae virulence genes containing SSRs have been summarized (38). However, no systematic attempt to assess the importance of this mechanism of genetic variation in a Gram-positive organism has been undertaken. In the present study, we attempted to understand the potential role of SSRs in Streptococcus agalactiae genome plasticity and phase variation. S. agalactiae is the leading cause of neonatal sepsis and meningitis and is an emerging pathogen causing invasive disease among the elderly (6, 32). The availability of three completely sequenced genomes and another five draft sequence genomes permitted us to perform a comprehensive comparative genomic analysis in order to identify SSRs and associated putative phase-variable genes. The presence of variation in such loci constitutes retrospective evidence of genome plasticity through slipped-strand mispairing, should the type of polymorphism be compatible with that mechanism. Such an approach has the advantage of largely avoiding preselection bias, in that a large number of SSR loci can be examined across available genomes, and the outcome can guide further and more targeted analysis. Phenotypes were then examined for selected loci, and attempts were made to detect phase variation in vitro.
MATERIALS AND METHODS
Bacterial strains.
Streptococcus agalactiae strains 2603V/R, NEM316, A909, 515, COH1, CJB111, H36B, and 18RS21 were used in the present study (capsular serotypes V, III, Ia, Ia, III, V, Ib, and II, respectively). Bacteria were grown in Todd-Hewitt liquid medium (THB) at 37°C in a water bath or on solid tryptic soy broth (TSB) or horse blood agar plates at 37°C with 5% CO2. The 2603V/R, 515, and COH1 unencapsulated mutants carry a deletion of the cpsE gene in the capsule locus (3) and were kindly provided by M. Cieslewicz.
Genome sequences.
The complete genome sequences of group B streptococcus (GBS) strains 2603V/R and NEM316 have been described (7, 37). The complete genome sequence of GBS strain A909 and the draft genome sequences of strains 515, COH1, CJB111, H36B, and 18RS21 have been described (36). For the draft genome sequences, enough random sequences were produced to achieve an average sequence coverage of at least 8× for each draft genome. Contigs were ordered and oriented based on their alignment to the completed strain 2603V/R, resulting in the assembly of pseudochromosomes.
SSR analysis.
For an overview of the workflow, see the supplemental material. Iterative DNA motifs (k-nucleotide repeats), including homopolymeric tracts, were searched in the 2603V/R genome sequence using the REPEATS program. The minimum length of homopolymeric tracts was 8 for A and T and 6 for G and C, based on the average G+C content of GBS (35.6%). At least four tandem copies of di- and trinucleotide repeats, three copies of 4 to 9 nucleotide motifs, and two imperfect copies of 10 to 15 nucleotide motifs were required. Overall, thresholds were set to the lowest possible copy number where there were phase variation precedents in the literature and which would still produce a total number of hits that were manageable to analyze. In the extended search for homopolymeric tracts only, the thresholds used were 7 for A and T and 5 for G and C.
Each repeat, plus flanking sequences 25 bp upstream and 25 bp downstream, was used as a query searching the other seven GBS genomes using the Smith-Waterman algorithm (33). The results were manually examined to (i) verify that the hit was of sufficient quality to assume that it represented the corresponding locus, (ii) identify gaps within the repeat region that could constitute either contraction or extension of repeat units, and in that case, (iii) examine the genome locus in detail to verify that adjacent ORFs were homologous and predict the consequences of the different genotypes for putative ON-OFF expression of proteins. The sequence quality was assessed for regions in which variants were found, and if ambiguous bases or the end of a contig were present within 200 bp, the variant was discarded.
For the O/E ratio (observed over expected ratios), the observed number of repeats for each type and length was obtained by using Artemis. The number of unique repeats of each length was calculated by subtracting all repeats contained within higher-order repeats (i.e., A8 contains 2 × A7, which were subtracted from the number of A7). The expected number of unique repeats was obtained by using the equation Pn × (1 − P) × G, where P is the frequency of the base in the genome, n is the repeat length, and G is the total number of base pairs in the genome.
The cluster analysis was performed by identifying the ORFs associated with the 1,850 homopolymeric repeats (found in the initial and extended search), applying an inclusion threshold of three repeats or more per ORF, and for each ORF by calculating the ratio between ORF length and the number of repeats relating to the ORF. This ratio is inversely related to repeat clustering. The ratio values were then grouped in intervals, thus yielding a frequency distribution histogram. The ORFs with the lowest ratio (highest repeat density) were examined.
Protein methods.
For preparation of GBS proteins, bacteria were grown overnight or to exponential phase (optical density at 600 nm = 0.4) in 10 ml of THB. Bacteria were pelleted by centrifugation at 12,000 × g for 10 min. Then, 1 ml of culture supernatant was precipitated with 10% trichloroacetic acid (TCA) for 1 h on ice. The precipitate was collected after centrifugation at 12,000 × g for 15 min. The pellet was resuspended in acetone and incubated on ice for 15 min. The sample was then centrifuged at 12,000 × g for 15 min, and the supernatant was discarded. The pellet was briefly air dried, resuspended in 40 μl of phosphate-buffered saline (PBS) and NuPAGE LDS sample buffer with reducing agent (Invitrogen), and incubated for 10 min at 95°C. The proteins obtained constituted the secreted fraction. In parallel, the cell pellet isolated from the initial step described above was used to extract proteins anchored to the peptidoglycan. The pellet was washed twice in PBS and then resuspended in 200 μl of protoplast buffer (0.1 M KPO4, 40% [wt/vol] sucrose [pH 6.2]). Enzymatic digestion of the peptidoglycan cell wall was performed by adding 300 U of mutanolysin (Sigma) and incubating the sample at 37°C for 90 min, with light agitation. Samples were centrifuged at 12,000 × g for 15 min, and the supernatant was collected. Proteins in the supernatant were then precipitated with TCA as described above, except that the final suspension was in 200 μl. These proteins represent the cell wall-attached fraction. Bacteria treated or not treated with mutanolysin were examined by light microscopy, and viable counts were made, which indicated only minor (<10%) lysis of cells. Proteins were separated by polyacrylamide gel electrophoresis under reducing conditions using precast 10% Bis-Tris gels (Invitrogen) and then transferred to nitrocellulose membranes by Western blotting. Membranes were blocked at 4°C in PBS plus 0.05% Tween 20 (PBST) and 5% skim milk (Difco) overnight. The membranes were next incubated for 1 h with PBST plus 1% bovine serum albumin (PBSTA) containing primary antibodies diluted 1:2,000, washed five times in PBST and incubated for 1 h with PBSTA containing secondary antibodies (goat anti-mouse horseradish peroxidase [HRP] conjugate; Bio-Rad) diluted 1:30,000; this was followed by another five washes in PBST. Protein bands reacting with the primary antibodies were visualized on film by using SuperSignal West Pico chemiluminescent substrate according to the manufacturer's instructions (Pierce).
Antisera.
Cloning, expression, and purification of recombinant His-tagged proteins corresponding to gene loci SAG1236, SAG2063, SAG0992, and SAG0416 in strain 2603V/R was performed, and these proteins were used to immunize mice and obtain the polyclonal antiserum used to detect the proteins. This work was part of another study (17) and is described in the supplementary material for that publication (www.sciencemag.org/cgi/content/full/309/5731/148/DC1).
In vitro phase variation screening.
