Abstract
The Methanococcus maripaludis energy-conserving hydrogenase B (Ehb) generates low potential electrons required for autotrophic CO2 assimilation. To analyze the importance of individual subunits in Ehb structure and function, markerless in-frame deletions were constructed in a number of M. maripaludis ehb genes. These genes encode the large and small hydrogenase subunits (ehbN and ehbM, respectively), a polyferredoxin and ferredoxin (ehbK and ehbL, respectively), and an ion translocator (ehbF). In addition, a gene replacement mutation was constructed for a gene encoding a putative membrane-spanning subunit (ehbO). When grown in minimal medium plus acetate (McA), all ehb mutants had severe growth deficiencies except the ΔehbO::pac strain. The membrane-spanning ion translocator (ΔehbF) and the large hydrogenase subunit (ΔehbN) deletion strains displayed the severest growth defects. Deletion of the ehbN gene was of particular interest because this gene was not contiguous to the ehb operon. In-gel activity assays and Western blots confirmed that EhbN was part of the membrane-bound Ehb hydrogenase complex. The ΔehbN strain was also sensitive to growth inhibition by aryl acids, indicating that Ehb was coupled to the indolepyruvate oxidoreductase (Ior), further supporting the hypothesis that Ehb provides low potential reductants for the anabolic oxidoreductases in M. maripaludis.
Hydrogenotrophic methanococci specialize in utilizing H2 as an electron donor, and these organisms possess six different Ni-Fe hydrogenases. These enzymes include two F420--reducing hydrogenases, two non-F420-reducing hydrogenases, and two membrane-bound hydrogenases (Eha and Ehb [5]). The F420-reducing hydrogenases reduce coenzyme F420, which subsequently reduces methenyltetrahydromethanopterin and methylenetetrahydromethanopterin, intermediates in the pathway of methanogenesis. In Methanococcus voltae, the F420-reducing hydrogenase is also reported to reduce the 2-mercaptoethanesulfonate:7-mercaptoheptanoylthreonine phosphate heterodisulfide formed in the final step of methanogenesis (2). In contrast, Methanothermobacter marburgensis utilizes the non-F420-reducing hydrogenase to reduce the heterodisulfide (22, 25).
The two membrane-bound hydrogenases couple the chemiosmotic energy of ion gradients to H2 oxidation and ferredoxin reduction. In the aceticlastic methanogen Methanosarcina barkeri, the homologous enzyme is called energy conserving hydrogenase or Ech and performs a variety of physiological functions, including the generation of a proton motive force during CO oxidation and concomitant proton reduction in aceticlastic methanogenesis and the generation of low potential electron donors for CO2 reduction to formylmethanofuran in the first step of methanogenesis and the reductive carboxylation of acetyl coenzyme A (acetyl-CoA) to pyruvate in carbon assimilation (11, 12). In the hydrogenotrophic methanogens, it is predicted that the two energy-conserving hydrogenases (Eha and Ehb) have distinct roles (26). The Ehb appears to reduce low potential electron carriers utilized in autotrophic CO2 fixation (16). Anabolic enzymes likely to be coupled to Ehb in this manner include (i) the carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) and the pyruvate oxidoreductase (Por), which catalyze the first two steps of carbon assimilation; (ii) the α-ketoglutarate oxidoreductase (Kor), which catalyzes the final step in the incomplete reductive tricarboxylic acid cycle; and (iii) the indolepyruvate oxidoreductase (Ior) and the 2-oxoisovalerate oxidoreductase (Vor), which are involved in amino acid biosynthesis from aryl and branched-chain acids, respectively. Support for these conclusions comes in large part from the phenotype of an M. maripaludis ehb gene replacement mutant S40, which was only capable of limited growth in the absence of acetate and amino acids (16). Furthermore, expression of CODH/ACS, Por, and Vor were significantly upregulated in the mutant, providing further evidence for a role of Ehb in these processes (16). In contrast, there is no direct evidence for the role of Eha. By analogy with the Methanosarcina Ech, it could be involved in generating reducing equivalents for the reduction of CO2 to formylmethanofuran. Alternatively, hydrogenotrophic methanogens may have an alternative method of CO2 reduction (27), and Eha could have another function entirely.
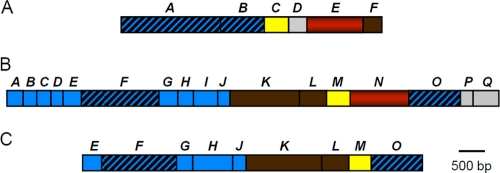
In spite of some functional similarities between the Ech of the aceticlastic methanogens and Eha or Ehb of hydrogenotrophs, the structures of their operons are very different (Fig. 1). Based upon sequence comparisons, all of these membrane-bound hydrogenases possess conserved large and small hydrogenase subunits, a 2[4Fe-4S] ferredoxin, and an integral membrane ion translocator (3, 8, 26). Otherwise, the structures are very different. The purified Ech from Methanosarcina barkeri contains six polypeptides encoded by the six genes of the ech operon (8, 11). The Eha and Ehb hydrogenases have never been purified. The eha and ehb operons from the hydrogenotrophic methanogen Methanothermobacter thermautotrophicus comprise 20 and 17 genes, respectively (23, 26). Most of these genes are predicted to encode transmembrane proteins, although there are also several polyferredoxins and hydrophilic proteins (26). Many of these genes are not homologous to the M. barkeri ech genes. The Methanococcus maripaludis genome contains homologs to the M. thermautotrophicus eha and ehb genes, although only nine of the ehb genes are contiguous on the genome (Fig. 1). In the present study, the Ehb from the hydrogenotrophic methanogen Methanococcus maripaludis was analyzed. M. maripaludis is a model organism that can be easily genetically modified. Furthermore, its genome has been sequenced, and many of its biochemical pathways have been characterized.
FIG. 1.
Genetic map of Methanosarcina barkeri ech (A), Methanothermobacter marburgensis ehb (MTH1235-1251) (B), and Methanococcus maripaludis ehb (MMP1631-1629) (C) operons. Genes encoding integral membrane proteins found only in Ehb are indicated in blue, integral membrane proteins conserved in both Ech and Ehb are blue with diagonal stripes, hydrogenase small subunits are yellow, hydrogenase large subunits are red, 4Fe-4S motif-containing proteins are brown, and other hydrophilic proteins present in Ehb but absent from Ech are gray. Notably, M. maripaludis contains homologs to all of the M. marburgensis ehb genes, but many are unlinked to the major gene cluster and not shown. Based upon references 5, 8, 11, and 26.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strains used in the present study are listed in Table 1 . Methanococcus maripaludis cultures were grown as described by Jones et al. (6). The media used included McA, a minimal medium containing 10 mM sodium acetate; McC, McA plus 0.2% (wt/vol) Casamino Acids; and McCV, McC plus 1% (vol/vol) vitamin mixture (29). M. maripaludis cultures were grown with 275 kPa of H2-CO2 (80:20 [vol/vol]). The antibiotics puromycin (2.5 μg ml−1) or neomycin (500 μg ml−1 in plates and 1 mg ml−1 in broth) were used where necessary. To reduce the selection for revertants, inocula for growth experiments with the mutants were taken from frozen stocks into prewarmed McC tubes. Cultures were grown to an absorbance at 600 nm of 0.4 to 0.5. To initiate the growth experiments, cultures were diluted in McA medium, and 105 cells were inoculated into the experimental medium.
