Abstract
The cutR gene was identified 314 bp upstream of the divergently oriented cutB1C1A1 operon encoding carbon monoxide (CO) dehydrogenase in Mycobacterium sp. strain JC1. Its deduced product was composed of 320 amino acid residues with a calculated molecular mass of 34.1 kDa and exhibits a basal sequence similarity to the regulatory proteins belonging to the LysR family. Using a cutR deletion mutant, it was demonstrated that CutR is required for the efficient utilization of CO by Mycobacterium sp. strain JC1 growing with CO as the sole source of carbon and energy. CutR served as a transcriptional activator for expression of the duplicated cutBCA operons (cutB1C1A1 and cutB2C2A2) and was involved in the induction of the cutBCA operons by CO. The cutBCA operons were also subjected to catabolite repression. An inverted repeat sequence (TGTGA-N6-TCACA) with a perfect match with the binding motif of cyclic AMP receptor protein was identified immediately upstream of and overlapping with the translational start codons of cutB1 and cutB2. This palindrome sequence was shown to be involved in catabolite repression of the cutBCA operons. The transcription start point of cutR was determined to be the nucleotide G located 36 bp upstream of the start codon of cutR. Expression of cutR was higher in Mycobacterium sp. strain JC1 grown with glucose than that grown with CO.
Carboxydobacteria are a group of bacteria that are able to grow on carbon monoxide (CO) as the sole source of carbon and energy under aerobic conditions (18, 24). Carboxydotrophic bacteria were identified in both Gram-negative and -positive bacteria (4, 15, 16, 18, 21, 24). We have previously shown that several mycobacteria, including Mycobacterium sp. strain JC1 and Mycobacterium tuberculosis H37Ra, were able to grow on CO as the sole source of carbon and energy (5, 27).
The key enzyme for the utilization of CO by carboxydobacteria is CO dehydrogenase (CO-DH), which catalyzes oxidation of CO to CO2, using H2O as the oxidant (18, 24). The electrons produced by the oxidation of CO are transferred to the respiratory electron transport chain to generate proton motive force, and CO2 is assimilated for the carbon source (24). The CO-DH has the quaternary structure of L2M2S2 with molybdopterin cytosine dinucleotide, flavin adenine dinucleotide (FAD), and two different [Fe-S] centers as cofactors (9, 15).
Mycobacterium sp. strain JC1, a nonpathogenic, fast-growing mycobacterium isolated in Korea, is a carboxydotrophic mycobacterium capable of growing aerobically on CO as the sole carbon and energy source (5, 37). This bacterium can also grow methylotrophically on methanol and heterotrophically using various organic compounds (32). The CO-DH of Mycobacterium sp. strain JC1 is similar in molecular weight, subunit structure, cofactor composition, and other properties to those of Gram-negative carboxydobacteria, but it differs in stability when exposed to oxygen (19). The CO-DH of this bacterium also has been shown to be immunologically unrelated with the enzymes of Gram-negative carboxydobacteria such as Oligotropha carboxidovorans and Pseudomonas carboxydohydrogena but related to other mycobacterial CO-DHs (19). It was demonstrated that the synthesis of heme oxygenase-1 is induced in mouse macrophages infected with M. tuberculosis, as well as at the site of infection in a mouse model (23). The resulting increase in the local concentration of CO (a by-product of heme oxygenase reaction) might affect negatively the growth of M. tuberculosis since CO inhibits heme-containing terminal oxidases of the respiratory electron transport chain. In this respect, the mycobacterial CO-DHs possibly serve as a CO defense system. Furthermore, the CO-DHs of Mycobacterium sp. strain JC1 and other mycobacteria possess the nitric oxide dehydrogenase (NO-DH) activity that may be involved in the protection of mycobacterial pathogens from nitrosative stress during infection, which is devoid in the Gram-negative carboxydobacterial CO-DHs (28). This unique property of mycobacterial CO-DHs supports the idea that the mycobacterial CO-DHs may have divergently evolved from the Gram-negative enzymes.
The genes encoding CO-DHs have been cloned from Pseudomonas thermocarboxydovorans, O. carboxidovorans, Hydrogenophaga pseudoflava, and Mycobacterium sp. strain JC1 and sequenced (4, 15, 34, 38). The open reading frames for the three subunits of CO-DH (either cutA, cutB, and cutC or coxL, coxM, and coxS for large, medium, and small subunits, respectively) form an operon in the transcriptional order of cutBCA (coxMSL). Two copies of the cutBCA operons encoding the structural genes of CO-DH occur in Mycobacterium sp. strain JC1 (38). The open reading frames encoding amino acid sequences similar to those of the CO-DH structural genes were also identified in the sequenced genomes of other mycobacterial species, which is in good agreement with the fact that the members of the genus Mycobacterium have an intrinsic ability of carboxydotrophic growth (20, 27, 38).
Although the CO-DHs were synthesized in Mycobacterium sp. strain JC1 and H. pseudoflava during growth on organic substrates even in the absence of CO, synthesis of the enzymes was further induced by CO (16, 31, 38). The regulatory molecule(s) and mechanism, by which the CO-DH genes are induced by CO or repressed in the presence of organic substrates, have not been reported yet. We first report cis- and trans-acting elements involved in the regulation of the cutBCA operons in Mycobacterium sp. strain JC1 and then discuss their implications in the regulation of the operons.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation conditions.