Bacteria from a frozen stock culture were streaked onto TSB plates. A single colony was picked and resuspended in PBS, and serial dilutions were performed. For screening purposes, 5 × 103 to 10 × 103 CFU of the resuspended bacteria were spread on a second TSB plate, which was incubated overnight. A nitrocellulose membrane was placed on the plate for 20 min at room temperature, and bacteria were fixed on the membrane by incubation at 80°C for 30 min. Blocking was performed overnight at 4°C using PBS-5% (wt/vol) skim milk. Filters were washed twice for 5 min each time in PBST. Filters were then incubated for 2 h with primary antibodies (mouse polyclonal antiserum) diluted 1:2,000 in 5 ml of PBS-1% bovine serum albumin (PBSA). Filters were washed twice for 5 min each time in PBST and then incubated for 1 h with secondary antibodies (goat anti-mouse IgG-HRP conjugate; Bio-Rad) diluted 1:5,000 in 5 ml of PBSA. The filters were then washed twice for 5 min each time in PBST. Colonies reacting with the primary antibodies were visualized colorimetrically using Opti4CN (Bio-Rad) according to the manufacturer's instructions. Typically, satisfactory signals were obtained after 5 to 10 min of incubation with substrate solution. The screening for slippage events aimed to detect unidirectional OFF-ON switches. When a positive signal was found, the corresponding area on the plate was located, colonies in that area were picked, and colony immunoblotting (CIB) was repeated at a lower CFU count, so that eventually a single positive colony could be picked.
For immunomagnetic enrichment of OFF-ON events, bacteria were grown in THB either overnight or to exponential phase (A600 = 0.4). Bacteria were then centrifuged at 3,000 × g for 10 min, washed in PBS, and recentrifuged, and 109 CFU were resuspended in 1 ml of PBSA, 10% normal rabbit serum, and a 1:1,000 dilution of primary antibodies (mouse polyclonal antiserum). The bacteria were incubated for 20 min on ice. Bacteria were pelleted by centrifugation for 5 min at 12,000 × g and washed three times with 1 ml of PBSA. The sample was then resuspended in 900 μl of PBSA-10% normal rabbit serum, and 100 μl of goat anti-mouse microbeads (Miltenyi Biotech) was added. Samples were incubated on ice for 20 min. Immunomagnetic separation was performed with LS separation columns and a MidiMACS separator (Miltenyi Biotec) according to the manufacturer's instructions, except that five column washes were performed instead of three. Eluted material, representing the positive fraction, was then analyzed by CIB.
For flow cytometry, bacteria were harvested after overnight growth or in exponential phase, and washed twice in PBSA. Each sample consisted of 100 μl of bacterial suspension, diluted to an A600 of 0.1. Bacteria were pelleted by centrifugation at 12,000 × g for 5 min, resuspended in 10 μl of normal calf serum, and incubated at room temperature for 20 min. Primary antibodies (polyclonal mouse sera) diluted 1:200 in 90 μl of PBSA were added to each sample, which was then incubated for 1 h on ice. Bacteria were washed twice, by centrifugation and resuspension in 500 μl of PBS-0.1% BSA. Bacteria were then resuspended in phycoerythrin-conjugated F(ab′)2 goat anti-mouse IgG (Jackson Immunoresearch) diluted 1:200 in 50 μl of PBS, 0.1% BSA, and 10% normal calf serum, followed by incubation on ice for 45 min. The bacteria were washed as described above and resuspended in 300 μl of PBS with (scan) or without (sorting) 1% paraformaldehyde. Flow cytometry was performed by using a Becton Dickinson LSRII, with DIVA software for data acquisition, and FlowJo (Tree Star, Inc.) for subsequent analysis. The fluorescence of the samples was compared to a control (primary antibodies) consisting of pooled mouse sera from nine control mice who had received immunizations with PBS alone. Fluorescence-activated cell sorting (FACS) was performed using the FACSAria cell-sorting system (Becton Dickinson). In brief, OFF-ON events were sorted by using an inclusion threshold set at ca. 0.1% of the total number of events. The subpopulation with the highest fluorescence was then plated, and CIB was performed. Potentially positive clones were isolated and subjected to flow cytometry again.
RESULTS
Comparative genomic analysis of simple sequence repeats.
Using permissive inclusion criteria based on the lengths of repeats and the presence of associated ORFs, we identified 650 simple sequence repeats (see Materials and Methods for inclusion criteria) in the complete genome sequence of GBS strain 2603V/R. These repeats comprised a wide variety of types, including homopolymeric tracts, repeats with long units but few repetitions, and degenerate repeats. Each of these repeats plus their flanking sequence was then used as a query in a search against each of seven other GBS genomes, in order to obtain comparative sequence data on the locus. The resulting alignments fell into several categories. A minor fraction of the alignments was poor, indicating that there was no corresponding locus in a genome or that the locus was highly divergent. The typical alignment indicated the presence of a corresponding locus and an identical SSR. In some cases there was “stabilization” of the repeat, meaning that point mutations had occurred which were likely to prevent a potential slipped strand mispairing event in the locus. Finally, there were alignments showing the loss or gain of repeat units, compatible with slipped-strand mispairing events. The corresponding loci were examined to verify the presence of homologous ORFs and assess the putative consequences of the variation, compared to the 2603V/R strain. Seventeen variable repeat loci were identified according to these criteria, where at least one other genome contained a variant (Table 1). Three SSRs were found upstream of genes: one involved a 2-bp change of the canonical distance between the −10 and −35 elements, another involved the substitution of a TATAAA for a TAAAAA, and the last was located between the ribosomal binding site (RBS) and start codon. The remainder of the SSRs were located in ORFs and resulted in frameshifts. All types of variations were identified, i.e., expansion, contraction, or both, compared to the reference strain. The number of comparison strains showing variation was from one to all. Eight of the putative seventeen proteins (47%) encoded are predicted to be surface localized, i.e., lipoproteins, cell wall-attached proteins, or proteins with membrane-spanning domains. Hypothetical proteins were the second most common group (six proteins). Homopolymeric tracts predominated among variants. Specifically, polyadenine (poly-A) accounted for 59% of the variants found. In comparison, poly-A constituted 18% of the original search set.
TABLE 1.
Simple sequence repeat variants in the seven S. agalactiae comparison genomes
| Genea | Annotationb | Repeatc | Positiond | Variantse | Commentsf |
|---|---|---|---|---|---|
| Sag0148 | Oligopeptide ABC transporter, lipoprotein | (TAAAAAAATG)3 | -103 | (−1); Cjb111 | Effect unknown |
| Sag0355 | CHP | (TA)4 | -40 | (−1); Coh1 | -10/-35 distance changes |
| Sag0466 | Thiolase | (A)8 | 116 | (−1); Nem316 | Truncation |
| Sag0972 | CHP, authentic frameshift | (A)9 | 145 | (−1); Nem316, Cjb111, H36b; and (−2); 515, Coh1 | Fusion in −1 |
| Sag0992 | Phosphate ABC transporter, lipoprotein | (A)8 | 2 | (−1); Coh1 | Truncation |
| Sag1033 | FtsK/SpoIIIE family protein | (G)7 | 13 | (+1); 515 | Truncation |
| Sag1133 | CHP, possible hydrolase | (AG)4 | 76 | (−1); 515, H36b | Truncation |
| Sag1236 | C5a peptidase, authentic frameshift | (A)8 | 3185 | (+1); 18rs21, Coh1 | LPXTG fusion |
| Sag1265 | Cadmium resistance transporter, putative | (T)8 | -1 | (−1); 18rs21 | Effect unknown |
| Sag1149 | Lipoprotein, putative | (A)8 | 11 | (−1); Coh1 | Truncation |
| Sag1555 | HP | (A)8 | 21 | (−1); 515, H36b, Coh1, Nem316 | Truncation |
| Sag1568 | Phosphoserine aminotransferase (SerC), frameshifted | (G)6 | 295 | (−1); Cjb111, H36b, Coh1, Nem316, 515 | Fusion |
| Sag1745 | HP | (A)8 | 18 | (−1); Cjb111 | Truncation |
| Sag1921 | Sensor histidine kinase | (A)9 | 3 | (−1); Cjb111 | Truncation |
| Sag2063 | BibA, CWP | (A)8 | 1120 | (−1); 515, Cjb111 | LPXTG truncation |
| Sag2094 | Competence/damage-inducible protein CinA, authentic frameshift | (T)8 | 301 | (−1); Cjb111, Coh1, H36b, Nem316 | Fusion |
| Sag2170 | CHP | (G)7 | 98 | (−1); A909 | Truncation |
That is, the gene identifier in the reference genome 2603v/r.