TABLE 1.
ehb homologs in Methanothermobacter thermautotrophicus ΔH and Methanococcus maripaludis S2
| Gene | Homolog |
% aa identitya | Predicted gene product | |
|---|---|---|---|---|
| M. thermautotrophicus | M. maripaludis | |||
| ehbA | mth1251 | mmp1469 | 57.7 | Transmembrane protein |
| ehbB | mth1250 | mmp1049 | 38.7 | Transmembrane protein |
| ehbC | mth1249 | mmp1073 | 46.7 | Transmembrane protein |
| ehbD | mth1248 | mmp1074 | 49.8 | Transmembrane protein |
| ehbE | mth1247 | mmp1629 | 52.0 | Transmembrane protein |
| ehbF | mth1246 | mmp1628 | 49.1 | Transmembrane protein, ion translocator |
| ehbG | mth1245 | mmp1627 | 41.6 | Transmembrane protein |
| ehbH | mth1244 | mmp1626b | 38.5 | Transmembrane protein |
| ehbI | mth1243 | mmp1626 | 38.5 | Transmembrane protein |
| ehbJ | mth1242 | mmp1625 | 38.9 | Transmembrane protein |
| ehbK | mth1241 | mmp1624 | 47.4 | Polyferredoxin |
| ehbL | mth1240 | mmp1623 | 49.6 | Ferredoxin |
| ehbM | mth1239 | mmp1622 | 61.8 | Hydrogenase small subunit |
| ehbN | mth1238 | mmp1153 | 52.1 | Hydrogenase large subunit |
| ehbO | mth1237 | mmp1621 | 50.0 | Transmembrane protein |
| ehbP | mth1236 | mmp0940 | 46.6 | Hydrophilic protein |
| ehbQ | mth1235 | mmp0400 | 57.1 | Hydrophilic protein |
Percent amino acid (aa) sequence identity.
mmp1626 is a gene fusion of mth1244 and mth1243.
Escherichia coli TOP10 strains utilized in genetic manipulations were obtained from Invitrogen (Carlsbad, CA). E. coli cultures were maintained in low-salt (0.5% [wt/vol]) Luria-Bertani medium containing 50 μg of ampicillin ml−1 when appropriate.
Construction of ehb plasmids.
Standard molecular biology techniques were used. M. maripaludis genomic DNA was purified by using a Wizard Genomic DNA purification kit (Promega, Madison, WI). Plasmids were purified using either the Wizard Plus Minipreps DNA purification system (Promega) for plasmids larger than 10 kb or the Qiaprep spin miniprep kit (Qiagen, Valencia, CA) for plasmids smaller than 10 kb. Gel extraction was performed by using a QiaQuick gel extraction kit (Qiagen).
The pWDK60 plasmid, which generated an ehbO::pac gene insertion into the M. maripaludis chromosomal DNA, was constructed as described by Lin and Whitman (9, 24). Briefly, amplification of genomic M. maripaludis DNA was performed by using a Ready-To-Go kit (Amersham Pharmacia Biotech, Piscataway, NJ), with the primers and PCR conditions as described by Major (10). Other ehb mutants were constructed by using the marklerless in-frame deletion method of Moore and Leigh (13). The pCRPrtNeo vector contained two multiple cloning regions for generating in-frame, markerless deletions in M. maripaludis. Upstream and downstream flanking regions were cloned in tandem on the pCRPrtNeo vector at one of these multiple cloning sites. Primers for DNA amplification of ehb flanking regions were described by Major (10). The open reading frames (ORFs) were reduced to four codons: the ATG start codon, either GGATCC (BamHI site) or TCTAGA (XbaI site), and the native stop codon. DNA was amplified by using Herculase enhanced DNA polymerase (Stratagene, La Jolla, CA) in a gradient thermocycler (Major [10]). The cycling program proceeded as follows. After 1 min of denaturation at 92°C, the following steps were performed for 15 cycles: a denaturation step was performed at 92°C for 30 s; a gradient extension step of 45 to 55°C was performed for 30 s, with an increase of 1°C per cycle; and a gradient annealing step was performed at 60 to 70°C for 1 min, with an increase of 0.5°C per cycle (10). The first 15 cycles were immediately followed by a second round of 25 cycles. The second round included a denaturation step at 92°C for 30 s, an extension step for 30 s at 52°C, and a gradient annealing step performed at 63 to 73°C for 1 min, with a 10 s increase in duration per cycle (10). Amplified DNA was subcloned with a TOPO TA cloning kit (Invitrogen) for ease of subsequent manipulation. The upstream and downstream ehb flanking regions were inserted into the pCRPrtNeo vector in a stepwise manner. Typically, the upstream flanking region was digested, ligated into the pCRPrtNeo vector, electroporated, and cloned. Correct constructs containing the upstream flanking region were then digested for insertion of the downstream flanking region.
Ehb mutants were complemented by using the pMEV2 vector containing the relevant ehb gene (9). Individual ehb genes were amplified as described above using the primers described by Major (10). The primers were constructed to contain an NsiI site at the 5′ start site of the gene and an XbaI site at or after the 3′ terminus of the gene to facilitate cloning into the pMEV2 vector.
Correct plasmid construction was confirmed by sequencing performed at the University of Michigan Sequencing Core Facility (Ann Arbor, MI). Primers used for sequencing are available upon request.
Transformations.
M. maripaludis chemical transformations were performed as previously described (28). Selection of in-frame deletion mutants was performed as described by Moore and Leigh (13). To avoid selection for second-site revertants, transformants were cultured on rich McCV medium. After the initial plating, well isolated colonies were restreaked for purity. After growth, well-isolated colonies were picked into McCV broth. A portion of the broth culture was used for preparation of glycerol stocks, and a portion was used for confirmation of the genotype. E. coli was transformed as described by Lin and Whitman (9).
Confirmation of ehb mutant genotypes.
Southern hybridization was used to confirm the ΔehbO::pac mutation. The DIG High Prime DNA Labeling and Detection Starter Kit I (Roche, Mannheim, Germany) was used for Southern blotting. The S40 probe was generated by digestion of pWDK60 using EcoRV/KpnI to generate the downstream flanking region of the ehbO gene (∼880 bp). Genomic DNA was digested using EcoRV. Markerless ehb mutants were confirmed via PCR screening and DNA sequencing.
Sample preparation for Blue Native PAGE.