The bacterial strains and plasmids used here are listed in Table 1. Mycobacterium sp. strain JC1 DSM 3803 was grown at 37°C in standard mineral base (SMB) medium with a gas mixture of 30% (vol/vol) CO-70% (vol/vol) air (SMB-CO) or SMB supplemented with 0.2% (wt/vol) glucose (SMB-glucose) as described previously (18, 27). For the construction of a mutant, Middlebrook 7H9 medium (Becton Dickinson, Sparks, MD) supplemented with 0.2% (wt/vol) glucose (7H9-glucose) was used to cultivate Mycobacterium sp. strain JC1. Escherichia coli strains were cultivated at 37°C in Luria-Bertani medium (LB). Ampicillin (50 μg/ml for E. coli) and hygromycin (70 μg/ml for Mycobacterium sp. strain JC1 and 200 μg/ml for E. coli) were added to culture medium when required.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Mycobacterium sp. strain JC1 | Wild type (DSM 3803) | 5, 37 |
| Mycobacterium sp. strain ΔCutR | cutR deletion mutant derived from Mycobacterium sp. strain JC1 | This study |
| E. coli DH5α | supE44 lac16 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 Δthi relA1 | 14 |
| Plasmids | ||
| pKO | Hygr; sacB, suicide vector | 7 |
| pNBV1 | Hygr; 5.8-kb plasmid derived from p16R1 | 12 |
| pCV77 | Kmr; promoterless lacZ | MediImmune |
| pNC | Hygr; promoterless lacZ | 38 |
| pTS19 | pBluescript II SK(+) containing the 995-bp cutR and cutB1 of Mycobacterium sp. strain JC1 | 38 |
| pTS49 | pBluescript II SK(+) containing the 873-bp cutB2 of Mycobacterium sp. strain JC1 | 38 |
| pSJ1 | pBluescript II KS(+)::1,304-bp EcoRI-BamHI fragment containing cutR | This study |
| pSJ2 | pBluescript II KS(+)::1,039-bp EcoRI-BamHI fragment containing the ΔcutR mutation | This study |
| pSJ3 | pKO::1,051-bp HindIII-BamHI fragment containing the ΔcutR mutation | This study |
| pCUTR | pNBV1::1,316-bp HindIII-BamHI fragment containing cutR | This study |
| pCUTB1LACZ2 | pNC::1,192-bp ClaI-XbaI fragment containing the cutB1 and cutR intergenic region | 38 |
| pCUTB2LACZ2 | pNC::872-bp ClaI-XbaI fragment containing the cutB2 and orf1 intergenic region | 38 |
| pCUTB2up461 | pNC::461-bp upstream region of cutB2 | This study |
| pCUTB2up82 | pNC::82-bp upstream region of cutB2 | This study |
| pCUTB2up58 | pNC::58-bp upstream region of cutB2 | This study |
| pCUTB2CBSPM | The same construct as pCUTB2up461 except point mutations in the putative CRP-binding site upstream of cutB2 | This study |
| pBScutB2 | pBluescript II KS(+)::899-bp BamHI-ClaI fragment from pCUTB2LACZ2 | This study |
Hygr, hygromycin resistance; Kmr, kanamycin resistance.
DNA manipulation and electroporation techniques.
Standard protocols or manufacturer's instructions were followed for recombinant DNA manipulations (33). The introduction of plasmids into Mycobacterium sp. strain JC1 was carried out as described elsewhere (36).
Construction of a cutR mutant.
For the construction of a deletion mutation of cutR, a 1,304-bp DNA fragment containing the cutR gene was amplified from pTS19 by PCR using PFU Turbo DNA polymerase and the primers cutREcoRI+ (5′-CCCGGAATTCCTCGTCTGATCCGCGGT-3′) and cutRBamHI− (5′-GCTAGGATCCGTTGCCGCCGCGATGG-3′). The PCR product was digested with EcoRI and BamHI and cloned into pBluescript II KS(+) to give the plasmid pSJ1. pSJ1 was digested with PstI and self-ligated to delete the 256-bp internal PstI fragment within cutR, yielding pSJ2. Finally, a 1,051-bp HindIII and BamHI fragment from pSJ2 was cloned into the suicide vector pKO1, which was restricted with the same enzymes. The resulting plasmid pSJ3 was introduced into Mycobacterium sp. strain JC1 by electroporation. Heterogenotes of the strain JC1, generated by a single recombination event, were selected for their hygromycin resistance, and homogenotes were obtained from the heterogenotes after a second recombination for sucrose resistance on 7H9-glucose plates containing 10% (wt/vol) sucrose. The allelic exchange in the homogenotes that produced isogenic cutR deletion (ΔCutR) mutants was verified by PCR.
Construction of plasmids. (i) pCUTR.
A 1,051-bp BamHI/HindIII fragment containing the cutR gene was isolated from pSJ1 and cloned into the shuttle vector pNBV1 digested with BamHI and HindIII, resulting in pCUTR, which was used to complement the ΔCutR mutant (ΔCutR mt).
(ii) pCUTB2up461, pCUTB2up82, pCUTB2up58, and pCUTB2CBSPM.
pCUTB2up461, pCUTB2up82, and pCUTB2up58 are the lacZ transcriptional fusion plasmids that contain the 5′ portion of cutB2, as well as 461-, 82-, and 58-bp DNA sequences upstream of the cutB2 start codon, respectively. In order to construct these plasmids, the promoter regions of the corresponding lengths were amplified by PCR using the forward primers with an XbaI restriction site and the reverse primers with a ClaI restriction site. pCUTB2LACZ2 was used as the template for PCR. The PCR products were restricted with XbaI and ClaI and cloned into pNC, resulting in the plasmids pCUTB2up461, pCUTB2up82, and pCUTB2up58. To construct pCUTB2CBSPM, site-directed mutagenesis was performed using the plasmid pBScutB2 as the template and the QuikChange site-directed mutagenesis kit (Stratagene). pBScutB2 was constructed by cloning of a 899-bp BamHI/ClaI fragment from pCUTB2LACZ2 into pBluescript II KS(+). Synthetic oligonucleotides, 33 bases long and containing TA in place of AC in the middle of their sequences, were used to mutagenize the putative CRP-binding site located upstream of cutB2. Mutation was verified by DNA sequencing. A 0.85-kb XbaI/ClaI fragment from the mutated pBScutB2 was cloned into pNC, resulting in the plasmid pCUTB2CBSPM, which has the same construct as pCUTB2up461 except for the point mutations.
RNA isolation and primer extension.
RNA isolation and primer extension were performed as described previously (38). The primer PER30 (5′-TACGTGCATTGATATTCCGG-3′), which is complementary to the nucleotide position 88 to 107 downstream of the cutR start codon, was used for both primer extension and sequencing.
Enzyme assays and protein determination.
