Abstract
The fatty acid synthase type II enzymatic complex of Mycobacterium tuberculosis (FAS-IIMt) catalyzes an essential metabolic pathway involved in the biosynthesis of major envelope lipids, mycolic acids. The partner proteins of this singular FAS-II system represent relevant targets for antituberculous drug design. Two heterodimers of the hydratase 2 protein family, HadAB and HadBC, were shown to be involved in the (3R)-hydroxyacyl-ACP dehydration (HAD) step of FAS-IIMt cycles. Recently, an additional member of this family, Rv0241c, was proposed to have the same function, based on the heterologous complementation of a HAD mutant of the yeast mitochondrial FAS-II system. In the present work, Rv0241c was able to complement a HAD mutant in the Escherichia coli model but not a dehydratase-isomerase deficient mutant. However, an enzymatic study of the purified protein demonstrated that Rv0241c possesses a broad chain length specificity for the substrate, unlike FAS-IIMt enzymes. Most importantly, Rv0241c exhibited a strict dependence on the coenzyme A (CoA) as opposed to AcpM, the natural acyl carrier protein bearing the chains elongated by FAS-IIMt. The deletion of Rv0241c showed that this gene is not essential to M. tuberculosis survival in vitro. The resulting mutant did not display any change in the mycolic acid profile. This demonstrates that Rv0241c is a trans-2-enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydratase that does not belong to FAS-IIMt. The relevance of a heterologous complementation strategy to identifying proteins of such a system is questioned.
In 2008, tuberculosis killed 1.8 million people, 30% of whom were human immunodeficiency virus positive. A total of 5% of all cases have multidrug-resistant tuberculosis that fails to respond to first-line drug therapy. Fighting this disease that affects mostly developing countries is one of the priorities of the World Health Organization (37). The development of new drugs effective against the resistant strains is particularly needed.
The tubercle bacillus, Mycobacterium tuberculosis, is protected by a highly impermeable envelope characterized by an outer membrane (14, 38) that holds unusual lipid components, mycolic acids. These very-long-chain α-alkylated β-hydroxylated fatty acids are the hallmark of the Corynebacterineae suborder. Each genus is typified by mycolic acids of different chain length. The simplest prototypes (C32 to C36), called “corynomycolic acids,” that result from an enzymatic condensation between two regular size fatty acids (C16 to C18) are found in Corynebacterium. The longest mycolates (C60 to C90) are biosynthesized by mycobacteria; they are the products of condensation between a very long meromycolic chain (C40 to C60) that has chemical functions at two main positions and a shorter α-chain (C20 to C24) (20). These molecules are essential to mycobacterial survival. Furthermore, they play a role in the virulence and persistence of M. tuberculosis within infected organisms (2, 8). The mycolic acid biosynthesis pathway represents a valuable source for potential new pharmacological targets (20).
Elongation cycles that include four main steps monitored by an acyl carrier protein (ACP)-dependent fatty acid synthase type II (FAS-II) system (3) lead to the formation of the meromycolic chain. FAS-II systems are found in plants, bacteria, parasites, and mitochondria (26). The dehydration step of FAS-II systems can be catalyzed by either a (3R)-hydroxyacyl-ACP dehydratase or a (3R)-hydroxyacyl-ACP dehydratase/isomerase that leads to the formation of unsaturated fatty acids due to its additional trans (E) Δ2-cis (Z) Δ3 isomerase activity (26). In E. coli, the corresponding enzymes are FabZ- and FabA-type enzymes, universally found in the known bacterial FAS-II systems (26). In contrast, the (3R)-hydroxyacyl-ACP dehydratase of yeast mitochondria is related to a different subfamily, the hydratases 2 (17). In both E. coli and yeast, the FAS-II system performs de novo fatty acid biosynthesis. The FAS-II system from Corynebacterineae is unique since it elongates long-chain fatty acids (C12 to C18) into very-long-chain fatty acids (3). In M. tuberculosis, we previously characterized two (3R)-hydroxyacyl-ACP dehydratases, HadAB and HadBC, involved in the third step of the cycle (28). These bacterial enzymes have the unique property of belonging to the hydratase 2 protein family. The latter also includes Rv0241c protein (5, 29), whose function is unknown and which has been annotated as a “conserved hypothetical protein” (6). Recently, however, it has been shown that Rv0241c gene was able to complement Δhtd2 (Δyhr067w) yeast mutant lacking a functional mitochondrial FAS-II dehydratase (9). Gurvitz et al. concluded that Rv0241c encodes a bona fide FAS-II-like 3-hydroxyacyl-thioester dehydratase (9).