To prepare whole-cell protein extracts, M. maripaludis was cultured in 5 ml of McA medium to an absorbance of ∼0.4. Proteins were prepared by a modification of the method of Schägger and von Jagow (21). All sample preparation steps were performed anaerobically. The cells were collected by centrifugation and lysed by osmotic shock in 0.5 ml of suspension buffer containing 50 mM NaCl, 5 mM 6-aminocaproic acid, 50 mM Bistris-HCl (pH 7.0), and 10% glycerol (19). To inhibit protease, 0.5 μl of 0.5 M phenylmethylsulfonyl fluoride (PMSF) in dimethyl sulfoxide was added. The cell lysate was then incubated with 5 U of RQ1 DNase (Promega) at room temperature for 1 h to digest DNA. To solubilize membrane proteins, 62.5 μl of 10% (wt/vol) laurylmaltoside was added. After 15 min of incubation at room temperature, the sample was centrifuged at 16,000 × g for 10 min. The supernatant was then transferred to a new microfuge tube and mixed with 10% glycerol.
The protein concentrations were determined with a BCA protein assay kit (Pierce, Rockford, IL). Before electrophoresis, 2.5 μl of a 5% (wt/vol) solution of Coomassie blue G in 500 mM 6-aminocaproic acid was added to 40 μl of sample containing 60 μg of protein, which was loaded in each lane.
Blue Native PAGE.
Blue Native gel electrophoresis was performed as described on 7% polyacrylamide gels at 80 V (15, 21). The gel buffer contained 0.5 M 6-aminocaproic acid and 50 mM Bis-Tris/HCl (pH 7.0). The anode buffer contained 50 mM Bis-Tris/HCl (pH 7.0). The beginning cathode buffer contained 50 mM Tricine, 15 mM Bis-Tris/HCl (pH 7.0), and 0.02% (wt/vol) Coomassie blue G. When proteins had migrated to the middle of the gels, the cathode buffer was replaced with the same buffer without the dye. The typical total running time was ∼4 h. Silver staining was then performed as described by Nesterenko et al. (14).
Western blotting of Blue Native gels.
The proteins were transferred by electroblotting from Blue Native gels to Immun-Blot polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA) using the Mini Trans-Blot system (Bio-Rad) at a constant voltage of 100 V for 1 h. The transfer buffer contained 25 mM Tris (pH 8.3), 192 mM glycine, and 10% methanol (vol/vol). After transfer, the membrane was washed with a solution of 30% methanol-10% acetic acid (vol/vol) overnight to destain the background.
For the preparation of antisera, multiple antigenic peptides (MAP) of the 15 amino acids of the N-terminal sequence of the hydrogenase large subunit (EhbN) were synthesized (Sigma-Genosys, St. Louis, MO) and injected into rabbits (Cocalico Biologicals, Reamstown, PA). Protein complexes containing EhbN were probed with rabbit polyclonal antiserum and a goat anti-rabbit secondary antibody (Bio-Rad). The secondary antibody was conjugated to alkaline phosphatase, and the bound antibodies were visualized with nitroblue tetrazolium chloride and BCIP (5-bromo-4-chloro-3-indolylphosphate).
In-gel hydrogenase activity assay.
For the hydrogenase activity assay, the Blue Native PAGE was performed anaerobically under an atmosphere of 95% N2-5% H2 in a Coy anaerobic chamber. The hydrogenase activity bands on the gels were located as previously described (1). The gels were immersed with 10 mM methylviologen in 25 mM potassium phosphate buffer (pH 7.5). An equal volume of 2,3,5-triphenyl tetrazolium chloride solution (2.5% [wt/vol]) was added after blue bands of hydrogenases appeared. The anaerobic incubation was continued until red bands were clearly formed.
Preparation of membrane fraction.
To prepare membrane fractions, M. maripaludis was cultured in 150 ml of McCV medium to an absorbance of ∼0.4. The cells were collected anaerobically by centrifugation at 10,000 × g for 30 min at 4°C and resuspended in 5 ml of buffer A containing 10 mM magnesium acetate, 30 mM KCl, 2 mM dithiothreitol, and 25 mM potassium PIPES (pH 7.0). To inhibit protease, 20 μl of 0.5 M PMSF in dimethyl sulfoxide was added. The cells were lysed by freezing at −20°C. Then, the cell lysate was incubated with 20 U of RQ1 DNase (Promega) at 37°C for 15 min to digest DNA. Unbroken cells were removed by centrifugation at 8,000 × g for 30 min at 4°C. Then, 0.5 ml of the supernatant was collected as whole-cell extract, and the rest was centrifuged at 200,000 × g for 1 h at 4°C. After ultracentrifugation, the supernatant was collected as the soluble fraction, and the pellet was resuspended in 0.2 ml of buffer A with 1 mM PMSF. The pellet suspension was loaded onto a 3.9 ml of 10 to 60% sucrose gradient in buffer A and centrifuged at 32,000 × g for 18 h at 20°C. The membrane fraction (∼1 ml) was collected at 30 to 50% sucrose.
Phylogenetic analysis.
The amino acid sequences of large hydrogenase subunits and relevant homologs were acquired from the NCBI database. Sequences were aligned within the MEGA3.1 program using the CLUSTAL X algorithm (7). Phylogenetic trees were generated by using the minimum evolution algorithm under default MEGA3.1 settings.
RESULTS AND DISCUSSION
Phylogenetic analysis of hydrogenotrophic methanogen ehb genes.
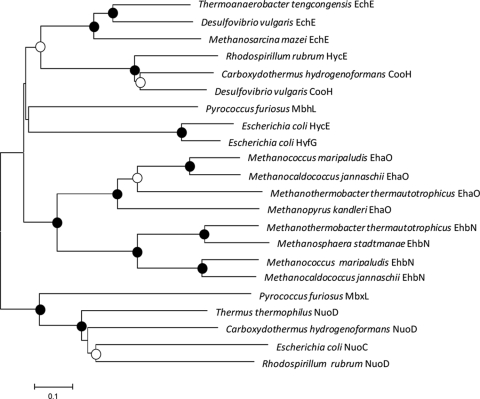
To identify the M. maripaludis ehb genes, the M. maripaludis genome was searched for homologs to the Methanothermobacter thermautotrophicus ehb genes by using BLAST (Table 1) (5, 26). M. thermautotrophicus has 17 ehb genes, which form one transcriptional unit (26). Although homologs to all of the M. thermautotrophicus ehb genes were found at various loci around the genome, only nine ehb genes clustered in M. maripaludis (Fig. 1). It was particularly noteworthy that the ORF mmp1153, which possessed the highest sequence similarity to the gene for the large hydrogenase subunit of Ehb or ehbN, was not part of the ehb gene cluster. Nevertheless, phylogenetic comparisons of the genes encoding homologs of the large hydrogenase subunits of Eha and Ehb from representative archaea, other hydrogenases, and the NADH:ubiquinone oxidoreductase support the hypothesis that mmp1153 is the likely ehbN in M. maripaludis (Fig. 2).
FIG. 2.