Cultures of Mycobacterium sp. strain JC1 were grown to an appropriate growth phase. Cells were harvested, resuspended in an appropriate buffer, and disrupted by passage through a French pressure cell twice. Cell-free crude extracts were obtained by centrifugation for 15 min at 20,000 × g.
CO-DH.
CO-DH activity was assayed spectrophotometrically by measuring the CO-dependent reduction of 2-(4-indophenyl)-3-(4-nitrophenyl)-2H-tetrazolium chloride (INT; ɛ496 = 17.981 mM−1 cm−1) by the method of Kraut et al. (22).
β-Galactosidase.
The β-galactosidase activity was assayed spectrophotometrically by determining the initial conversion rate of the substrate analog ONPG (o-nitrophenyl-β-d-galactopyranoside) to o-nitrophenol at 420 nm and 30°C for 1 min with a spectrophotometer (26).
Protein concentration was determined by the bicinchoninic acid protein assay (Pierce, Rockford, IL), with bovine serum albumin as a standard.
Activity staining of CO-DH.
Staining by activity of CO-DH was performed as described previously (17).
Immunoblotting analysis.
SDS-PAGE and Western blotting with antiserum to the CO-DH of Mycobacterium sp. strain JC1 (19) were performed as described elsewhere (25).
RESULTS
Identification of the cutR gene.
Two copies of the structural genes (cutB1C1A1 and cutB2C2A2) encoding CO-DH have been identified in Mycobacterium sp. strain JC1 (38). The cutB, cutC, and cutA genes code for the medium, small, and large subunits of CO-DH, respectively, and form a transcriptional unit. The open reading frame designated cutR was identified to be located 314 bp upstream of cutB1 with the transcriptional orientation opposite to the cutB1C1A1 operon. Its deduced protein product consists of 320 amino acid residues with a calculated molecular mass of 34.1 kDa and an isoelectric point of 9.72. A BLAST search using the deduced amino acid sequence of CutR showed the highest overall resemblances (73 to 74% identity and 82 to 84% similarity) to the CutR homologs from Mycobacterium bovis, Mycobacterium marinum, M. tuberculosis, Mycobacterium smegmatis, and Mycobacterium ulcerans. CutR is a member of the family of LysR-type transcriptional regulators (LTTRs). A plausible helix-turn-helix (HTH) motif, which represents the DNA-binding domain, occurs in the NH2-terminal part (amino acids 1 to 60) of CutR. The COOH-terminal part (amino acids 90 to 290) of CutR exhibited some homology to the substrate-binding domain of the LTTR (35).
Phenotypic features of a cutR mutant.
To investigate the function of the cutR gene in Mycobacterium sp. strain JC1, a mutant carrying a deletion within the cutR (ΔCutR) was constructed. The ΔCutR mutant (ΔCutRmt) grew heterotrophically with glucose or mixotrophically with glucose and CO at the approximately same rate as the wild-type (WT) strain JC1. However, under lithoautotrophic conditions where CO was supplied as the sole source of carbon and energy, the doubling time of the ΔCutRmt (58.7 h) was considerably longer than that of the WT strain (25.5 h). This result suggests that CutR is required for efficient utilization of CO by Mycobacterium sp. strain JC1.
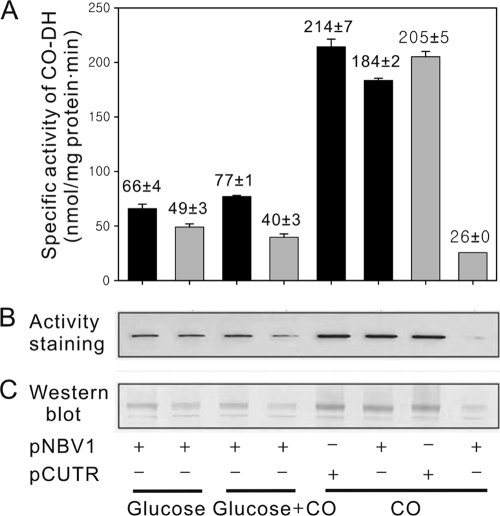
The possible role of CutR in the production of CO-DH was investigated by determining the activity and amount of CO-DH synthesized in the WT and ΔCutRmt strains grown on various conditions, since CO-DH is the key enzyme for CO utilization. As shown in Fig. 1, the WT strain harboring the empty vector pNBV1 (WT-pNBV1), which was grown lithoautotrophically with CO, showed a 2.8-fold increase in CO-DH activity, compared to WT-pNBV1 grown on glucose. When WT-pNBV1 was grown mixotrophically with glucose and CO, CO-DH activity detected in WT-pNBV1 was marginally increased relative to that in WT-pNBV1 grown on glucose. The levels of the CO-DH protein determined by activity staining and immunoblotting were well correlated with CO-DH activity detected in the strain grown under the corresponding growth conditions. This result indicates that the synthesis of CO-DH is induced in the presence of CO and repressed by glucose. The ΔCutRmt carrying pNBV1 (ΔCutRmt-pNBV1) grown heterotrophically or mixotrophically exhibited lower CO-DH activities than WT-pNBV1 grown under the same growth conditions. It is especially noteworthy that the synthesis of CO-DH was not induced in the ΔCutRmt-pNBV1 grown lithoautotrophically. Introduction of the plasmid-borne cutR gene into the ΔCutRmt (ΔCutRmt-pCUTR) led to restoration of CO-DH activity to the level detected in WT-pCUTR, indicating that incapability of the ΔCutRmt in the induction of CO-DH synthesis by CO is caused by the inactivation of the cutR gene. Taken together, these results indicate that the CutR protein is involved in the induction of CO-DH synthesis in the presence of CO.
FIG. 1.