Abbreviations: HP, hypothetical protein; CHP, conserved hypothetical protein; CWP, cell wall-attached protein.
The repeat unit is shown in parenthesis, and the number of consecutive units is indicated.
The location of the SSR respective to the start codon of the closest ORF. SSRs with positive numbers are found in the coding sequence, whereas negative numbers indicate an upstream location.
The number of repeat units compared to the reference strain is given in parentheses, and the strain(s) where the variants are found are indicated.
The predicted effect of the variation relative to the reference strain.
Long homopolymeric tracts.
A factor commonly seen to influence the frequency of slipped strand mispairing is the length of a repeat tract (5, 25). We examined the longest homopolymeric tracts (n = 49) in the 2603V/R strain (see Table S2 in the supplemental material). The poly-A/T tracts (n = 33) were fairly evenly divided between intergenic regions or ORFs. The longest tract found was a 15-bp poly-A located intergenically, for which there is experimental evidence of variation in the range of 8 to 15 bp, resulting in diverse expression of the alpha C protein (22). The remainder of the long poly-A/T tracts were 9 to 10 bp long, and within ORFs there were only poly-A tracts. The longest poly-G/C tracts (n = 16) were all located within ORFs, 7 bp long, and most were poly-G (n = 13). The ORFs associated with these repeats included membrane proteins, transcriptional regulators, two sortases, two extracellular proteases, and three lipoproteins. Notably, some genes or operons (i.e., the vex locus) were associated with multiple long SSRs.
Extended analysis of homopolymeric tracts.
Since homopolymeric tracts were overrepresented among the examples of variation found in our screening, a second comparative genomic analysis was performed including only this type of SSR. The inclusion threshold was lowered by 1 bp (7 for A/T and 5 for G/C), and 1,583 such repeats (in or upstream of ORFs) were identified in strain 2603V/R. Subsequent comparative genomic analysis identified 39 SSR showing variation in the other genomes (Table 2). With respect to the previous search, a larger proportion of the SSR variants were located in intergenic regions (n = 17/39 versus 3/17). Two of these were located within putative promoters and thus changed the canonical distance between the −10 and −35 elements. Curiously, the typical intergenic variant was constituted by an SSR located between an RBS and the start codon, or upstream of the RBS. Among the identified 14 variants associated with an RBS, notable genes were cpsA, encoding a regulator of capsule expression; a putative exfoliative toxin (Sag1215); and several transcriptional regulators. The consequences, if any, of variation in these intergenic regions are unclear. However, the phase variation in expression of UspA1, a surface-associated protein in Moraxella catarrhalis, was shown to be related to expansion/contraction of a poly-G tract located 30 bp upstream of the start codon, and 168 bp downstream of the transcriptional start site (14). The difference of a single guanine residue had pronounced effects on protein expression.
TABLE 2.
Additional homopolymeric tract variations in seven S. agalactiae comparison genomes
| Genea | Annotationb | Repeatc | Positiond | Variantse | Commentsf |
|---|---|---|---|---|---|
| Sag0003 | Diacylglycerol kinase domain, putative | (C)5 | 427 | (−1); Cjb111 | Truncation (74) |
| Sag0078 | Preprotein translocase SecY subunit | (A)7 | 1024 | (−1); 18rs21 | Truncation (21) |
| Sag0157 | DNase related, frameshift | (A)7 | 146 | (+1); A909, Nem316, 515, Cjb111, H36b | Fusion |
| Sag0203 | Polyribonucleotide nucleotidyltransferase | (A)7 | -20 | (−1); Coh1 | Unknown (NR) |
| Sag0232 | HP | (A)7 | 158 | (−1); Cjb111 | Truncation (71) |
| Sag0412-3 | Reg. protein RecX and RNA methyl transferase | (T/A)7 | -50/-55 | (−1); 515 | Unknown/in put. promoter |
| Sag0416 | CspA, serine protease, CWP | (A)7 | 1325 | (+1); A909; and (−1); Coh1 | Truncation (64) |
| Sag0679 | CHP | (A)7 | 714 | (−1); H36b | Truncation (29) |
| Sag0708 | Alpha amylase family protein | (T)7 | -11 | (−1); 515 | Unknown (NR) |
| Sag0723 | RNase III | (A)7 | 0 | (−1); Coh1, 515 | Unknown (NR) |
| Sag0771 | CWP | (A)7 | 594 | (−1); Coh1 | Truncation (61) |
| Sag0792-3 | CHP/glycerate kinase 2 | (T/A)7 | -120/-39 | (−1); Coh1 | -10 promoter element/unknown (NR) |
| Sag0817 | Probable thiamine transporter | (A)7 | -11 | (−2); Coh1 | Unknown (NR) |
| Sag0820 | Ribonucleoside-diphosphate reductase 2 | (A)7 | -13 | (+1); Coh1 | Unknown (NR) |
| Sag0833 | HP | (A)7 | 4 | (−1); Coh1 | Truncation (99) |
| Sag0835 | CHP | (A)7 | 16 | (−1); 515 | Truncation (98) |
| Sag0858 | ATP synthase F0, subunit | (A)7 | -13 | (−1); Coh1 | Unknown (NR) |
| Sag0878 | Acetoin dehydrogenase | (T)7 | -10 | (−1); 515 | Unknown (NR) |
| Sag0913 | Chloramphenicol acetyltransferase | (A)7 | 14 | (−1); Nem316, Cjb111 | Truncation (98) |
| Sag1003 | Putative permease, MP | (A)7 | 744 | (−1); Coh1 | Truncation (74) |
| Sag1041 | HP | (A)7 | -43 | (−1); Coh1, Cjb111 | Unknown (NR) |
| Sag1063 | Flavoprotein-related protein | (A)7 | 670 | (+1); Coh1 | Truncation (1) |
| Sag1086 | Xanthine phosphoribosyl transferase | (T)7 | -26 | (+2); Coh1 | In put. promoter |
| Sag1127 | Conserved domain protein | (A)7 | 1068 | (+1); A909, H36b, 515 | None (prior independent truncation) |
| Sag1175 | CpsA | (T)7 | -18 | (−1); A909, 18rs21, 515 | Unknown (NR) |
| Sag1183 | Ribose 5-phosphate isomerase | (T)7 | 92 | (+2); 18rs21 | Truncation (85) |
| Sag1215 | Putative exfoliative toxin A | (T)7/(T)7 | 133/-12 | (−1/0); 515 | Truncation (85) |
| (0/-1); A909, H36b | Unknown (NR) | ||||
| Sag1221 | Glycerophosphoryl diester phosphodiesterase, inactivated by PM | (A)6 | 71 | (+1); A909, H36b | Truncation (96), PM generates new start downstream |
| Sag1409 | RogB transcriptional reg., frameshift | (T)7 | 957 | (+1); Cjb111, 18rs21, H36b | Fusion |
| Sag1465 | Protease, putative | (T)7 | 173 | (−1); Cjb111 | Truncation (80) |
| Sag1503 | HP | (T)7 | 7 | (+1): Cjb111, H36b | Truncation (94) |
| Sag1554 | HP | (A)7 | 0 | (−1); Coh1, Cjb111, H36b | Unknown (NR) |
| Sag1576 | Transposase IS30 family, frameshift | (G)5 | 366 | (−1); A909, Coh1, 18rs21 | Fusion |
| Sag1734 | Transporter, putative | (A)7 | -13 | (−1); A909, NeM316, Coh1, 515, Cjb111, H26b | Unknown (NR) |
| Sag1756 | CHP/threonin aldolase | (T)7 | -21 | (+1); NeM316, 515 | Unknown (NR) |
| Sag1896 | RegR, sugar-binding transcriptional reg. | (A)7 | -32 | (−1); Cjb111 | Unknown (NR) |
| Sag1933 | PTS system, IIC component, putative | (A)7 | 357 | (+2); Coh1 | Truncation (75) |
| Sag2072 | Uridine phosphorylase | (A)7 | -28 | (+1); A909, H36b; and (+2); Coh1 | Unknown (NR) |
| Sag2170 | CHP | (G)7 | 98 | (−1); A909 | Truncation |
That is, the gene identifier in the reference genome 2603v/r.