In order to determine whether Rv0241c behaved as a 3-hydroxyacyl-thioester dehydratase or as a 3-hydroxyacyl-thioester dehydratase-isomerase, we achieved complementation experiments in another model, E. coli, which holds both types of enzyme (36). Only the function of dehydratase and not that of dehydratase-isomerase could be restored by Rv0241c in E. coli mutants. To investigate the effective involvement of the Rv0241c protein in the M. tuberculosis FAS-II (FAS-IIMt) system, we performed a detailed analysis of its substrate specificity with respect to the chain length and the nature of the acyl chain carrier using purified enzyme. Furthermore, the essentiality of the gene was studied by knocking it out in M. tuberculosis. The implication of Rv0241c in mycolic acid biosynthesis was examined through a phenotypic analysis of the deletion mutant.
MATERIALS AND METHODS
Sequence analyses.
Analyses of genome sequences were done via the TubercuList (6), National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/genomes/lproks.cgi), Wellcome Trust Sanger Institute (www.sanger.ac.uk/), and Comprehensive Microbial Resource (cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi) (24) web servers. Sequence alignments were performed by using BLAST algorithms (1) with default parameters.
Heterologous complementation assays.
The Rv0241c gene was amplified by PCR from the BAC-Rv329 DNA (Institut Pasteur, Paris, France [4]) with the oligonucleotides 5′-TAAGCCGTCTAGATGACTCAACCCAGCGGCCTGAAGA-3′ and 5′-GATAACTCTCGAGCGTGGCCCGGCTATAGACC-3′, using the Pfu polymerase (Promega, Madison, WI). The PCR product was then cloned into the pBluescript II SK(+) (pBSK+) expression vector (Stratagene/Agilent Technologies, La Jolla, CA) between the XbaI and XhoI restriction sites downstream of a lactose-inducible promoter. Competent bacteria of E. coli CY50 (fabA-thermosensitive [Ts]) strain (E. coli Genetic Stock Center, New Haven, CT) were transformed by heat shock in the presence of either pBSK+ vector, the pBSK::Rv0241c plasmid, or the pDM5 plasmid (pBR322::wild-type E. coli fabA [fabAEc], kindly provided by D. de Mendoza) and screened on LB agar supplemented with carbenicillin at 30°C. At least 10 single colonies of each recombinant strain were used in the complementation assays. Each colony was cultivated in four different conditions: on rich broth (RB) agar supplemented with 50 μg of carbenicillin/ml either in the presence or in the absence of sodium oleate (0.1% [vol/vol]) and at either 30°C (permissive temperature) or 42°C (non permissive temperature). Assays were performed in the presence or in the absence of lactose (0.5% [vol/vol]), a positive regulator of the lac promoter in pBSK+; similar data were obtained in both conditions. Competent bacteria of E. coli HW7 (DY330 fabZ::kan/pBAD24::fabZ Clostridium acetobutylicum) strain (kindly provided by H. Wang [35]) were transformed by heat shock in the presence of either pBSK+ vector, pBSK::Rv0241c, or pBSK::fabZEc and cultivated for 90 min in LB supplemented with 0.5% (vol/vol) l-arabinose at 30°C. After extensive washing with LB, a suspension of each recombinant strain was used to inoculate RB agar medium supplemented with 50 μg of carbenicillin/ml, 15 μg of kanamycin/ml, and either 0.5% (vol/vol) l-arabinose or 0.1% (vol/vol) d-fucose. The cultures were incubated at 30°C.