Synthesis of CO-DH in the WT and ΔCutRmt strains of Mycobacterium sp. strain JC1 grown on various growth substrates. The strains were grown to an OD600 of 0.7 in SMB medium supplemented with 0.2% (wt/vol) glucose (glucose), 0.2% (wt/vol) glucose plus 30% (vol/vol) CO (glucose + CO), or 30% (vol/vol) CO (CO). The ΔCutRmt with pNBV1 grew with CO as the sole source of carbon so slowly that the mutant was grown to an OD600 of 0.3. (A) CO-DH activities. The specific activity indicates nanomoles of INT reduced per milligram protein per minute. The black and gray bars indicate the enzyme activities detected in the WT and ΔCutRmt strains, respectively. The error bars indicate the standard deviation of the values obtained from two independent determinations. (B) Activity staining of CO-DH. Crude extracts (10 μg) were subjected to nondenaturing PAGE on 7.5% (wt/vol) acrylamide gel, and the gel was subsequently subjected to activity staining. (C) Western blot analysis with a polyclonal CO-DH antibody. Crude extracts (10 μg) were subjected to denaturing SDS-PAGE on 12.5% (wt/vol) acrylamide gel. The bands represent the large subunit of CO-DH. “+” and “-” indicate the presence or absence, respectively, of the corresponding plasmids in Mycobacterium sp. strain JC1 strains.
Transcriptional regulation of the cutBCA operons.
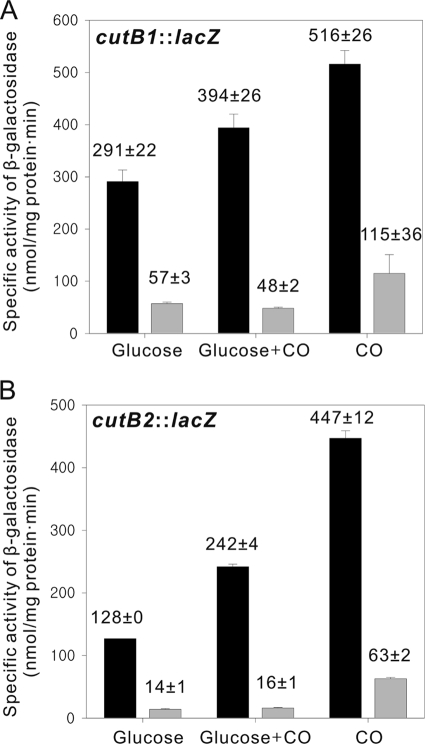
In order to elucidate the control of each of the cutBCA operons at the transcriptional level, the cutB1::lacZ and cutB2::lacZ transcriptional fusions in pCUTB1LACZ2 and pCUTB2LACZ2, respectively, were used to study the expression of the operons in WT and ΔCutRmt strains grown under various growth conditions (Fig. 2). The WT and ΔCutRmt strains harboring the empty vector pNC served as the negative controls, and virtually no β-galactosidase activity was detected in these strains grown heterotrophically, mixotrophically, and lithoautotrophically (data not shown). The considerable levels of the promoter activities of cutB1 and cutB2 were detected in the WT strain grown heterotrophically with glucose even in the absence of CO. In agreement with increased activity and protein levels of CO-DH in the lithoautotrophically grown cells, the promoter activities of cutB1 and cutB2 in the WT strain grown lithoautotrophically with CO were increased by 1.8- and 3.5-fold, respectively, compared to those in the strain grown on glucose. When grown under mixotrophic conditions with glucose and CO, the WT strain showed the intermediate promoter activities of cutB1 and cutB2 between those detected in WT grown heterotrophically and lithoautotrophically, indicating that the expression of cutB1 and cutB2 is induced in the presence of CO and repressed by glucose. In the ΔCutRmt the promoter activities of both cutB1 and cutB2 were significantly lower than those observed for WT grown under the corresponding growth conditions. Interestingly, the ΔCutRmt strain grown mixotrophically showed the induction of neither cutB1 nor cutB2, compared to the ΔCutRmt grown heterotrophically. However, induction of both cutB1 and cutB2 was observed in the ΔCutRmt grown lithoautotrophically with CO, when their promoter activities were compared to those in the mutant grown heterotrophically. All of the results presented in Fig. 2 suggest the following: (i) CutR serves as an activator for expression of both the cutBCA operons; (ii) CutR is involved in the induction of the cutBCA operons by CO; and (iii) in addition to CutR, a regulatory pathway independent of CutR is involved in the induction of the cutBCA operons in the strain grown lithoautotrophically with CO.
FIG. 2.
Expression of the cutBCA operons in the WT and the ΔCutRmt strains of Mycobacterium sp. strain JC1 grown on various growth substrates. For the determination of the promoter activities for the cutB1 (A) and cutB2 (B) genes, the strains harboring pCUTB1LACZ2 and pCUTB2LACZ2, respectively, were grown as described in Fig. 1, and the β-galactosidase activities were determined. The black and gray bars indicate the enzyme activities detected in the WT and ΔCutRmt strains, respectively. All values provided are the means of two independent determinations. Error bars represent the standard deviations from the mean.
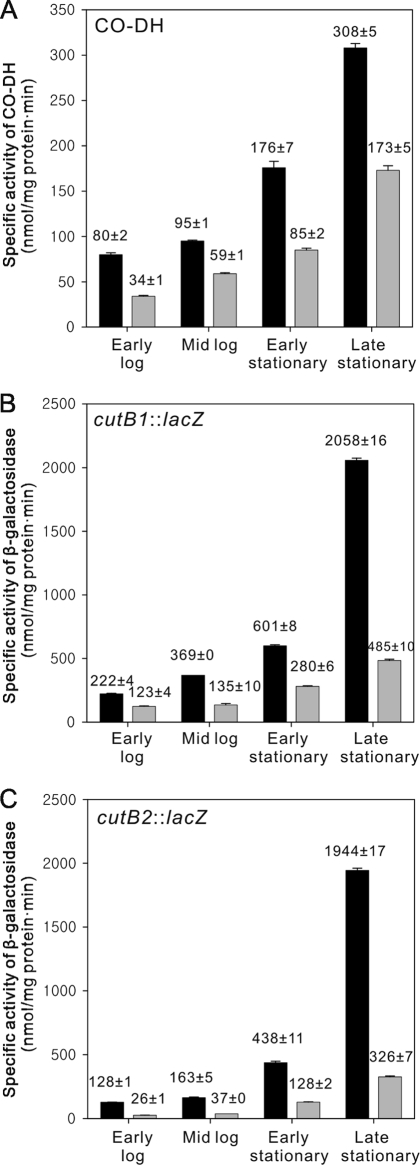
As shown in Fig. 3 A, CO-DH activity was increased as the growth phase proceeded, when the WT and ΔCutRmt strains were grown heterotrophically with glucose. However, the activity detected in the ΔCutRmt was lower than that in the WT strain. The highest activities of CO-DH in both strains were observed in the late stationary phase. In good agreement with this observation, the promoter activities of cutB1 and cutB2 in the WT and ΔCutRmt strains grown on glucose were increased in a growth-dependent manner, although the expression of both genes was significantly impaired in the ΔCutRmt strain (Fig. 3B and C). This result implies that the induction of the cutBCA operons in the stationary phase in the absence of CO is mediated by a regulatory system other than CutR and that CutR is required for optimal expression of the cutBCA operons.