Abbreviations: HP, hypothetical protein; CHP, conserved hypothetical protein; CWP, cell wall-attached protein; reg., regulator.
The repeat unit is shown in parenthesis, and the number of consecutive units is indicated. When the SSR is located between two ORFs on different strands, the form “(A/T)” “is used. When two separate SSRs relate to the same ORF, the form” “(T)/(T)” is used.
The location of the SSR respective to the start codon of the closest ORF. SSRs with positive numbers are found in coding sequence, whereas negative numbers indicate an upstream location.
The number of repeat units compared to the reference strain, and the strain(s) where variants are found.
The predicted effect of the variation relative to the reference strain. Percentage values are indicated in parentheses. NR, near RBS. put., putative.
Among the SSRs located in the coding sequence, the poly-A tracts were again the most numerous, with 18 loci showing variation. Seven poly-T loci were also found, as well as one each of poly-C and poly-G. The variations included expansion, contraction or both compared to the reference genome, and the phenotypic consequences included both fusions and truncations. For example, RogB, a transcriptional regulator of several putative virulence factors (9) contains an authentic frameshift that inactivates the gene in the reference strain. The gene contains two SSRs: a poly-A and a poly-T. In three genomes the poly-T is expanded by one thymine residue compared to the reference genome, which results in a complete ORF. Another strain shows contraction of the poly-A tract, which leaves the gene truncated.
Analysis of frequency, position, and clustering of homopolymeric tracts.
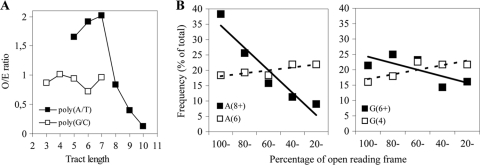
The apparent homogeneity of homopolymeric tracts prompted us to examine the overall frequency of these elements. A recurring argument is that tracts that are statistically unlikely (i.e., a high observed over expected [O/E] ratio) are particularly prone to vary. All homopolymeric repeats of various tract lengths, irrespective of location, were counted. The result was compared to the theoretically expected frequency in a random genome with the same %G+C content (Fig. 1 A). For poly-G/C tracts, the observed frequencies were comparable to the expected frequencies for different tract lengths, whereas poly-A/T showed a modest overrepresentation of medium long tracts and a pronounced underrepresentation of long tracts (O/E = 0.12 for 10-bp tracts). An identical analysis was performed on the NEM316 genome with similar results (O/E = 0.08 for 10-bp tracts). The sole exception was the above-mentioned A15 tract (O/E = 12.2). In the eight genomes, this is the only homopolymeric tract longer than 10 bp.
FIG. 1.
Frequency and position of homopolymeric tracts. (A) The observed frequency of homopolymeric repeat tracts is compared to the statistically expected number of such tracts in a random genome. Symbols indicate the O/E ratio for a particular tract length. (B) The position of homopolymeric adenine (left) and guanine (right) tracts in ORFs. The longest tracts are represented by filled boxes, while medium long tracts are indicated by empty boxes. Percentages indicate the extent to which an ORF is affected in case of a frameshift at the SSR locus.
The location of a SSR in an ORF can be important for the impact of a slipped-strand mispairing event. In case of variation, a SSR located in the 5′ region of an ORF is bound to have a profound effect on the protein encoded. In comparison, it is more difficult to predict the effects of minor truncations, which could leave the protein functionally uncompromised. Our analysis of the longest homopolymeric tracts seemed to suggest a tendency of such tracts to be preferentially located toward the 5′ ends of ORFs, but the total number of such tracts was too low to permit any definite conclusion. We hypothesized that if homopolymeric tracts were important for genome plasticity in S. agalactiae, there could be a preferential bias of such tracts not only in terms of presence or absence within certain genes but also of the position within the gene. We analyzed the position of the longer homopolymeric tracts (8 or more for A/T and 6 or more for G/C) within ORFs, and compared it to a control set of randomly selected tracts of the same type but (2 bp) shorter (Fig. 1B). For the long poly-A tracts, there was an apparent skew in the distribution. Such tracts were preferentially located toward the 5′ ends of ORFs and were progressively less common toward the 3′ ends, e.g., ∼40% of the long poly-A tracts are located in the first fifth of ORFs. In comparison, the shorter poly-A tracts were evenly distributed across the length of ORFs, as would be expected for a random distribution. The long poly-G tracts also showed a discrete skew, but it was not sufficiently different from the short poly-G distribution and theoretical random distribution. Long poly-T and poly-C tracts were unusual in coding sequence, and the low numbers did not permit a conclusive analysis.
Although S. agalactiae does not contain the improbable (high O/E ratio) SSRs that are found in some Gram-negative bacteria, our results thus far indicated that homopolymeric tracts have the capacity to contribute to diversifying the phenotype. We investigated the possibility that clustering of such SSRs was occurring in selected loci, thereby increasing the probability of variation. The ORFs related to the 1,583 repeats identified in the extended search were analyzed, examining the ratio between ORF length and the number of associated repeats. ORFs containing two or less repeat tracts were excluded. The ratio showed a positively skewed distribution, and the ORFs with the lowest ratio (high density of repeat tracts) were examined. These 17 ORFs were generally of average size (404 to 1,091 bp), and most were annotated with a putative function (Table 3). Only two ORFs contained more than five repeat tracts. One of them was cpsH (cpsV) in the capsule biosynthesis locus, containing six different SSRs. This gene encodes a sugar transferase involved in capsule synthesis and is associated with capsular serotype V. The corresponding locus in strain A909 (type Ia strain) contains only three homopolymeric tracts, but instead also has (AT)9, which is a uniquely long dinucleotide repeat in that genome. The second ORF was a sortase containing seven separate SSRs. This sortase is part of a genetic locus that was recently reported to be responsible for the assembly of pilus structures (15). Interestingly, the housekeeping sortase srtA, responsible for covalent attachment of proteins to the cell wall, also showed some clustering. Although not pronounced enough to pass the inclusion threshold, it ranked in the top 6.5% for clustering. Among the selected ORFs, two transcriptional regulators were found, as well as the only phage exclusion system (abiG) in the genome. Concomitantly, the selection included a prophage ORF, encoding a putative repressor protein.