Protein production and purification.
The Rv0241c gene was amplified by PCR from the total DNA of M. tuberculosis H37Rv by using the primer pair 5′-GCAACGTCATTCGTGTCTG-3′ (forward) and 5′-GATAACGTCGGATGGCTGAC-3′ (reverse) and the high-fidelity polymerase PfuUltra (Stratagene/Agilent Technologies). In a second PCR, an N-terminal His6 tag was introduced by using an alternative forward primer, 5′-ATG GCT CAT CAT CAT CAT CAT CAT GGT ACT CAA CCC AGC GGC CTG AAG AAC-3′. After the addition of a 3′A overhang by incubation with Taq polymerase (New England Biolabs, Ipswich, MA), the DNA fragment was ligated to the pCR T7 TOPO vector (Invitrogen, Carlsbad, CA). Cloning was performed in E. coli TOP10 (Invitrogen), and the correctness of the isolated gene was verified by DNA sequencing. Expression was assessed in E. coli BL21(DE3) (Invitrogen). The cells were cultured in LB medium supplemented with 50 μg of ampicillin/ml at 37°C. At an optical density at 600 nm of 0.4 to 0.7, the temperature was lowered to 24°C, and expression of the target gene was induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 to 4 h. After harvesting by centrifugation, the cell pellet was resuspended in buffer A (50 mM NaH2PO4, 300 mM NaCl, 10% [vol/vol] glycerol; pH 8.0) supplemented with 10 mM imidazole, 0.5% (vol/vol) Triton X-100, 1 mg of lysozyme/ml, 10 μg of RNase A/ml, 20 μg of DNase I/ml, and 1 mM phenylmethylsulfonyl fluoride and lysed using a OneShot cell disruptor (Constant Systems, Ltd., Northants, United Kingdom). The soluble fraction was loaded onto a 1-ml preequilibrated Ni Sepharose FF column (GE Healthcare, Uppsala, Sweden). After being washed with 20 column volumes of buffer A plus 20 mM imidazole, the protein was eluted with 5 column volumes of 250 mM imidazole in buffer A. Fractions containing the protein were identified by SDS-PAGE, pooled, and further purified on a HiLoad 16/60 Superdex 75 prep-grade column (GE Healthcare) equilibrated with buffer A. The protein fractions identified by SDS-PAGE analysis were pooled and stored at −20°C after the addition of 50% (vol/vol) glycerol.
Synthesis of fatty acid derivatives.
The trans-2-enoyl-coenzymes A (CoAs) were synthesized as described previously (25, 29). trans-2-octenoyl-ACPEc and trans-2-acyl-AcpM were prepared enzymatically from trans-2-octenoic acid (Acros Organics, Geel, Belgium) and either E. coli holo-ACP (ACPEc, Sigma-Aldrich, St. Louis, MO) or M. tuberculosis AcpM using the E. coli acyl-ACP synthase as previously reported (27). The purification of trans-2-octenoyl-ACPEc was done as described previously (16) except that a Poros 10 HQ column (Applied Biosystems, Inc., Foster City, CA) was used instead of the DEAE-cellulose column. The purification of trans-2-acyl-AcpM was performed according to the same procedure used for the purification of holo-AcpM (see below) except that a first step of elution through a Hi-Trap Blue HP column (1 ml; GE Healthcare) with Tris 50 mM (pH 8) plus 0.5% Triton X-100 (wt/vol) was performed to eliminate the acyl-ACP synthase. Recombinant C-terminal His-tagged acyl-ACP synthase was produced and purified from E. coli C41(DE3) strain (kindly provided by J. E. Walker, [22]) transformed by the plasmid pAasH (kindly provided by J. Shanklin) as previously described (33), except that a His-Trap HP column (GE Healthcare) was used. Holo-AcpM was produced and purified from an E. coli C41(DE3) transformed by a pET-15b::acpM plasmid (kindly provided by Y.-M. Zhang) as described previously (19). The concentration of octenoyl-ACP was determined by measuring the ΔA at 340 nm in the presence of purified InhA protein and a saturating concentration of NADH as previously reported (25) or by both BCA protein assay and urea-PAGE. The gels were made of a stacking gel (4% [wt/vol] polyacrylamide, 125 mM Tris-HCl [pH 6.8], 0.5 mM EDTA, and 2.5 M urea) and a resolving gel (12% [wt/vol] polyacrylamide, 375 mM Tris-HCl [pH 9.0], 0.5 mM EDTA, and 2.5 M urea). The electrophoresis running buffer (pH 8.3) contained 25 mM Tris and 192 mM glycine. Samples were applied in a loading buffer containing 62.5 mM Tris-HCl (pH 6.8), 20% (vol/vol) glycerol, and 0.05% (wt/vol) bromophenol blue. After electrophoresis (at 30 mA/gel), the proteins were stained with Coomassie blue.