FIG. 3.
Effect of the growth phase on the expression of the cutBCA operons in the WT and the ΔCutRmt strains of Mycobacterium sp. strain JC1. The WT (black bar) and ΔCutRmt (gray bar) strains were grown to an OD600 of 0.3 (early log), 0.6 (mid log), and 2.3 (early stationary) in SMB medium supplemented with 0.2% (wt/vol) glucose. For the late stationary culture, cells which had reached the early stationary phase, were further grown for additional 50 h. (A) CO-DH activities; (B) promoter activities of cutB1; (C) promoter activities of cutB2. All values provided are the means of two independent determinations. Error bars represent the standard deviationd from the mean.
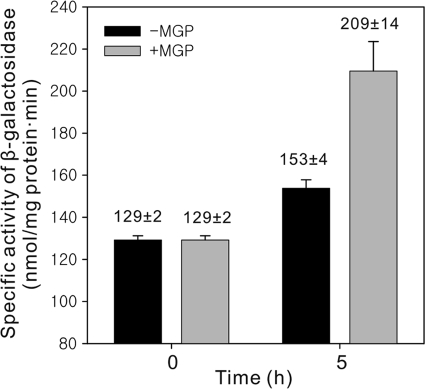
The induction of the cutBCA operons in cells growing in the stationary phase in the absence of CO can result from low level of oxygen or nutritionally poor environments in cultures. To ascertain whether the induction of the cutBCA operons was brought about in response to low oxygen tensions, we determined the CO-DH activity in the WT strain subjected to low aeration (for 15 h). The CO-DH activity was not increased in the strain exposed to hypoxic conditions compared to that detected in the WT strain grown aerobically (data not shown). However, when the WT strain grown on 0.2% (wt/vol) glucose was further grown on 0.002% (wt/vol) glucose for 5 h, the CO-DH activity was increased by 1.6-fold relative to that detected in the same strain grown on 0.2% (wt/vol) glucose for additional 5 h as a control (data not shown), indicating that the induction of CO-DH synthesis in the stationary phase in cells growing with glucose is related to glucose depletion in the growth medium. When the WT strain grown heterotrophically on 0.1% (wt/vol) glucose was treated with a 100-fold excess amount (10%) of methyl-α-d-glucopyranoside (MGP), which is a nonmetabolizable structural homolog of glucose, the promoter activity of cutB1 was 1.4-fold higher than that in the control group (Fig. 4), reinforcing the possibility that glucose starvation can lead to the induction of expression of the cutBCA operons.
FIG. 4.
Expression of the cutB1 gene in Mycobacterium sp. strain JC1 under glucose-limiting conditions. The WT strain carrying pCUTB1LACZ2 was grown in SMB medium supplemented with 0.1% (wt/vol) glucose to an OD600 of 0.5. The culture was divided into two groups. The control group (black bar) was grown for additional 5 h under the same conditions. In the case of the experimental group (gray bar), methyl-α-d-glucopyranoside (MGP), a nonmetabolizable glucose homolog, was added to the culture to a final concentration of 10% (wt/vol), and the culture was further grown for additional 5 h. The β-galactosidase activities in cells grown to an OD600 of 0.5 (0 h) and for 5 h thereafter (5 h) were measured to determine expression of cutB1. Error bars indicate the standard deviations of the results obtained from two independent determinations.
Determination of the transcriptional start point of cutR.
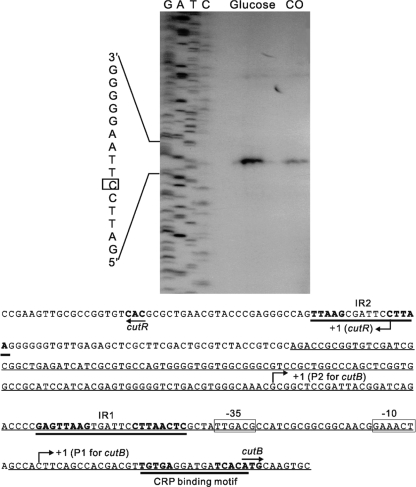
The transcriptional start point of the cutR gene was determined by primer extension analysis, which was performed with total RNA isolated from glucose- and CO-grown cells of Mycobacterium sp. strain JC1. As shown in Fig. 5, an extension product corresponding to the nucleotide G located 36 bp upstream of the start codon of cutR was detected. The extension analysis also indicated that expression of cutR was higher in cells grown under heterotrophic conditions with glucose than that grown under CO-lithoautotrophic conditions, which was confirmed by promoter assay using a cutR::lacZ transcriptional fusion. The expression level of cutR in cells grown under glucose conditions was 3.5 times as high as that grown under CO conditions (data not shown).
FIG. 5.
Mapping of the 5′ ends of the cutR gene transcripts by primer extension and the nucleotide sequence of the intergenic region between cutR and cutB1. The total RNA isolated from Mycobacterium sp. strain JC1 grown on CO or glucose was subjected to primer extension analysis. The boxed base (+1) indicates the location of the identified transcriptional start point. The positions of the identified transcriptional start sites are marked by “+1”. The putative −35 and −10 regions of CO-induced P1 are boxed. The translational starts (ATG and GTG in boldface) of cutB and cutR are indicated by the arrows with the gene names. Three inverted repeat sequences (IR1, IR2, and CRP site) are marked by thick underlining. The 233-bp upstream region of cutB1 is identical to that of cutB2, and the corresponding nucleotide sequence is underlined.