TABLE 3.
Clustering of SSRs in coding sequences
| Genea | Annotationb | Repeatsc | No. of SSRsd | ORF length (bp) |
|---|---|---|---|---|
| Sag0218 | Transcriptional regulator, Cro/CI family | 2× A7, T7 | 3 | 473 |
| Sag0224 | Replication initiation protein, putative | C5, 3× A7, G5 | 5 | 995 |
| Sag0231 | HP | 2× A8, A7, T7 | 4 | 404 |
| Sag0232 | HP | A8, 3× A7 | 4 | 557 |
| Sag0245 | Protein of unknown function/lipoprotein, putative | 2× A7, G5 | 3 | 455 |
| Sag0548 | Prophage LambdaSa1, repressor protein, putative | 3× A7, G5 | 4 | 794 |
| Sag0848 | GtrA family protein | 2× A7, T7 | 3 | 452 |
| Sag0994 | Inositol monophosphatase family protein | 2× G5, 2× A7 | 4 | 761 |
| Sag1064 | Flavoprotein-related protein | T7, C5, 2× A7 | 4 | 683 |
| CpsH | Polysaccharide biosynthesis protein CpsH(V) | 3× T8, 2× T7, T7 | 6 | 1,091 |
| AbiGI | Abortive infection protein AbiGI | G6, A7, A8 | 3 | 587 |
| Sag1406 | Sortase family protein | 5× A7, A9, G5 | 7 | 878 |
| Sag1723 | Signal peptidase I | T7, 2× A7 | 3 | 590 |
| Sag1758 | Ribosomal-protein-alanine acetyltransferase, putative | 2× A7, T7 | 3 | 404 |
| Sag1910 | Transcriptional regulator, MarR family | 3× A7 | 3 | 422 |
| Sag2170 | CHP | 2× A7, G7, T7, G5 | 5 | 869 |
That is, the gene identifier in the reference genome 2603v/r.
Abbreviations: HP, hypothetical protein; CHP, conserved hypothetical protein.
The repeat unit and the number of consecutive units is indicated. The notation “n×” indicates multiple occurrences of the same repeat.
The total number of SSRs present in the ORF.
Comparison with Gram-negative bacteria.
A comparison of the SSR repertoire in S. agalactiae and Gram-negative bacteria reported to exhibit phase variation was made (Table 4). The analysis aimed to compare the presence and type of unusually long repeats. Repeats were identified with the Microsatellite Analysis Server (35). Campylobacter jejuni contains numerous long poly-G/C repeats and no other long repeat type. Phase variation in C. jejuni has been reported to involve poly-G/C tracts (20, 44). Helicobacter pylori has several types of long repeats, the most frequent again being poly-G/C tracts. There are nearly as many long poly-A/T tracts and seven long dinucleotide repeat tracts. H. pylori has the highest total number of long repeats among these organisms. The literature reports more poly-G/C tracts and dinucleotide tracts than homopolymeric A/T tracts as likely to be phase variable (1, 27, 30, 40). In Haemophilus influenzae the long repeat tracts are predominantly tetranucleotide repeats, and there are two examples of pentanucleotide repeat tracts of unusual length. Indeed, reported phase variation mainly involves tetranucleotide repeats (11). Neisseria meningitidis has the broadest repertoire of long repeats, including all repeat types except for dinucleotide tracts. Among the tracts previously reported as phase variable, the poly-G/C tracts are the most numerous, followed by tetranucleotide repeats (18, 29, 34). Some poly-A tracts are also reported. In comparison, our reference S. agalactiae strain contains only two repeats comparable in length to those mentioned above. The long poly-A tract has been experimentally shown to vary, while the role of the long dinucleotide tract is unclear (22). The locus is present in six other S. agalactiae genomes (data not shown), but the dinucleotide repeats are replaced with a poly-A tract. Our comparative genomic analysis suggests that among the different repeat types examined homopolymeric adenine tracts have the highest propensity for variation in S. agalactiae. The polymorphisms we have described are usually found in poly-A tracts of 7 to 10 bp. Although other repeat types predominate among reported phase-variable loci in the Gram-negative bacteria, there are several examples of poly-A tract polymorphisms in H. pylori and N. meningitidis where tract length is in the size range of 5 to 11 bp (1, 29).
TABLE 4.
Comparison of unusually long SSRs in S. agalactiae and Gram-negative bacteria
| Species | Genome size (Mb) | % G+C | Type of repeat unit(s)a |
Phase variationb | Source or reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| Poly-A/T | Poly-G/C | Di | Tetra | Penta | |||||
| S. agalactiae | 2.16 | 35.6 | 1×(15) | 0 | 1×(8) | 0 | 0 | A > G > Di | 22; this study |
| C. jejuni | 1.64 | 30.6 | 0 | 26×(9-12), 9 | 0 | 0 | 0 | G/C | 20, 44 |
| H. pylori | 1.67 | 39 | 19×(11-16), 14 | 23×(9-15), 12 | 7×(8-11), 9 | 0 | 0 | G/C > Di >A | 1, 27, 30, 40 |
| H. influenzae | 1.83 | 38 | 0 | 0 | 0 | 12×(6-37), 22 | 2×(4-12) | Tetra | 11 |
| N. meningitidis | 2.27 | 51.5 | 10×(10-14), 10.5 | 13×(10-14), 11 | 0 | 5×(5-20), 9 | 5×(10-16), 13 | G/C > Tetra > A | 18 |
For each type of repeat unit, “n×” represents the number of occurrences of SSRs with the range of lengths in parentheses, followed by the median length in base pairs.
Previously reported phase variation and the types of repeats involved in numerical order of importance.
Variant proteins have different subcellular localization and are no longer accessible to specific antibodies.
Three SSR-associated genes were chosen for further analysis at the protein level. The proteins were chosen based on the availability of antiserum, in-house data supporting expression, putative function, and the predicted surface exposure. Sag1236 (or ScpB) is a C5a peptidase, covalently anchored to the cell wall through an LPXTG motif (4, 31). By degrading C5a, the enzyme interferes with the chemotactic gradient responsible for neutrophil recruitment (12). In the reference strain 2603V/R, an authentic frameshift in the 3′ end of the gene results in a small truncation mediating apparent loss of the cell wall attachment signal. In the other strains, the gene is complete. Sag0416 or CspA also belongs to the family of subtilisinlike proteases and has a cell wall attachment signal. The protease cleaves fibrinogen and several chemokines (2a, 9a). Two SSR variants were found, representing both expansion and contraction of a poly-A tract with respect to the reference strain, in both cases resulting in C-terminal truncation (64% deletion) of the protein, including the loss of the cell wall anchor. Sag2063 is another cell wall-attached protein, recently described as an immunogenic adhesin with antiphagocytic properties and named BibA (28). The strains Cjb111 and 515 both contain an expansion of a poly-A tract in the coding sequence, resulting in a frameshift and truncation (40%) of the protein.