Phylogenetic relationships of NiFe large hydrogenase subunits and NADH:ubiquionone oxidoreductase. Amino acid sequence alignments and trees were generated by using the MEGA3.1 program. Alignments were generated by using the CLUSTAL × algorithm. The phylogenetic tree was generated by using the minimum evolutionary distance matrix method, although similar trees were found by using the neighbor-joining and maximum-parsimony methods (data not shown). The bootstrap values are indicated by circles at the branch points: •, >70%; ○, 50 to 70%; unlabeled, <50%. The scale bar is 0.1 expected amino acid substitutions per site. The accession numbers for the amino acid sequences (from the top to the bottom of the tree) are NP_621827, AAS94913, AAM32020, YP_426513, NC_007503, AAS96764, AAL81566, AAC75763, AAC75540, CAF31018, NP_247493, CAB52770, NP_613748, CAB52791, YP_448457, CAF30709, NP_248021, AAL81558, YP_005886, YP_360254, AAC75346, and YP_426645.
The phylogenetic analyses also illustrated the uniqueness of Ehb. For instance, EchE, which encodes the large hydrogenase subunit of the methanosarcinal Ech, is more similar to the bacterial enzyme from Desulfovibrio than the archaeal Eha and Ehb enzymes. Likewise, the Eha and Ehb hydrogenase large subunits are no more similar to the archaeal MbhL from Pyrococcus than the bacterial EchE (Fig. 2). These observations are consistent with the large differences seen in operon structure and support the hypothesis that the Ehb and Eha of the hydrogenotrophic methanogens represent a separate family of energy-conserving hydrogenases (8, 26).
A search of the M maripaludis genome also revealed that the hyp and other hydrogenase maturation genes were not linked to the ehb or other hydrogenase operons. Apparent homologs were (numbers reflect the relative position of their genes on the genome): hypF, MMP0140; hypE, MMP0274; hypD, MMP0289; hypA, MMP0301; hycI, MMP1185; hypC, MMP1330; and hypB, MMP1520. In addition, the genome possessed MMP0291, which was annotated as “hydrogenase expression/formation protein related,” and MMP1337, which was annotated as “hydrogenase maturation protease, related.” This result is not unusual in M. maripaludis, where functionally related genes are not often linked unless they are subunits of a larger protein complex. Because the genome encodes six different Ni-Fe hydrogenases, the maturation proteins are probably active with multiple enzyme systems.
Functional characterization of ehb deletion mutants.
Because many hydrogenases are abundant in M. maripaludis, it is difficult to evaluate Ehb activity biochemically (16). An alternative, albeit qualitative, method for monitoring physiological function is to measure growth rates in minimal medium. Since Ehb plays an important role in acetate and amino acid biosynthesis, ehb mutants grow poorly in the absence of acetate and amino acids (16). However, unlike some strains of M. maripaludis, the wild-type strain S2 grows poorly in minimal medium without acetate as a carbon source. When the inoculum size is small, cultures in minimal medium frequently fail to grow at all. For this reason, the mutants were tested in medium containing acetate. Moreover, preliminary experiments indicated that cultures of ehb mutants frequently recovered the wild-type phenotype after several transfers in minimal medium, presumably through the accumulation of mutations at other loci. Therefore, mutants were stored as frozen stocks immediately after isolation from rich medium.
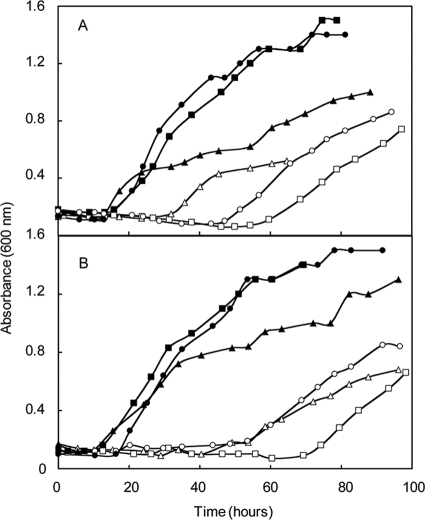
EhbF is a hypothetical transmembrane protein with sequence similarity to ion translocators and is hypothesized to be an essential component of Ehb (16). S40, a mutant where the ehbF gene was replaced with the antibiotic-resistant cassette pac, was used to evaluate its role in Ehb (16). As expected, S40 grew poorly in minimal medium with only acetate (Fig. 3A). Although amino acids were stimulatory, growth in rich medium was still much poorer than that of the wild type. For instance, the mean growth rates and lag times with standard deviations (n = 3) of the wild type in minimal and rich media were 0.28 ± 0.01 h−1 and 11 ± 0 h versus 0.31 ± 0.02 h−1 and 7 ± 0 h, respectively. In contrast, for the mutant S40 the values were 0.22 ± 0.01 h−1 and 59 ± 11 h in minimal medium versus 0.27 ± 0.01 h−1 and 10 ± 0 h in rich medium, respectively. The poor growth of the mutant in minimal medium confirmed the importance of EhbF in carbon assimilation. For comparison, ehbO, which encodes a small hydrophobic protein and is at the 3′ end of the ehb gene cluster, was replaced with the pac cassette to generate strain S42. Growth of this strain was greatly improved in rich medium, but it still did not grow as well as wild type (Fig. 3B). For instance, the mean growth rate and lag times with standard deviations (n = 3) of the mutant S42 in minimal and rich media were 0.28 ± 0.07 h−1 and 23 ± 2 h versus 0.24 ± 0.03 h−1 and 10 ± 0 h, respectively. Because the pac cassette is found in both mutants, the major growth deficit of S40 cannot be attributed to expression of the puromycin transacetylase per se. Therefore, the differences in growth must be attributed to either differences in the roles of EhbF and EhbO or the effects of polar expression of different regions of the gene cluster. Growth of S42 was also impaired in minimal medium but not as severely as the ehbF mutant. Thus, EhbO appeared to play a role in Ehb, but its function was not essential.
FIG. 3.
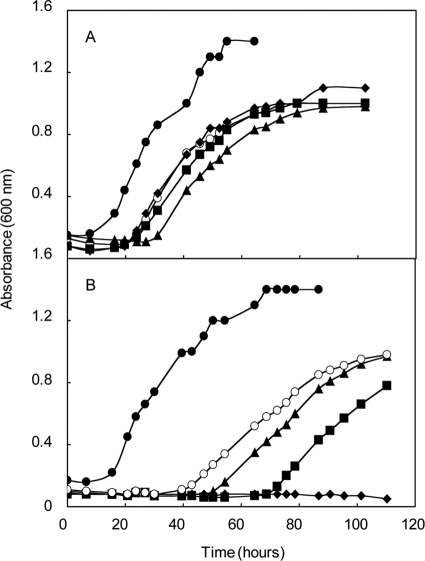
Evaluation of gene replacement (ΔehbF::pac and ΔehbO::pac) and deletion (ΔehbF) mutants of the conserved integral membrane proteins in rich (McCA, closed symbols) and minimal (McA, open symbols) media. (A) Growth of the ΔehbF::pac strain S40 (triangles) and the parental strain S2 (circles). (B) Growth of the ΔehbO::pac strain S42 (squares) and the parental strain S2 (circles). For panels A and B, the experiment was begun with ∼2 × 106 cells upon inoculation into prewarmed media. (C) Growth of the ΔehbF strain S940 (triangles), strain S943 containing the ΔehbF mutation complemented with pMEV2+ehbF (squares), and the parental strain S900 (circles). In these experiments, prewarmed McA media were inoculated with ∼105 cells. Growth curves are representative of experiments that were performed at least twice in duplicate.