Identification of cis-control elements upstream of the cutBCA operons.
It has been demonstrated that the cutB genes have two transcriptional start points on the basis of two extension products identified by primer extension analysis (38). One (P1) is strongly induced under lithoautotrophic growth conditions with CO, and the other (P2) is constitutively expressed regardless of the presence or absence of CO (Fig. 5). Sequence regions resembling the mycobacterial −35 and −10 regions were identified upstream of P1. An inverted repeat sequence (IR1, GAGTTAAG-N6-CTTAACTC) is located immediately upstream of the putative −35 region. A similar inverted repeat sequence (IR2, TTAAG-N6-CTTAA) was also identified 274 bp upstream of the translational start of cutB1 but not cutB2. Since expression of both cutB1 and cutB2 are regulated in the identical manner, the presence of IR2 appears not to be necessary for the regulation of cutB1 expression by CO and glucose. There is another inverted repeat sequence (TGTGA-N6-TCACA) with a perfect match with the binding motif of cyclic AMP (cAMP) receptor protein (CRP), which is located immediately upstream of the translational starts of cutB1 and cutB2.
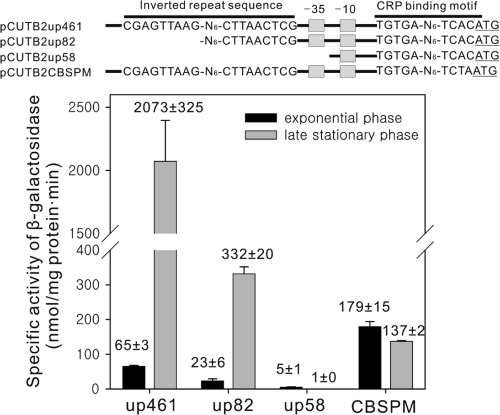
Using several cutB2::lacZ transcriptional fusions, we next investigated the role of the identified cis-acting regulatory elements (promoter elements, IR1, and CRP-binding site) in the expression of cutB2 and its induction in late stationary phase (Fig. 6). The cutB2 gene was chosen for the experiment since only one IR1 sequence occurs upstream of cutB2, in contrast to cutB1 whose upstream sequence contains both IR1 and IR2. As a positive control, the WT strain harboring pCUT2up461 (WT-pCUT2up461) was used, which contains 461 bp of the upstream sequence of cutB2. Expression of cutB2 in WT-pCUT2up461 was shown to be strongly induced, when it was grown to the late stationary phase. The transcriptional fusion plasmid pCUTB2up82 contains an 82-bp upstream sequence of cutB2 with the 5′-half portion of IR1 removed. Expression of cutB2 was induced in the WT strain with pCUTB2up82 (WT-pCUTB2up82) grown to the late stationary phase in comparison to that in WT-pCUTB2up82 grown to the exponential phase, although the expression levels of cutB2 in WT-pCUTB2up82 were significantly lower than those in WT-pCUTB2up461. Deletion of the putative −35 region (pCUTB2up58) completely abolished the expression of cutB2 in the WT strain grown to exponential or late stationary phase, indicating that the region between 58 and 82 bp upstream of cutB2 contains the promoter element(s). Interestingly, expression of cutB2 was not induced in late stationary phase when the CRP-binding site was point mutated (pCUTB2CBSPM). Furthermore, the WT strain with pCUTB2CBSPM (WT/pCUTB2CBSPM) grown to exponential phase showed increased levels of promoter activity compared to the WT-pCUT2up461 grown to the same growth phase. These results suggest that a regulatory transcription factor binding to the putative CRP binding site rather than to IR1 is responsible for the induction of the cutB genes in the late stationary phase and that the CRP-like transcription factor serves as an activator in the late stationary phase.
FIG. 6.
Promoter activities from the serially deleted and point-mutated promoters of cutB2 in Mycobacterium sp. strain JC1 grown on glucose. The strains harboring the corresponding transcriptional fusion plasmids were grown on SMB medium with 0.2% (wt/vol) glucose either to mid-exponential phase (black bar) or to late stationary phase (gray bar) as described in Fig. 3. pCUTB2up461, pCUTB2up82, and pCUTB2up58 contain the 461-, 82-, and 58-bp upstream sequences of cutB2, respectively. pCUTB2CBSPM has the same construct as pCUTB2up461 except that the CRP-binding sequence is point mutated. The β-galactosidase activities in cells were measured to determine expression of cutB2. Error bars indicate the standard deviations of the results obtained from two independent determinations.
DISCUSSION
CutR is a transcriptional activator for the induction of the cutBCA operons.
The cutR gene is located divergently to the cutB1C1A1 operon. The modular structure (HTH motif in the NH2-terminal domain and LysR substrate binding domain in the central and COOH-terminal domains) and the overall sequence similarity of CutR to those of known LTTRs suggest that CutR belongs to the LTTR family (35). Both cutBCA operons were not induced in the CutRmt grown mixotrophically with glucose and CO compared to the mutant grown heterotrophically with glucose, indicating that CutR is responsible for the induction of the cutBCA operons in the presence of CO. Furthermore, inactivation of CutR led to decreased expression of the cutBCA operons irrespective of the presence or absence of CO. Both observations allow us to suggest that CutR is an activator for the expression of the cutBCA operons even in the absence of CO and that it mediates the induction of the cutBCA operons by CO. Two very similar, 16-bp long sequence segments (IR1 and IR2) occur between cutB1 and cutR. They share the consensus sequence 5′-TTAAG(C/T)GATTCCTTAA-3′ and contain the pentamers (underlined) of inverted repeats on both ends. IR1 is located at positions −58 to −43 relative to the transcriptional start points of cutB1 and cutB2. IR2 is located far upstream of cutB1 and is not present upstream of cutB2. In the case of σ70-dependent promoters, transcriptional activators bind predominantly to positions between −80 and −30 relative to the transcriptional start points (29). Since both cutB1 and cutB2 are regulated in the same way in terms of their induction in the presence of CO and in the stationary phase, IR2 appears not to be required for the regulation of the cutBCA operons. In addition to the cutR genes, the conserved IR1 sequences (GAGTTAAG-N6-CTTAACTC) are present upstream of all mycobacterial cutBCA operons (Fig. 7). This finding strongly indicates that the IR1 and IR2 sequences might be the binding sites for the CutR activator, which is corroborated by the presence of a LysR motif (T-N11-12-A) within the IR1 and IR2 sequences (11). A binding assay of CutR to IR1 and IR2 is necessary to prove that the IR sequences serve as the CutR binding sites.