Enzyme assays and steady-state kinetics.
Reactions were performed in a quartz cuvette in a total volume of 700 μl, at 25°C, in 100 mM sodium phosphate buffer (pH 7.0) in the presence of substrate; after equilibration of the baseline, reactions were started by adding a defined amount of enzyme, and measurements were performed for 1.5 to 3 min. Hydratase or dehydratase activities were monitored by spectrophotometry at 263 nm in the presence of trans-2-enoyl-CoA, trans-2-enoyl-ACP, trans-2-enoyl-AcpM, or 3-hydroxyacyl-CoA substrates (ΔA of 0.67 for a variation of concentration of 100 μM) using a thermostated Uvikon 923 spectrophotometer (Kontron Instruments, Milan, Italy). Assays in the presence of short-chain substrates (C4), crotonoyl-CoA (trans-2-butenoyl-CoA), and 3-hydroxybutyryl-CoA (Sigma-Aldrich) were performed using 25 μM substrate and 8 or 80 nM enzyme, respectively. Measurements of the specific activities obtained for the different-chain-length trans-2-enoyl-CoAs (C4 to C20) or the different acyl chain carrier (CoA, AcpM, and ACP) were performed at fixed concentrations of substrate (2.5 μM) and enzyme (8 nM). Control experiments lacking the enzyme were also performed.
Construction of an M. tuberculosis deletion mutant of Rv0241c.
M. tuberculosis was grown in Middlebrook 7H9 liquid medium containing 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC) supplement (Becton Dickinson, Franklin Lakes, NJ) and 0.05% (wt/vol) Tween 80 or on Middlebrook 7H10 agar containing 10% (vol/vol) OADC. Concentrations of 100 μg of hygromycin/ml, 20 μg of kanamycin/ml, 10 μg of gentamicin/ml, 50 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml, and 2% (wt/vol) sucrose were used where appropriate. The Rv0241c gene deletion delivery vector was constructed by amplifying the regions upstream and downstream of the gene by using the primer pairs EUF11 (5′-AAGCTTGGGCTGCACGAGTTCTTTAC-3′) and EUR11 (5′-GGATCCATGGAAAAGTCAACGCGAAC-3′); and EUF12 (5′-GGATCCAATATCGAAGCCCGTTTTCC-3′) and EUR12 (5′-GGTACCGGTATCGGCCACTGAGATGT-3′) and cloned into p2NIL (23) digested with HindIII-KpnI using the underlined restriction sites (HindIII-BamHI and BamHI-KpnI, respectively). The PacI cassette containing lacZ, sacB, and hyg from pGOAL19 (23) was introduced to construct the final vector pTACK0241. The plasmid was electroporated into M. tuberculosis and single crossovers were isolated. Double crossovers were isolated from the single crossover strain as previously described (23). Colonies were screened for the presence of the wild-type (wt) or deletion alleles by PCR using primers 0241D1 (5′-CATTCGCGCATTAGATTGAA-3′) and 0241D2 (5′-AATTCGCCACTCGAAACATC-3′). The deletion mutant was confirmed by Southern blotting.
Analysis of the lipid content of M. tuberculosis ΔRv0241c.