An ΔehbF in-frame deletion strain (S940) without pac was generated to more clearly identify the effect of the ehbF deletion in the absence of polarity from the pac insertion. This mutation was constructed in a derivative of S2 called S900. This strain is a Δhpt mutant that contains a wild-type ehb locus (13). Growth of S900 was the same as S2 in minimal and rich media (data not shown). Growth of S940 was indistinguishable from the parental strain in rich medium, confirming that the growth defects of the gene replacement strain S40 in rich medium were not due to the ehbF mutation (Fig. 3C). Presumably, they resulted from overexpression of the remainder of the ehb gene cluster in S40. Moreover, S940 growth in minimal medium was severely impaired, which confirmed the importance of EhbF in Ehb function and carbon assimilation. Small inocula were utilized in these experiments to eliminate the possibility that the long lags were due to selection for low levels of second-site revertants (see above). Complementation of ΔehbF with pMEV2+ehbF in strain S943 restored the ability to grow in minimal medium. Therefore, the poor growth of S940 in minimal medium was due to ΔehbF and not a secondary mutation.
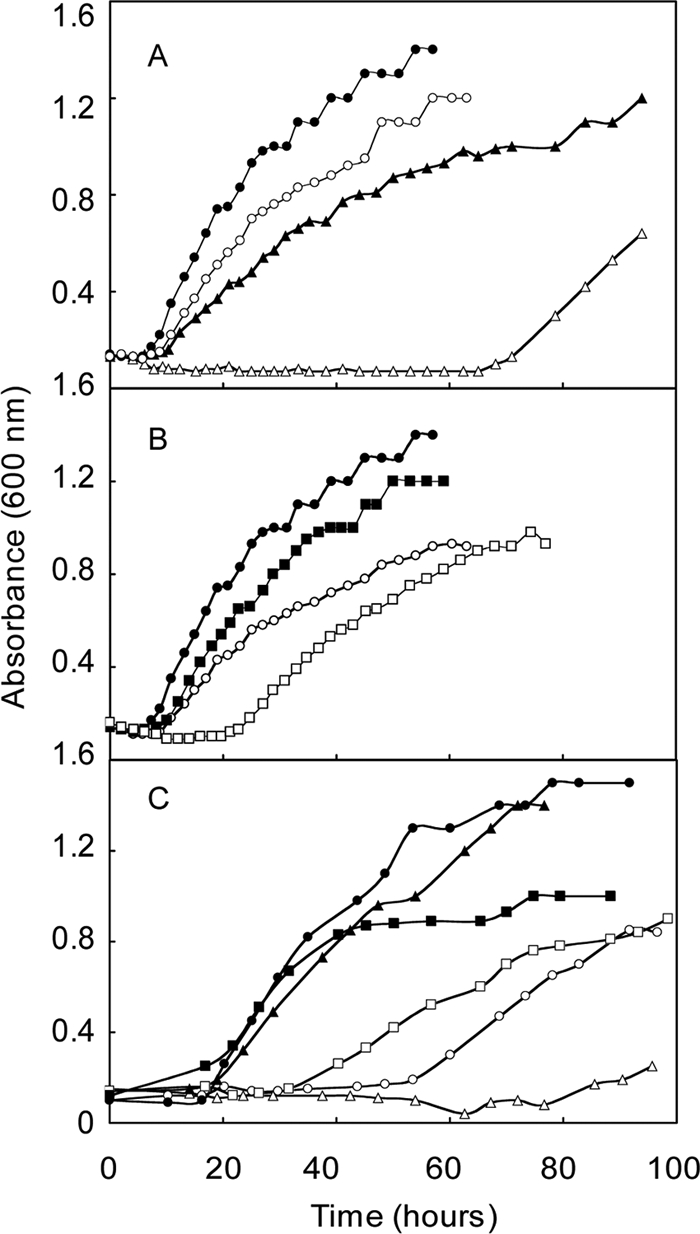
In-frame deletions were constructed for additional ehb genes. Phylogenetic analyses predicted that the large and small subunits of the hydrogenase component, EhbN and EhbM, were encoded by mmp1153 and mmp1622, respectively. Since EhbN was not encoded within the ehb gene cluster, it was of special interest to confirm its role in Ehb. Mutants with deletions in either ehbM or ehbN grew poorly in minimal medium (Fig. 4). The growth of the ΔehbN mutant was comparable to that of ΔehbF, suggesting that both proteins played major roles in Ehb function and confirming the annotation of this ORF. In contrast, growth of the ΔehbM mutant (S960) was less severely impaired. It was unclear why this mutation caused a less severe phenotype because the small hydrogenase subunit is also essential for hydrogenase activity. In addition, the growth phenotypes of the complementation strains for both mutants were complex. Although growth in minimal medium always improved, it never equaled that of the wild-type strain, and growth in rich medium was also impaired.
FIG. 4.
Growth of deletion mutants of the small hydrogenase subunit (ΔehbM) and the large hydrogenase subunit (ΔehbN) in rich (McCA, closed symbols) and minimal (McA, open symbols) media. (A) Growth of the ΔehbM strain S960 (squares), strain S963 containing the ΔehbM mutation complemented with pMEV2+ehbM (triangles), and the parental strain S900 (circles). (B) Growth of the ΔehbN strain S965 (squares), strain S968 containing the ΔehbN mutation complemented with pMEV2+ehbN (triangles), and the parental strain S900 (circles). For other experimental conditions, see the legend to Fig. 3.
In the Methanosarcina barkeri Ech, 4Fe-4S clusters play central roles in electron transfer and, possibly, proton translocation (4). In M. maripaludis, EhbK and EhbL contain the 4Fe-4S motifs CX2CX2CX3C and are predicted to be a polyferredoxin and ferredoxin, respectively. To determine the importance of EhbL in Ehb function, the deletion mutant ΔehbL (S955) was generated. In minimal medium, the growth of this mutant was inhibited but not as severely as the ΔehbF and ΔehbN mutants (Fig. 5). Upon complementation of the mutation with the expression vector for ehbL (strain S945), growth was actually better than the wild type in minimal medium. Similarly, in rich medium the complemented strain possessed a shorter growth lag but longer generation time. Thus, expression of EhbL from the strong promoter in the pMEV vector had complex consequences under both growth conditions.
FIG. 5.
Growth phenotypes of deletion mutants of the ferredoxin (ΔehbL) and polyferredoxin-ferredoxin (ΔehbKL) in rich (McCA, closed symbols) and minimal (McA, open symbols) media. (A) Growth of the ΔehbL strain S955 (squares), strain S958 containing the ΔehbL mutation complemented with pMEV2+ehbL (triangles), and the parental strain S900 (circles). (B) Growth of the ΔehbKL strain S945 (squares), strain S948 containing the ΔehbKL mutation complemented with pMEV2+ehbL (triangles), and the parental strain S900 (circles). For other experimental conditions, see the legend to Fig. 3.