FIG. 7.
Inverted repeat sequences identified upstream of the cutB genes of various mycobacteria. The conserved inverted repeats are indicated in boldface. The consensus sequences of the conserved inverted sequences are given below the aligned sequences. The start codons of the cutB genes are underlined. Abbreviations: JC1, Mycobacterium sp. strain JC1; JLS, Mycobacterium sp. strain JLS; KMS, Mycobacterium sp. strain KMS; MCS, Mycobacterium sp. strain MCS; Msm, M. smegmatis; Mtb, M. tuberculosis H37Rv; BCG, M. bovis BCG; Mbo, M. bovis; Mma, M. marinum; Mul, M. ulcerans.
The cutBCA operons are subjected to catabolite repression.
The presence of glucose in the growth medium suppressed the expression of the cutBCA operons in the presence of CO. The fact that the excess amount of MGP relative to glucose derepressed the expression of cutB1 strongly suggests that the cutBCA operons are subjected to catabolite repression (8). We determined the promoter activity of cutB2 in the WT strain of Mycobacterium sp. strain JC1 grown to early log phase (optical density at 600 nm [OD600] of 0.3) in SMB medium containing either 0.2% glucose or 0.2% glycerol. The expression level of cutB2 in cells grown in glycerol-SMB medium was 2.18-fold higher than that detected in cells grown in glucose-SMB medium, confirming catabolite repression of the cutBCA operons by glucose (data not shown). The strong induction of the cutBCA operons in Mycobacterium sp. strain JC1 grown lithoautotrophically with CO appears to result from the combinatory effect of both CO induction by CutR and the abolishment of catabolite repression in the absence of glucose. Intriguingly, an inverted repeat sequence (TGTGA-N6-TCACA) with a perfect match to the binding motif of CRP (13) was found to be centered at the −7 position with respect to the start codons of both cutB1 and cutB2. The 3′-end portion of the putative CRP-binding site overlaps with the start codons of cutB1 and cutB2. The CRP-binding motifs also occur immediately upstream of the cutB gene homologs of fast-growing mycobacteria such as M. smegmatis and Mycobacterium sp. strains JLS, KMS, and MCS, whereas they are not found in the upstream control regions of the cutB homologs of M. tuberculosis, M. bovis, M. marinum, and M. ulcerans (Fig. 7). The mutation of TGTGA-N6-TCACA to TGTGA-N6-TCTAA completely abolished the induction of cutB2 in late stationary phase, indicating that this consensus CRP-binding site might be related to catabolite repression.
The genes for the CRP homologs, rv3676 and mb3700, were identified in the genomes of M. tuberculosis and M. bovis, respectively (6, 10). The Rv3676 protein was shown to bind to the consensus sequence of E. coli CRP-binding site (TGTGA-N6-TCACA) (2). cAMP enhanced the binding affinity of Rv3676 to its target DNA sequences and led to allosteric alterations in its conformation (2, 3). The Rv3676 binding motifs were identified in 73 promoter regions regulating 114 genes in the M. tuberculosis genome (2). It is noteworthy that 34 genes involved in nutrient starvation response have the Rv3676 binding motifs in their upstream control regions, implying that Rv3676 might be important in regulation of the starvation response. It has been also demonstrated that Rv3676 plays an important role in the proliferation of M. tuberculosis in macrophages and in mice (30). We recently identified and cloned the gene encoding the Rv3676 homolog in Mycobacterium sp. strain JC1 (J. H. Kim and Y. M. Kim, unpublished data). The nucleotide sequence of Rv3676 homolog of Mycobacterium sp. strain JC1 showed 96% identity to that of M. tuberculosis Rv3676. The binding of CRP homolog to the promoter region of the CO-DH genes was already predicted by computational analysis on the M. smegmatis genome (1). Although we present here the first indication that the Rv3676 homolog is involved in the regulation of the cutBCA operons in Mycobacterium sp. strain JC1, experimental validation with an Rv3676 homolog knockout mutant will be required to provide direct evidence.
Acknowledgments
This study was supported by a research grant from the Korea Science and Engineering Foundation (R01-2003-000-10007-0).
Footnotes
Published ahead of print on 28 May 2010.