Lipids were extracted, separated, and analyzed using previously reported protocols (8). The same procedure was applied to the parent strain M. tuberculosis H37Rv as a control. Briefly, extractable lipids were recovered by a series of three extractions with CHCl3/CH3OH (at 1:2, 1:1, and 2:1 ratios) and analyzed by thin-layer chromatography (TLC; silica gel G-60) using two types of eluent (CHCl3/CH3OH/H2O, 60/16/2; CHCl3/CH3OH, 9/1). Delipidated bacilli and lipid extracts were saponified. Mycolic acids were extracted with diethyl ether, methylated with diazomethane, analyzed by analytical TLC, and purified by preparative TLC (silica gel G-60 plates; eluent, petroleum ether-diethyl ether [9/1]; five passages). Each type of mycolic acid was analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) (see below).
MALDI-TOF MS.
MS analyses were performed in reflectron mode, on a 4700 Proteomics Analyzer (Voyager DE-STR; Applied Biosystems, Framingham, MA) equipped with an Nd:YAG laser (355-nm wavelength, 500-ps pulses. and 200-Hz frequency). A total of 2,500 shots were accumulated in positive ion mode, and MS data were acquired by using the instrument default calibration. Lipid samples were dissolved in chloroform and directly spotted onto the target plate, mixed with 0.5 μl of matrix solution (10 mg of 2,5-dihydroxybenzoic acid [Sigma-Aldrich]/ml in CHCl3/CH3OH, 1/1 [vol/vol]), and allowed to crystallize at room temperature.
RESULTS
Conservation of Rv0241c protein among the Corynebacterineae.
Rv0241c protein has a 280-amino-acid sequence and a theoretical mass of 30,163 Da. It is a putative (R)-specific enoyl hydratase/3-hydroxyacyl dehydratase belonging to the hydratase 2 protein family and it holds a well-conserved hydratase 2 motif, including the catalytic residues “D-x(4)-H” (5, 29). It is predicted to have a “double hot dog fold” structure (5, 29), where each of the two structural domains corresponds to a hot dog fold (mainly a long α-helix wrapped into a five-stranded antiparallel β-sheet [13]).
Sequence alignments using Rv0241c protein as a probe showed that the latter is conserved among all of the genomes (41 genomes) of mycobacteria sequenced thus far, a finding in agreement with earlier observations on a smaller number of mycobacterial genomes (12 genomes) (29). An orthologous protein was also found in other Corynebacterineae, namely, Rhodococcus, Nocardia, Gordonia, Tsukamurella, and Brevibacterium (data not shown). These data emphasize the importance of Rv0241c protein and its orthologs in the physiology of mycobacteria and related genera. All of these organisms contain a FAS-II system because of their production of mycolic acids holding a meromycolic chain longer than a regular size fatty acid (>C18). To reinforce the putative correlation between the occurrence of Rv0241c and the production of mycolates, the presence of an Rv0241c ortholog was sought in the Corynebacterium genus that has the particularity to be devoid of FAS-II system. Indeed, no ortholog of FAS-IIMt enzymes have been detected in Corynebacterium so far (28), a finding consistent with the fact that the meromycolic chain of its mycolic acids corresponds to a regular size fatty acid (mainly C16 to C18). Strikingly, we observed that about half of the 15 genomes examined coded for an ortholog of Rv0241c (Table 1). Thus, these data questioned the potential involvement of Rv0241c in mycolic acid biosynthesis and especially in FAS-IIMt cycles.
TABLE 1.