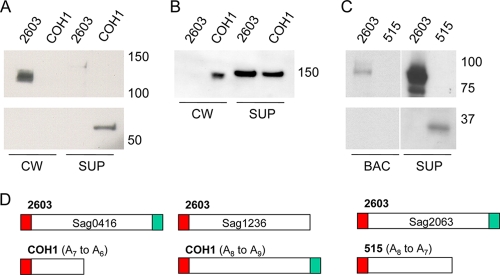
To investigate the presence and localization of these proteins, pairs of strains containing a complete and a frameshifted gene encoding each of the three proteins were selected and subjected to analysis of the cell wall fraction and the secreted fraction using antiserum against recombinantly expressed proteins (Fig. 2). Whenever gene variants contained frameshifts, the corresponding protein was no longer found in the cell wall fraction but as a truncated form in the supernatant. Antiserum against Sag2063 reacted with multiple bands (with an estimated molecular weight equal to and below that of the mature protein) in the cell wall fraction (data not shown). This protein seems susceptible to intracellular proteases released during the enzymatic digestion of the bacterial cell wall with a degree of concomitant lysis (I. Santi, unpublished data). If whole bacteria were boiled without digestion of the cell wall, a discrete single band of the right size was identified with anti-Sag2063 antibodies. Two of the proteins, Sag2063 and Sag1236, were consistently found in the secreted fraction even when the genes were complete, and this may represent a degree of shedding, mis-sorting, and/or lysis.
FIG. 2.
Western blot immunodetection of secreted and surface-associated bacterial proteins. Proteins from culture supernatant (SUP), bacterial cell wall extracts (CW), or whole boiled bacteria (BAC) were separated by electrophoresis and transferred to a membrane. The strains used to prepare the extracts are indicated above the panels. Mouse antisera against recombinant proteins Sag0416 (A), Sag1236/ScpB (B), and Sag2063/BibA (C) were used to detect reactive species in the extracts. (D) Schematics of the predicted peptides in the various strains, with signal peptide shown in red and a cell wall attachment motif shown in green. The SSR genotype compared to the reference strain is shown.
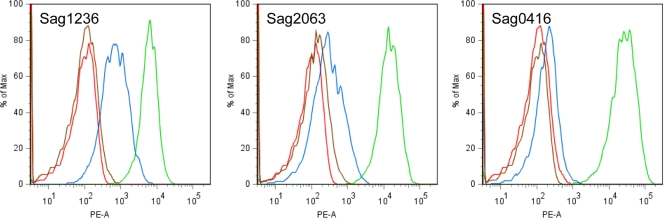
We hypothesized that the apparent change in subcellular localization would influence antigen recognition at the bacterial surface. Flow cytometry analysis was performed on the above strain pairs using polyclonal antiserum against the different proteins (Fig. 3). Strains containing complete genes encoding cell wall-attached proteins showed a shift in fluorescence intensity when using antiserum compared to using preimmune serum. Strains in which genes were frameshifted showed no such shift or less of a shift. In the case of Sag2063 and Sag0416, it would appear that the proteins are no longer present in a recognizable form on the bacterial surface. Surface-associated Sag1236 was still recognized by specific antibodies in the strain with an SSR-mediated frameshift, although to a significantly lesser extent. The frameshift causes a minor truncation with loss of the C-terminal cell wall-attachment anchor. The near-complete protein may thus be recognized transiently, during secretion and/or translocation. Thus, SSR variation of the kind described above is likely to prevent or modify binding of specific antibodies to the bacterial surface. Among the three strains examined, every strain was positive for at least one protein and negative for another, thus reducing the likelihood that strain-specific factors independent of the genetic loci influenced the results.
FIG. 3.
SSR variation influences antigen recognition by specific antibodies. Flow cytometry was performed using specific primary antibodies against the indicated protein on strains that were genotypically ON (green line) or OFF (blue line). Background controls were with pooled serum from animals immunized with adjuvant only, using the same ON (orange line) or OFF (red line) strains for each respective protein. The ON/OFF strains were Coh1/2603 for Sag1236 (ScpB), 2603/515 for Sag2063 (BibA), and 2603/Coh1 for Sag0416.
Screening for slippage events.
CIB has been the method of choice when screening for phase variants. Attempts to use the method for Streptococcus agalactiae with the positive and negative strains above failed to reproducibly confirm the phenotypes we had observed with other methods. The difference in signal between supposedly negative and positive colonies was small, and the signal intensity in general was low, irrespective of whether alkaline phosphatase or HRP was used (data not shown). In an attempt to exclude that capsular polysaccharide was contributing to background and/or masking antigens, three isogenic cpsE knockout mutants (3) derived from the parent strains used above were obtained and subjected to the assay. Colonies from strains that had complete genes generally showed binding of antibodies raised against the corresponding proteins, while strains in which genes were frameshifted had negative colonies (data not shown). Screening for phase variation in vitro was performed by starting with a strain genotypically negative for a protein and trying to identify positive colonies. Thus, unencapsulated strains 2603V/R (Sag1236), COH1 (Sag0416), and 515 (Sag2063) were screened repeatedly, using an average of 2,000 colonies per plate. Potentially positive colonies were picked, and the procedure repeated at a lower colony density to verify the phenotype and isolate clones. No phase variant with a consistently positive signal was obtained for any of the strains and proteins.
Immunomagnetic separation has been used successfully to isolate streptococci with specific antigenic properties starting from a mixed culture (8). We attempted a similar enrichment strategy to increase the fraction of potential OFF-ON bacteria among the original OFF bacteria. Up to 109 bacteria were incubated with polyclonal mouse antiserum against Sag1236, Sag0416, or Sag2063. Superparamagnetic beads coated with goat anti-mouse antibodies were then incubated with the bacteria, and bacteria decorated with beads were retained on a column through magnetic force. These bacteria, representing the positively selected fraction, were then plated and screened by CIB as described above. No phase variants were found for any of the proteins. However, colony counts of the selected fractions were surprisingly high, suggesting that the carryover of nonspecifically retained bacteria was substantial. Control experiments using antiserum against a highly expressed surface protein (pilus backbone protein) were performed. Starting with a mixed culture where the ratio of negative (knockout strain) to positive (overexpressing strain) was known (10,000 and 1,000), the degree of enrichment obtained with immunomagnetic separation was on the order of 10-fold (data not shown). The efficiency in the experiments above could be lower, since the surface proteins in question are likely to have lower expression levels than an overexpressing strain, and the pilus is particular in that it extends far out from the bacterial surface.
We also attempted isolating OFF-ON events by FACS. Starting with OFF strains, single bacteria that had an aberrantly high fluorescence intensity, one comparable to the mean fluorescence intensity of an ON strain, were sorted, plated, and subjected to CIB. The fraction of bacteria thus selected was on the order of 0.1% of the total number of bacteria (100,000 in two experiments for each OFF strain). Colonies that were subsequently positive in CIB were picked and expanded to prepare frozen stocks, which were then grown and analyzed by one or several of the above-mentioned methods, followed by sequencing of the relevant locus when there were indications of OFF-ON switching. We did not find any events where restoration of a phenotype was matched with the expected SSR variation in the sequenced locus. Since more than 300,000 bacteria were screened, the mutation frequency in the selected loci is likely less than 10−5.