REFERENCES
- 1.Akhter, Y., S. Yellaboina, A. Farhana, A. Ranjan, N. Ahmed, and S. E. Hasnain. 2008. Genome scale portrait of cAMP-receptor protein (CRP) regulons in mycobacteria points to their role in pathogenesis. Gene 407:148-158. [DOI] [PubMed] [Google Scholar]
- 2.Bai, G., L. A. McCue, and K. A. McDonough. 2005. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J. Bacteriol. 187:7795-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai, G., M. A. Gazdik, D. D. Schaak, and K. A. McDonough. 2007. The Mycobacterium bovis BCG cyclic AMP receptor-like protein is a functional DNA binding protein in vitro and in vivo, but its activity differs from that of its M. tuberculosis ortholog, Rv3676. Infect. Immunol. 75:5509-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, G. W., C. M. Lyons, E. Williams, J. Colby, M. Kehoe, and C. O'Reilly. 1990. Cloning and expression of the carbon monoxide dehydrogenase genes from Pseudomonas thermocarboxydovorans strain C2. FEMS Microbiol. Lett. 58:249-254. [DOI] [PubMed] [Google Scholar]
- 5.Cho, J. W., H. S. Yim, and Y. M. Kim. 1985. Acinetobacter isolate growing with carbon monoxide. Korean J. Microbiol. 23:1-8. [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.David, R. S., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. U. S. A. 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutscher, J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87-93. [DOI] [PubMed] [Google Scholar]
- 9.Dobbek, H., L. Gremer, O. Meyer, and R. Huber. 1999. Crystal structure and mechanism of CO dehydrogenase, a molybdo iron-sulfur flavoprotein containing S-selanylcysteine. Proc. Natl. Acad. Sci. U. S. A. 96:8884-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garnier, T., K. Eiglmeier, J.-C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. U. S. A. 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goethals, K., M. Van Montagu, and M. Holsters. 1992. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc. Natl. Acad. Sci. U. S. A. 89:1646-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard, N. S., J. E. Gomez, C. Ko, and W. R. Bishai. 1995. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166:181-182. [DOI] [PubMed] [Google Scholar]
- 13.Jansen, C., A. M. Gronenborn, and G. M. Clore. 1987. The binding of the cyclic AMP receptor protein to synthetic DNA sites containing permutations in the consensus sequence TGTGA. Biochem. J. 246:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jessee, J. 1986. New subcloning efficiency competent cells: >1 × 106 transformants/μg. Focus 8:9. [Google Scholar]
- 15.Kang, B. S., and Y. M. Kim. 1999. Cloning and molecular characterization of the genes for carbon monoxide dehydrogenase and localization of molybdopterin, flavin adenine dinucleotide, and iron-sulfur centers in the enzyme of Hydrogenophaga pseudoflava. J. Bacteriol. 181:5581-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiessling, M., and O. Meyer. 1982. Profitable oxidation of carbon monoxide and hydrogen during heterotrophic growth of Pseudomonas carboxydoflava. FEMS Microbiol. Lett. 13:333-338. [Google Scholar]
- 17.Kim, Y. M., and G. D. Hegeman. 1981. Purification and some properties of carbon monoxide dehydrogenase from Pseudomonas carboxydohydrogena. J. Bacteriol. 148:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, Y. M., and G. D. Hegeman. 1983. Oxidation of carbon monoxide by bacteria. Int. Rev. Cytol. 81:1-32. [DOI] [PubMed] [Google Scholar]
- 19.Kim, K. S., Y. T. Ro, and Y. M. Kim. 1989. Purification and some properties of carbon monoxide dehydrogenase from Acinetobacter sp. strain JC1 DSM 3803. J. Bacteriol. 171:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King, G. M. 2003. Uptake of carbon monoxide and hydrogen at environmentally relevant concentrations by mycobacteria. Appl. Environ. Microbiol. 69:7266-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King, G. M., and C. F. Weber. 2007. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5:107-118. [DOI] [PubMed] [Google Scholar]
- 22.Kraut, M., I. Hugendieck, S. Herwig, and O. Meyer. 1989. Homology and distribution of CO dehydrogenase structural genes in carboxydotrophic bacteria. Arch. Microbiol. 152:335-341. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, A., J. S. Deshane, D. K. Crossman, S. Bolisetty, B. Yan, I. Kramnik, A. Agarwal, and A. J. C. Steyn. 2008. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283:18032-18039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, O., and H. G. Schlegel. 1983. Biology of aerobic carbon monoxide-oxidizing bacteria. Annu. Rev. Microbiol. 37:277-310. [DOI] [PubMed] [Google Scholar]
- 25.Mouncey, N. J., and S. Kaplan. 1998. Redox-dependent gene regulation in Rhodobacter sphaeroides 2.4.1(T): effects on dimethyl sulfoxide reductase (dor) gene expression. J. Bacteriol. 180:5612-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh, J. I., and S. Kaplan. 1999. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 38:2688-2696. [DOI] [PubMed] [Google Scholar]
- 27.Park, S. W., E. H. Hwang, H. Park, J. A. Kim, J. Heo, K. H. Lee, T. Song, E. Kim, Y. T. Ro, S. W. Kim, and Y. M. Kim. 2003. Growth of mycobacteria on carbon monoxide and methanol. J. Bacteriol. 185:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, S. W., T. Song, S. Y. Kim, E. Kim, J. I. Oh, C. Y. Eom, and Y. M. Kim. 2007. Carbon monoxide dehydrogenase in mycobacteria possesses a nitric oxide dehydrogenase activity. Biochem. Biophys. Res. Commun. 362:449-453. [DOI] [PubMed] [Google Scholar]
- 29.Raibaud, O., and M. Schwarz. 1984. Positive control of transcription initiation in bacteria. Annu. Rev. Genet. 18:173-206. [DOI] [PubMed] [Google Scholar]
- 30.Rickman, L., C. Scott, D. M. Hunt, T. Hutchinson, M. C. Menendez, R. Whalan, J. Hinds, M. J. Colston, J. Green, and R. S. Buxton. 2005. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 56:1274-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ro, Y. T., I. K. Chung, J. Lee, D. Kim, J. W. Suh, S. W. Kim, and Y. M. Kim. 1995. Activity and expression of carbon monoxide dehydrogenase in Acinetobacter sp. strain JC1 growing with different growth substrates. Microorganisms Ind. 21:252-260. [Google Scholar]
- 32.Ro, Y. T., J. G. Seo, J. H. Lee, D. Y. Kim, I. K. Chung, T. U. Kim, and Y. M. Kim. 1997. Growth on methanol of a carboxydobacterium, Acinetobacter sp. strain JC1 DSM 3803. J. Microbiol. 35:30-39. [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Santiago, B., U. Schübel, C. Egelseer, and O. Meyer. 1999. Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene 236:115-124. [DOI] [PubMed] [Google Scholar]
- 35.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 36.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 37.Song, T., H. Lee, Y. H. Park, E. Kim, Y. T. Ro, S. W. Kim, and Y. M. Kim. 2002. Reclassification of a carboxydobacterium, Acinetobacter sp. strain JC1 DSM3803, as Mycobacterium sp. strain JC1 DSM 3803. J. Microbiol. 40:237-240. [Google Scholar]
- 38.Song, T., S. W. Park, S.-J. Park, J. H. Kim, J. Y. Yu, J.-I. Oh, and Y. M. Kim. 2010. Cloning and expression analysis of the duplicated genes for carbon monoxide dehydrogenase of Mycobacterium sp. strain JC1 DSM 3803. Microbiology 156:1009-1018. [DOI] [PubMed] [Google Scholar]