Orthologs of Rv0241c protein in Corynebacterium
| Straina | Proteinb | Identity (%) | Similarity (%) | Length of alignment (aa)c | E-valued |
|---|---|---|---|---|---|
| C. aurimucosum ATCC 700975 | Cauri_2166 | 42 | 58 | 236 | 8e-47 |
| C. genitalicum ATCC 33030 | ZP_05706810.1 | 43 | 59 | 283 | 5e-56 |
| C. jeikeium ATCC 43734 | ZP_05845933.1 | 43 | 56 | 259 | 2e-50 |
| C. jeikeium K411 | jk0821 | 43 | 56 | 244 | 4e-47 |
| C. kroppenstedtii DSM 44385 | Ckrop_0818 | 41 | 56 | 272 | 3e-51 |
| C. lipophiloflavum DSM 44291 | ZP_03978914.1 | 40 | 54 | 282 | 2e-46 |
| C. striatum ATCC 6940 | ZP_03936418.1 | 39 | 53 | 271 | 8e-50 |
| C. urealyticum DSM 7109 | Cur_1137 | 42 | 59 | 278 | 2e-57 |
Only strains holding an ortholog of the Rv0241c protein are displayed. Seven other Corynebacterium genomes were considered: C. amycolatum SK46 (incomplete genome), C. diphtheriae NCTC 13129, C. efficiens YS-314, C. glutamicum ATCC 13032 Bielefeld, C. glutamicum ATCC 13032 Kitasato, C. matruchotii ATCC 14266 (incomplete genome), and C. tuberculostearicum SK141 (incomplete genome).
The protein name or NCBI reference sequence number.
The number of amino acid (aa) residues aligned by the BLAST algorithm (1).
“Expect value” calculated by the BLAST algorithm.
Heterologous complementation experiments of E. coli fabZ and fabA mutants using the Rv0241c gene.
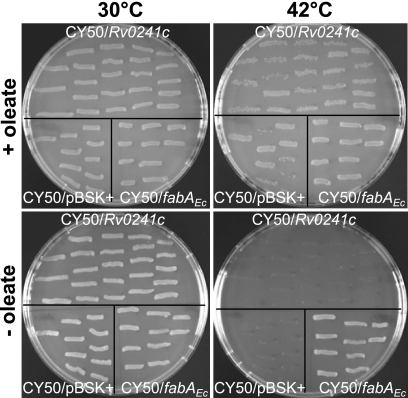
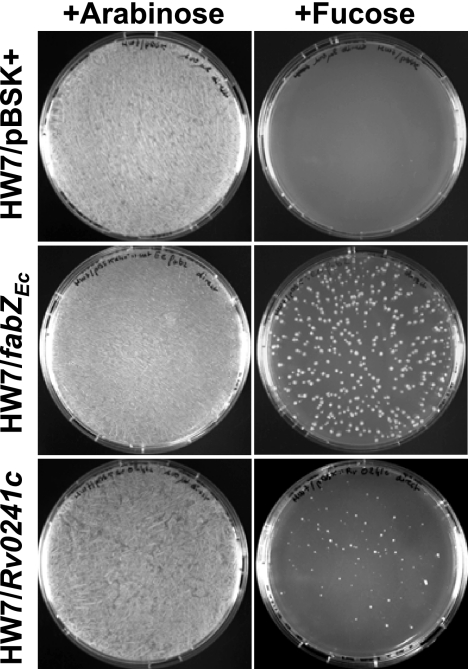
Heterologous complementation assays were realized in parallel using a fabZ knockout (KO) mutant, E. coli HW7, and a fabA Ts mutant, E. coli CY50. E. coli CY50 that has a Ts point mutation in fabA is able to grow at 30°C (permissive temperature) but becomes auxotrophic for unsaturated fatty acids, such as oleate (C18:1Δ9 acid), at 42°C (7). E. coli CY50 was transformed by Rv0241c carried by an expression plasmid (pBSK+) under the control of a lactose- and IPTG-inducible promoter. At a permissive temperature, the recombinant strain grew both in the absence and in the presence of oleate in the culture medium (Fig. 1). However, it was not able to grow at a nonpermissive temperature in the absence of oleate, whereas E. coli CY50 transformed by wt fabAEc actively grew in such conditions (Fig. 1). In E. coli HW7, the essential fabZ gene has been disrupted by a kanamycin resistance cassette. To keep the viability of the strain, the mutation has been complemented by the wt fabZ gene of Clostridium acetobutylicum (fabZCa) under the control of an inducible arabinose promoter (35). Thus, this mutant grows in the presence of arabinose but is not viable in the absence of arabinose or when the promoter is repressed by fucose (Fig. 2). After transformation of E. coli HW7 by Rv0241c, we observed that the recombinant strain grew in the presence of either arabinose or fucose. This means that Rv0241c was able to restore the (3R)-hydroxyacyl-ACP dehydration (HAD) function, although less efficiently than the wt fabZEc gene itself (Fig. 2). For comparison, heterologous complementation assays were also performed using a yeast mutant, Saccharomyces cerevisiae Δhtd2, deficient in the mitochondrial FAS-II (3R)-hydroxyacyl-ACP dehydratase Htd2 related to the hydratase 2 family (17). This mutant displays a respiratory deficiency phenotype; it can grow in the presence of glucose (both fermentable and respirable carbon source) but not of glycerol (usable only through the respiratory metabolism). Consistent with a previous report (9), we observed that Rv0241c also complemented this mutation, since it allowed the recovering of the respiratory ability of the strain (data not shown).