Although the double-mutant ΔehbKL (S945) was isolated, an ΔehbK mutant was not isolated after numerous attempts. Growth of the ΔehbKL mutant in minimal medium was intermediate between that of the wild type and ΔehbF and ΔehbN mutants and, in that regard, resembled that of the ΔehbL mutant. Complementation of the ΔehbKL with the ehbL gene restored growth to wild-type levels in minimal medium but did not yield the growth stimulation observed in the ΔehbL background. In rich medium, growth was somewhat impaired but not to the same extent as in the ΔehbL background (Fig. 5). These results suggest that EhbK is necessary for proper Ehb function even with EhbL overexpression.
The complex phenotypes of the ΔehbL and ΔehbKL mutants and the ehbL overexpression strains suggest complex interactions for EhbL and EhbK in the cell. Although sufficient information for definitive conclusions is lacking, this circumstantial evidence suggests that EhbL and EhbK interact. The effects of the overexpression of EhbL were greatly attenuated in the double-deletion mutant ΔehbKL, suggesting that EhbK mitigates the effects of EhbL. Second, ΔehbK mutants were only isolated in the presence of a ΔehbL mutation. If the ΔehbK mutation is in fact lethal in the presence of normal levels of EhbL, as these results seem to suggest, it could be evidence for functional coupling where EhbL becomes toxic in the absence of EhbK. Although it is premature to speculate on the details, interactions of EhbK and EhbL, either with the Eha homologs or with other enzyme systems, could be responsible for these complex phenotypes. In conclusion, although EhbL and EhbK both appear to be important in Ehb function, it is not possible to assign their roles without additional experimentation.
Utilization of aryl acids by S965.
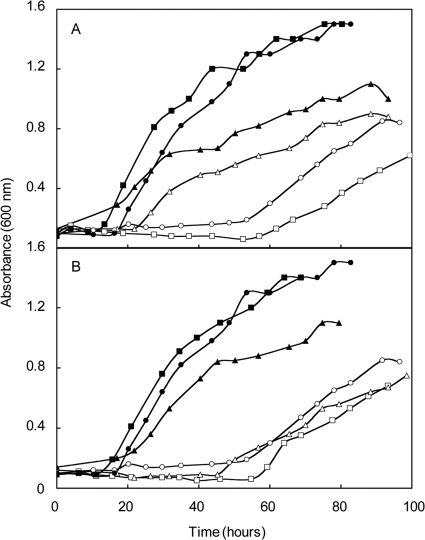
In the functional model of Porat et al. (15), Ehb was proposed to generate the low potential electron donor to indolepyruvate oxidoreductase (Ior), which is required for one of the two pathways of aromatic amino acid biosynthesis in M. maripaludis (17, 18). The deletion mutant ΔiorA2 was unable to utilize the aryl acids phenylacetate, indoleacetate, and p-hydroxyphenylacetate to make the aromatic amino acids. In addition, the aryl acids also inhibited growth of the mutant, presumably because they downregulated the expression of the de novo aromatic amino acid biosynthetic pathway (18). If Ehb produces the electron donor for Ior, an ehb mutant should behave like an ior mutant and aryl acids should be inhibitory. To test this hypothesis, the ΔehbN strain S965 was cultured in the presence of aryl acids and aromatic amino acids (Fig. 6). In minimal medium, aryl acids completely inhibited growth of the ΔehbN mutant. In contrast, aryl acids did not inhibit the growth of either S2 or S900 under these conditions (Fig. 6 and data not shown). The further addition of the aromatic amino acids largely restored growth of the mutant, indicating that inhibition was due to an effect of aromatic acid biosynthesis. These results support the hypothesis that ehb mutants are unable to use the Ior pathway of aromatic amino acid biosynthesis and that Ehb is the electron donor to this pathway.
FIG. 6.
Requirement for Ehb during conversion of the aryl acids to amino acids. The growth of wild-type strain S2 (A) and the ΔehbN mutant S965 (B) was evaluated. Media included complex McCA medium (•), McA medium containing aryl acids (⧫, 1 mM phenylacetate, indoleacetate, and p-hydroxyphenylacetate), and aromatic amino acids (▴, 1 mM tyrosine, phenylalanine, and tryptophan), both amino and aryl acids (▪), or no additions (○). For other experimental conditions, see the legend to Fig. 3.
Analyses of the proteome of the S40 strain had suggested that Vor was upregulated and that Ehb was involved in branched-chain amino acid biosynthesis (16). Therefore, a similar growth experiment using branched-chain fatty acids was performed. However, a difference between growth in the presence of the fatty acids or amino acids was not detected (data not shown). Presumably, branched-chain fatty acids do not inhibit the de novo biosynthetic pathway in the same manner as the aryl acids.
Structural characterization of Ehb.
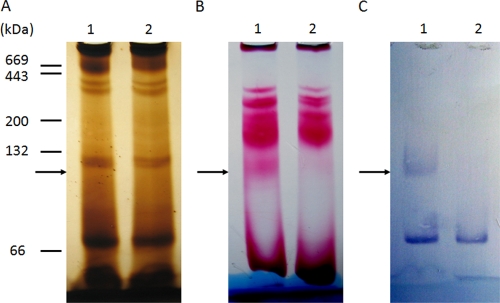
Originally identified by homology to other membrane-bound hydrogenases and genetic screens (26, 30), the physical structure of Ehb has not been characterized. Purified membranes of M. maripaludis contained high levels of hydrogenase activity, but this activity was not diminished in S40, the ehbF::pac mutant (16). Although there are a number of possible explanations, one likely possibility is that Ehb represented only a small fraction of the total hydrogenase activity. To test this possibility, whole protein extracts of M. maripaludis were fractionated by Blue Native PAGE (Fig. 7). EhbN, which encodes the large subunit of the hydrogenase, was identified in wild-type cells by its cross-reactivity with anti-EhbN antibodies (Fig. 7C). Hydrogenases were identified by activity staining, and a minor band of activity had the same electrophoretic mobility as EhbN from wild-type but not ΔehbN mutant cells (Fig. 7B). Moreover, staining with silver or Coomassie blue failed to reveal the protein complex even at the fairly large amounts of protein necessary for the activity staining and Western blotting (Fig. 7A and data not shown). Therefore, Ehb was not an abundant protein complex and represented only a minor component of the hydrogenase activity. This result was consistent with an anabolic role for Ehb in these hydrogenotrophs.
FIG. 7.
Hydrogenase activity comigrates with Ehb. A 7% Blue Native PAGE gel was prepared anaerobically and loaded with triplicate cell extracts of the wild-type S2 and the ΔehbN mutant S965 (60 μg of protein in each lane). Lane 1, S2; lane 2, S965. Standard proteins (5 μg of each), including albumin from bovine serum (66 kDa, monomer; 132 kDa, dimer), β-amylase (200 kDa), and apoferritin (443 kDa), were used as molecular mass markers. After electrophoresis, the gel was sliced into three pieces for silver staining (A), in-gel hydrogenase activity staining (B), and Western blotting with antibody targeting EhbN (C). The position of the Ehb complex (marked by arrows) is indicated by the band detected by Western blotting in lane 1 of panel C.