In a final attempt to detect slippage events, we chose to sequence selected loci in strains that had been subjected to recent in vivo passage. The 2603V/R strain was subjected to serial passage in the mouse, for a total of 19 times. The three SSR loci described above were then sequenced in the passaged strain. The SSRs were identical to those in the starting strain (data not shown). Moreover, four clinical isolates from patients with invasive GBS disease were obtained (kindly provided by Carol Baker). The strains were atypical in that no capsular polysaccharide was detected with antibodies against type V capsule, although molecular genotyping indicated the presence of a typical type V capsule locus (C. Baker, unpublished data). The cpsH (cpsV) locus described above, containing seven SSRs, was sequenced in the four strains. All four strains contained the locus, and in three cases the locus was identical to that of the 2603V/R strain with respect to the seven SSRs (data not shown). In one case, there was a 450-bp insertion in the cpsH gene, representing the mobile genetic element IS1381.
DISCUSSION
Phase variation can occur in different ways, including slipped-strand mispairing of SSRs during DNA replication, general and site-specific recombination, excision/insertion of mobile genetic elements such as transposons and insertion sequences, and epigenetic regulation through differential methylation (42). Over the years, SSRs have emerged as a common and important mechanism for phase variation both of particular genes/proteins and on a genome scale (41). The present study is one of the first comprehensive attempts to understand the importance of SSRs for phase variation in a Gram-positive pathogen, S. agalactiae. Through comparative genomic analysis of eight bacterial genomes representing the most frequently isolated serotypes, we present evidence of genotypic variation indicative of slipped-strand mispairing in SSR regions.
In a first stringent screening, a wide variety of SSRs were examined across the eight genomes. The results suggested that the repeat type that has the highest propensity to vary is the homopolymeric tract, especially the poly-A tract. With an extended screening of homopolymeric tracts, additional polymorphisms were identified, and this reinforced the potential importance of poly-A as a means to drive phase variation. In addition, the longest homopolymeric tracts identified were examined, and gene loci with clustered SSRs were identified. Moreover, a previously undescribed positional bias of poly-A tracts within ORFs was observed. The longest tracts were preferentially located in the 5′ ends of ORFs. Should a poly-A tract within a coding sequence alter and a frameshift ensue, it may be advantageous that the resulting product is a short peptide. A major truncation is more likely to efficiently inactivate the protein and may avoid inadvertent effects due to misfolded or functionally altered proteins. Moreover, the bioenergetic cost of translation is proportional to the length of the product. Interestingly, while poly-A tracts seem to be the most slippage-prone among the SSRs, the longest such tracts are statistically underrepresented in the genome, implying an evolutionary selection against such sequence elements. An alternative means of repressing potential variation are point mutations within SSRs (stabilization), of which several examples were noted during screening procedures.
Overall, among the loci identified there are many examples of genes encoding surface proteins or secreted proteins or of genes that indirectly affect such proteins. Examples include cell wall attached proteins, lipoproteins, membrane proteins, and sortase, the latter of which could influence a range of cell wall attached proteins. Such surface structures constitute rather typical examples of potential phase variation. Less typical is the observed SSR polymorphisms in transcriptional regulators and two-component systems. It would seem disadvantageous to override regulation through the genotypic inactivation of regulatory components. However, in a recent study it was shown that homopolymeric tract polymorphisms in a C. jejuni response regulator cause ON/OFF phase variation of both flagellar biosynthesis and its regulator (10). The gene encoding the regulator is variable through a cluster of five poly-A and one poly-T tracts, which are comparable in length to the tracts discussed in the present study.
The inclusion of draft genomes in the analysis represents an increased risk to include sequencing errors. Steps were taken to exclude hits where sequence quality was in doubt, such as in the vicinity of contig ends (see Materials and Methods). Although only a subset of the variant loci was verified through manual scrutiny of trace file quality, there is little to suggest that overall genome sequence quality had a major impact on our results. Overall, no clear relationship between the degree of fragmentation (number of contigs) and the number or type of hits in our screening was noted. A limited number of loci were selected for resequencing. Nine loci, representing poly-A or poly-T and showing variation in at least one draft genome compared to the reference genome, were resequenced in those genomes and SSR polymorphisms were confirmed (data not shown). Nonetheless, it cannot be ruled out that selected indels represent sequencing errors.
The comparative genomic analysis and experiments performed here suggests that the phase variation mechanism involving SSR slippage is at play in S. agalactiae and has resulted in antigenic differentiation between strains. However, despite considerable efforts and the use of three different methodologies, it was not possible to detect an intrastrain event of SSR slippage for any of the three proteins that were experimentally investigated. Compared to other bacterial species, S. agalactiae mostly lacks the unusually long repeats responsible for high-frequency phase variation (5). Moreover, the preference for poly-A tracts is atypical. Although there are several reports of genotypic switching through poly-A tract slippage (of comparable length), none of these suggest a high frequency. It has been suggested that poly-A tracts pose a delicate problem for bacteria, in that RNA polymerase (transcriptional) slippage seems to occur in addition to replicational slippage, and this may be a reason why some microorganisms show an underrepresentation of long poly-A/T tracts (2). Our difficulties in isolating genotypic switching in vitro may derive from the methodological issues associated with screening a sample representing a large enough number of bacteria. Moreover, an ON-OFF switching event may confer a selective advantage in vivo that is not evident in vitro, if the relevant proteins are surface exposed, immunogenic, and expendable. For practical purposes, our screening was biased, in that OFF-ON events were targeted. In our case, such events would have involved the expansion of SSRs. Mutation frequencies may differ significantly between ON-OFF and OFF-ON events, and contractions seem to be more frequent than expansions (5).
A recent publication (23) describes eight atypical clinical isolates (vaginal/rectal colonization) of S. agalactiae that were unencapsulated. Among various polymorphisms with little or no phenotypic impact, three of the eight strains contained deletion of an adenine in the cpsG locus, resulting in a frameshift and truncation of the protein with likely loss of function, and thus compromised capsule biosynthesis. Upon closer examination of the sequence, we note that the deletion is located in a poly-A tract, and represents a SSR change from A8 to A7. This suggests that, in vivo, SSR slippage in a poly-A tract may constitute a significant way to modify capsular biosynthesis. We describe extreme repeat clustering in cpsH, but did not find evidence of variation in four unencapsulated invasive clinical isolates. In another attempt to approach the in vivo situation, the three selected genes of interest were sequenced in a strain that had undergone repeated mouse passages, and all three genes remained unchanged with respect to the starting inoculum. Thus, in vivo growth in naive mice does not seem to involve a selective pressure for variation in these loci.
In the strict sense, the lack of experimental in-strain variation means that we have no direct evidence of SSR-mediated phase variation. Nevertheless, we believe that S. agalactiae uses SSRs as an adaptive strategy but in a significantly different role compared to that in Gram-negative bacteria. Bet-hedging as an evolutionary strategy comes at a cost, but it can improve fitness when environmental circumstances change frequently enough (26). Moreover, generalist bacteria with large genomes and considerable genome redundancy may sustain genotype switching better than specialists. S. agalactiae, by preferentially using homopolymeric A tracts, selecting against long repeats in general, and using damage control by positioning of SSRs, is ensuring that mutation frequency is kept in check. Nevertheless, SSRs are used, either for long-term adaptation or possibly during the uncommon invasive infections. This represents a cautious bet-hedging strategy suitable for a specialized commensal and occasional opportunist. Genome plasticity, rather than phase variation, may be the adequate term for this process.