In Blue Native PAGE, the native Ehb complex migrated with an apparent molecular mass of ∼110 kDa (Fig. 7). However, upon summing the masses of all of the subunits listed in Table 1, a molecular mass of ∼360 kDa was expected. Because the migration in Blue Native PAGE is sensitive to the isoelectric point of the proteins in addition to charge (20), the apparent low molecular weight may be misleading. To test whether EhbF, EhbK, EhbL, and EhbM were part of the complex, the formation of the Ehb complex was examined in the ehb mutants. On sodium dodecyl sulfate (SDS) gels, two proteins were readily detected by Western blotting of extracts of wild-type cells with anti-EhbN antibodies (Fig. 8). However, only the lower-molecular-weight protein, whose size was identical to that expected for EhbN, was absent in extracts of the ΔehbN mutant. Therefore, this protein was identified as the authentic EhbN subunit, and the higher-molecular-weight band was a cross-reacting protein. Presumably, this protein was not observed in Western blotting of the Blue Native PAGE because its native form did not cross-react with the antibodies. In any case, ehbN is the only gene in M. maripaludis with sufficient sequence similarity to the MAP peptide to be detected by BLAST searches. Therefore, it is unlikely that the cross-reacting material is the subunit of another hydrogenase. EhbN was readily detected in cell extracts of all of the other ehb mutants. Thus, the levels of EhbN were not affected, even though the growth was severely impaired in some cases. However, only the wild-type cells contained the Ehb membrane complex identified by Blue Native PAGE (Fig. 8B). Therefore, either the complex failed to form or was unstable during electrophoresis in these mutants.
FIG. 8.
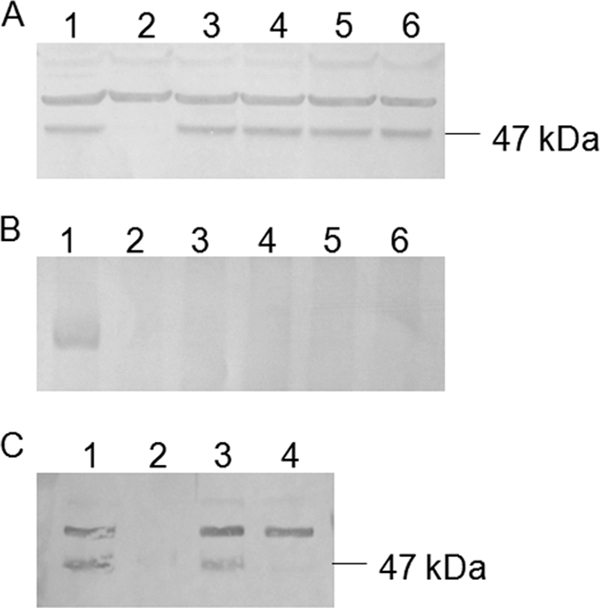
Formation of the Ehb membrane complex in ehb mutants. Whole-cell extract samples were separated in an SDS-12% PAGE gel (20 μg of protein each lane) (A) and 7% Blue Native PAGE gela (B) (120 μg of protein each lane). Lane 1, S2 (WT); lane 2, S965 (ΔehbN); lane 3, S940 (ΔehbF); lane 4, S955 (ΔehbL); lane 5, S945 (ΔehbKL); lane 6, S960 (ΔehbM). (C) Whole-cell extract, soluble fraction, and membrane fraction samples were separated in SDS-12.5% PAGE gel (20 μg of protein each lane). Lane 1, whole-cell extract of S2 (WT); lane 2, soluble fraction of S2 (WT); lane 3, membrane fraction of S2 (WT); lane 4, whole-cell extract of S965 (ΔehbN). Proteins were then transferred to PVDF membranes. EhbN and protein complexes containing EhbN were subsequently detected with rabbit polyclonal antiserum and a secondary goat anti-rabbit alkaline phosphatase conjugate. The position of a 47-kDa marker protein in panels A and C is given on the right.
The presence of Ehb in membranes was also determined by Western blotting of the membrane fraction with anti-EhbN antibodies. EhbN was pelleted by centrifugation at 200,000 × g for 1 h, and only ca. 3% of the initial cross-reactive material remained in the soluble fraction of cell extracts (Fig. 8C). Upon centrifugation of the membrane pellet in sucrose gradients, cross-reactive material was recovered in the membrane fraction.
These results provide direct evidence of the formation of a multisubunit Ehb membrane complex. However, they do not distinguish between the possibilities that the Ehb proteins either participate as subunits or are required for biosynthesis or assembly of the complex. For instance, EhbK and EhbL might be required for insertion of key Fe-S clusters into an apoenzyme, which would be unable to form a stable complex in their absence. If this were true, they might perform a similar role in other enzyme systems, which could explain the complex phenotype of their deletion mutants. If these proteins are components of the complex, the molecular mass would be expected to exceed ∼360 kDa, the minimum size of a complex comprised of one of each of these subunits.
It is unclear how a Δehb mutant grows autotrophically if the Ehb is required to produce low potential electrons for several oxidoreductases. Because these organisms possess a limited capacity to assimilate organic substrates such as pyruvate, these mutations might be expected to be lethal (9, 16, 31). If mutations in the large subunit of hydrogenase or the ion translocator completely inactivate Ehb as expected, then another enzyme system or pathway must compensate. Possibly, Eha possesses low levels of Ehb functionality. Alternatively, Eha subunits might function in heterologous complexes with Ehb. For instance, the intermediate phenotype of the ΔehbM mutation could be explained if the small subunit of the Eha hydrogenase was capable of forming an active enzyme with EhbN. This hybrid complex might have low Ehb activity and be unstable during electrophoresis.
A variety of energy-conserving hydrogenases have been described in both bacteria and archaea, such as Escherichia coli (Hyc and Hyf), the sulfate-reducing bacterium Desulfovibrio gigas (Ech and Coo), the phototroph Rhodospirillum rubrum (Coo), the hyperthermophilic archaeon Pyrococcus furiosus (Mbh and Mbx), the aceticlastic methanoarchaeon Methanosarcina barkeri (Ech), and the hydrogenotrophic methanoarchaeaon Methanococcus maripaludis (Eha and Ehb). The functions of these enzymes are diverse: the oxidation of formate or carbon monoxide in bacteria, the oxidation of glyceraldehyde 3-phosphate in Pyrococcus, or the oxidation of acetate and the reduction of carbon dioxide in methanogenesis in Methanosarcina. To group enzymes with such diverse functions would suggest that there is a great deal of structural or sequence similarity. However, these enzyme systems vary widely in the number of genes (from 21 in the Methanococcus eha to 6 in the Desulfovibrio ech) and the percent sequence similarity between various protein homologs. Many energy-conserving hydrogenases also possess subunits for which there are no homologs in the other systems. Furthermore, the hydrogenotrophic methanogens have developed a unique form of energy-conserving hydrogenase that couples energy transduction to autotrophic anabolism. The primary function of the other energy conserving hydrogenases appears to be energy transduction via catabolism. From this general synopsis, it seems that the energy-conserving hydrogenases are loosely grouped. As more energy-conserving hydrogenases are identified and characterized, understanding of the genetic, structural, and functional diversity of this group of enzymes is sure to increase.