Supplementary Material
Acknowledgments
We are indebted to Michael Cieslewicz for the gift of unencapsulated S. agalactiae strains. We also thank R. Moxon and coworkers for helpful discussions in the initial stages of the project and for critical reading of the manuscript. Simona Tavarini and Sandra Nuti provided technical expertise during flow cytometry experiments. We thank C. Donati and V. Raguzzi for assistance with the design and execution of search algorithms.
During this work, R.J. was initially funded by a Marie-Curie intra-European fellowship (FP5), and for a minor part, by the Swedish Research Council.
Footnotes
Published ahead of print on 21 May 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Baranov, P. V., A. W. Hammer, J. Zhou, R. F. Gesteland, and J. F. Atkins. 2005. Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 6:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Bryan, J. D., and D. W. Shelver. 2009. Streptococcus agalactiae CspA is a serine protease that inactivates chemokines. J. Bacteriol. 191:1847-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139-146. [DOI] [PubMed] [Google Scholar]
- 4.Cleary, P. P., J. Handley, A. N. Suvorov, A. Podbielski, and P. Ferrieri. 1992. Similarity between the group B and A streptococcal C5a peptidase genes. Infect. Immun. 60:4239-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Bolle, X., C. D. Bayliss, D. Field, T. van de Ven, N. J. Saunders, D. W. Hood, and E. R. Moxon. 2000. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 35:211-222. [DOI] [PubMed] [Google Scholar]
- 6.Edwards, M. S., and C. J. Baker. 2005. Group B streptococcal infections in elderly adults. Clin. Infect. Dis. 41:839-847. [DOI] [PubMed] [Google Scholar]
- 7.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 8.Gottschalk, M., S. Lacouture, and L. Odierno. 1999. Immunomagnetic isolation of Streptococcus suis serotypes 2 and 1/2 from swine tonsils. J. Clin. Microbiol. 37:2877-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutekunst, H., B. J. Eikmanns, and D. J. Reinscheid. 2003. Analysis of RogB-controlled virulence mechanisms and gene repression in Streptococcus agalactiae. Infect. Immun. 71:5056-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Harris, T. O., D. W. Shelver, J. F. Bohnsack, and C. E. Rubens. 2003. A novel streptococcal surface protease promotes virulence, resistance to opsonophagocytosis, and cleavage of human fibrinogen. J. Clin. Investig. 111:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrixson, D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646-1659. [DOI] [PubMed] [Google Scholar]
- 11.Hood, D. W., M. E. Deadman, M. P. Jennings, M. Bisercic, R. D. Fleischmann, J. C. Venter, and E. R. Moxon. 1996. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. U. S. A. 93:11121-11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji, Y., L. McLandsborough, A. Kondagunta, and P. P. Cleary. 1996. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect. Immun. 64:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan, P., L. A. Snyder, and N. J. Saunders. 2003. Diversity in coding tandem repeats in related Neisseria spp. BMC Microbiol. 3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in group B streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 16.Levinson, G., and G. A. Gutman. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4:203-221. [DOI] [PubMed] [Google Scholar]
- 17.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. Nardi Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B streptococcus vaccine by multiple genome screen. Science 309:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, P., T. van de Ven, N. Mouchel, A. C. Jeffries, D. W. Hood, and E. R. Moxon. 2003. Experimentally revised repertoire of putative contingency loci in Neisseria meningitidis strain MC58: evidence for a novel mechanism of phase variation. Mol. Microbiol. 50:245-257. [DOI] [PubMed] [Google Scholar]
- 19.Moxon, R., C. Bayliss, and D. Hood. 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40:307-333. [DOI] [PubMed] [Google Scholar]
- 20.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 21.Pericone, C. D., D. Bae, M. Shchepetov, T. McCool, and J. N. Weiser. 2002. Short-sequence tandem and nontandem DNA repeats and endogenous hydrogen peroxide production contribute to genetic instability of Streptococcus pneumoniae. J. Bacteriol. 184:4392-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puopolo, K. M., and L. C. Madoff. 2003. Upstream short sequence repeats regulate expression of the alpha C protein of group B streptococcus. Mol. Microbiol. 50:977-991. [DOI] [PubMed] [Google Scholar]
- 23.Ramaswamy, S. V., P. Ferrieri, L. C. Madoff, A. E. Flores, N. Kumar, H. Tettelin, and L. C. Paoletti. 2006. Identification of novel cps locus polymorphisms in nontypeable group B streptococcus. J. Med. Microbiol. 55:775-783. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen, M., and L. Bjorck. 2001. Unique regulation of SclB: a novel collagen-like surface protein of Streptococcus pyogenes. Mol. Microbiol. 40:1427-1438. [DOI] [PubMed] [Google Scholar]
- 25.Richardson, A. R., and I. Stojiljkovic. 2001. Mismatch repair and the regulation of phase variation in Neisseria meningitidis. Mol. Microbiol. 40:645-655. [DOI] [PubMed] [Google Scholar]
- 26.Salathe, M., J. W. Van Cleve, and M. W. Feldman. 2009. Evolution of stochastic switching rates in asymmetric fitness landscapes. Genetics 182:1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salaun, L., B. Linz, S. Suerbaum, and N. J. Saunders. 2004. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiology 150:817-830. [DOI] [PubMed] [Google Scholar]
- 28.Santi, I., M. Scarselli, M. Mariani, A. Pezzicoli, V. Masignani, A. Taddei, G. Grandi, J. L. Telford, and M. Soriani. 2007. BibA: a novel immunogenic bacterial adhesin contributing to group B Streptococcus survival in human blood. Mol. Microbiol. 63:754-767. [DOI] [PubMed] [Google Scholar]
- 29.Saunders, N. J., A. C. Jeffries, J. F. Peden, D. W. Hood, H. Tettelin, R. Rappuoli, and E. R. Moxon. 2000. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 37:207-215. [DOI] [PubMed] [Google Scholar]
- 30.Saunders, N. J., J. F. Peden, D. W. Hood, and E. R. Moxon. 1998. Simple sequence repeats in the Helicobacter pylori genome. Mol. Microbiol. 27:1091-1098. [DOI] [PubMed] [Google Scholar]
- 31.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 32.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 34.Snyder, L. A., S. A. Butcher, and N. J. Saunders. 2001. Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology 147:2321-2332. [DOI] [PubMed] [Google Scholar]
- 35.Sreenu, V. B., G. Ranjitkumar, S. Swaminathan, S. Priya, B. Bose, M. N. Pavan, G. Thanu, J. Nagaraju, and H. A. Nagarajaram. 2003. MICAS: a fully automated web server for microsatellite extraction and analysis from prokaryote and viral genomic sequences. Appl. Bioinform. 2:165-168. [PubMed] [Google Scholar]
- 36.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 39.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 40.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 41.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Woude, M. W., and A. J. Baumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Ham, S. M., L. van Alphen, F. R. Mooi, and J. P. van Putten. 1993. Phase variation of Haemophilus influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell 73:1187-1196. [DOI] [PubMed] [Google Scholar]
- 44.Wassenaar, T. M., J. A. Wagenaar, A. Rigter, C. Fearnley, D. G. Newell, and B. Duim. 2002. Homonucleotide stretches in chromosomal DNA of Campylobacter jejuni display high-frequency polymorphism as detected by direct PCR analysis. FEMS Microbiol. Lett. 212:77-85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.