Acknowledgments
This study was supported by grant DE-FG02-05ER15709 from the U.S. Department of Energy.
Footnotes
Published ahead of print on 28 May 2010.
REFERENCES
- 1.Adams, M. W. W., and D. O. Hall. 1979. Purification of the membrane-bound hydrogenase of Escherichia coli. Biochem. J. 183:11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodersen, J., G. Gottschalk, and U. Deppenmeier. 1999. Membrane-bound F420H2-dependent heterodisulfide reduction in Methanococcus voltae. Arch. Microbiol. 171:115-121. [DOI] [PubMed] [Google Scholar]
- 3.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, J. F. Weidman, J. L. Fuhrmann, D. Nguyen, T. R. Utterback, J. M. Kelley, J. D. Peterson, P. W. Sadow, M. C. Hanna, M. D. Cotton, K. M. Roberts, M. A. Hurst, B. P. Kaine, M. Borodovsky, H.-P. Klenk, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 4.Forzi, L., J. Koch, A. M. Guss, C. G. Radosevich, W. W. Metcalf, and R. Hedderich. 2005. Assignment of the [4Fe-4S] clusters of Ech hydrogenase from Methanosarcina barkeri to individual subunits via the characterization of site-directed mutants. FEBS J. 272:4741-4753. [DOI] [PubMed] [Google Scholar]
- 5.Hendrickson, E. L., R. Kaul, Y. Zhou, D. Bovee, P. Chapman, J. Chung, E. Conway de Macario, J. A. Dodsworth, W. Gillett, D. E. Graham, M. Hackett, A. K. Haydock, A. Kang, M. L. Land, R. Levy, T. J. Lie, T. A. Major, B. C. Moore, I. Porat, A. Palmeiri, G. Rouse, C. Saenphimmachak, D. Söll, S. Van Dien, T. Wang, W. B. Whitman, Q. Xia, Y. Zhang, F. W. Larimer, M. V. Olson, and J. A. Leigh. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186:6956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones, W. J., M. J. B. Paynter, and R. Gupta. 1983. Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch. Microbiol. 135:91-97. [Google Scholar]
- 7.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 8.Künkel, A., J. A. Vorholt, R. K. Thauer, and R. Hedderich. 1998. An Escherichia coli hydrogenase-3-type hydrogenase in methanogenic archaea. Eur. J. Biochem. 252:467-476. [DOI] [PubMed] [Google Scholar]
- 9.Lin, W. C., and W. B. Whitman. 2004. The importance of porE and porF in the anabolic pyruvate oxidoreductase of Methanococcus maripaludis. Arch. Microbiol. 179:444-456. [DOI] [PubMed] [Google Scholar]
- 10.Major, T. A. 2006. The role of the energy conserving hydrogenase B in autotrophy and the characterization of sulfur metabolism in Methanococcus maripaludis. Ph.D. dissertation. University of Georgia, Athens.
- 11.Meuer, J., S. Bartoschek, J. Koch, A. Künkel, and R. Hedderich. 1999. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 265:325-335. [DOI] [PubMed] [Google Scholar]
- 12.Meuer, J., H. C. Kuettner, J. K. Zhang, R. Hedderich, and W. W. Metcalf. 2002. Genetic analysis of the archaeon Methansarcina barkeri fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. U. S. A. 99:5632-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore, B. C., and J. A. Leigh. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187:972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesterenko, M. V., M. Tilley, and S. J. Upton. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28:239-242. [DOI] [PubMed] [Google Scholar]
- 15.Nijtmans, L. G., S. M. Artal, L. A. Grivell, and P. J. Coates. 2002. Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods 26:327-334. [DOI] [PubMed] [Google Scholar]
- 16.Porat, I., W. Kim, E. L. Hendrickson, Q. Xia, Y. Zhang, T. Wang, F. Taub, B. C. Moore, I. J. Anderson, M. Hackett, J. A. Leigh, and W. B. Whitman. 2006. Disruption of the operon encoding Ehb hydrogenase limits anabolic CO2 assimilation in the archaeon Methanococcus maripaludis. J. Bacteriol. 188:1373-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porat, I., M. Sieprawska-Lupa, Q. Teng, F. J. Bohanon, R. H. White, and W. B. Whitman. 2006. Biochemical and genetic characterization of an early step in a novel pathway for the biosynthesis of aromatic amino acids and p-aminobenzoic acid in the archaeon Methanococcus maripaludis. Mol. Microbiol. 62:1117-1131. [DOI] [PubMed] [Google Scholar]
- 18.Porat, I., B. W. Waters, Q. Teng, and W. B. Whitman. 2004. Two biosynthetic pathways for aromatic amino acids in the archaeon Methanococcus maripaludis. J. Bacteriol. 186:4940-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schägger, H. 2001. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65:231-244. [DOI] [PubMed] [Google Scholar]
- 20.Schägger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 21.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 2:223-231. [DOI] [PubMed] [Google Scholar]
- 22.Setzke, E., R. Hedderich, S. Heiden, and R. K. Thauer. 1994. H2:heterodisulfide oxidoreductase complex from Methanobacterium thermautotrophicum: composition and properties. Eur. J. Biochem. 220:139-148. [DOI] [PubMed] [Google Scholar]
- 23.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stathopoulos, C., W. Kim, T. Li, I. Anderson, B. Deutsch, S. Palioura, W. Whitman, and D. Söll. 2001. Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis. Proc. Natl. Acad. Sci. U. S. A. 98:14292-14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stojanowic, A., G. J. Mander, E. C. Duin, and R. Hedderich. 2003. Physiological role of the F420-non-reducing hydrogenase (Mvh) from Methanothermobacter marburgensis. Arch. Microbiol. 180:194-203. [DOI] [PubMed] [Google Scholar]
- 26.Tersteegen, A., and R. Hedderich. 1999. Methanobacterium thermoautotrophicum encodes two multisubunit membrane-bound [NiFe] hydrogenases. Eur. J. Biochem. 264:930-943. [DOI] [PubMed] [Google Scholar]
- 27.Thauer, R. K., A.-K. Kaster, H. Seedorf, W. Buckel, and R. Hedderich. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579-590. [DOI] [PubMed] [Google Scholar]
- 28.Tumbula, D. L., R. A. Makula, and W. B. Whitman. 1994. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol. Lett. 121:309-314. [Google Scholar]
- 29.Whitman, W. B., J. S. Sheih, S. H. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterization of 22 mesophilic methanococci. Syst. Appl. Microbiol. 7:235-240. [Google Scholar]
- 30.Whitman, W. B., D. L. Tumbula, J.-P. Yu, and W. Kim. 1997. Development of genetic approaches for the methane-producing archaebacterium Methanococcus maripaludis. BioFactors 6:37-46. [DOI] [PubMed] [Google Scholar]
- 31.Yang, Y. L., J. Gluska, and W. B. Whitman. 2002. Intracellular pyruvate flux in the methane-producing archaeon Methanococcus maripaludis. Arch. Microbiol. 178:493-498. [DOI] [PubMed] [Google Scholar]