FIG. 1.
Heterologous complementation assays of the E. coli fabA Ts mutant. E. coli CY50 was transformed by either the Rv0241c gene, the expression vector (pBSK+) alone, or wt fabAEc and grown on RB agar in the presence or absence of oleate, at either 30 or 42°C, as indicated. The photographs are representative of three independent experiments.
FIG. 2.
Heterologous complementation assays of E. coli fabZ KO mutant. In E. coli HW7, the lethal disruption of fabZEc has first been complemented by fabZCa under the control of an arabinose promoter that is induced by arabinose and repressed by fucose (35). The strain was then cotransformed by either the Rv0241c gene, the expression vector (pBSK+) alone, or wt fabZEc and grown on RB agar supplemented with arabinose or fucose, as indicated. The photographs are representative of three independent experiments.
These data strongly suggested that Rv0241c protein is able on the one hand to work as an HAD in vivo by using the substrates available and processed by the E. coli and mitochondrial FAS-II systems, i.e., short- to long-chain ACP derivatives (C4-C16) (11, 12), and on the other hand to perform coupled reactions with enzymes of these FAS-II systems.
Purification of Rv0241c protein and activity assays.
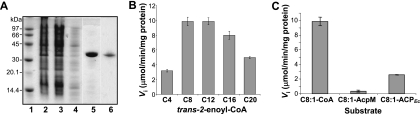
The enzymatic activity of Rv0241c was studied in order to evaluate its putative contribution to the FAS-IIMt system. For this purpose, Rv0241c gene was cloned into the pCR T7 TOPO expression vector, and the protein was produced in E. coli BL21(DE3) in fusion with a N-terminal polyhistidine tag (total molecular mass, 31.1 kDa) by induction with IPTG. Bacteria were lysed using a cell disruptor in the presence of lysozyme. The purification consisted in a two-step chromatography procedure over a Ni Sepharose FF column, using an imidazole gradient, and a Superdex 75 gel filtration column (Fig. 3 A).
FIG. 3.
Purification and enzymatic study of Rv0241c. (A) SDS-PAGE analysis of the purification steps. Lane 1, molecular weight markers; lane 2, cleared cell lysate; lanes 3 to 5, Ni Sepharose FF column (flowthrough, wash, and elution of Rv0241c with 250 mM imidazole); lane 6, Superdex 75 gel filtration column (elution fraction containing Rv0241c). Gels (20% polyacrylamide) stained with Coomassie blue are shown. The dividing lines separate different parts of the gel. (B) Chain length specificity profile. (C) Acyl chain carrier specificity. In panels B and C, the specific activities of Rv0241c were measured at fixed concentrations of substrate (2.5 μM) and enzyme (8 nM) in 100 mM sodium phosphate buffer (pH 7.0). The data are means ± the standard deviation.
Enzymes belonging to the (R)-specific enoyl hydratase/hydroxyacyl dehydratase family preferentially catalyze the hydration reaction when they are isolated from their enzymatic complex (26). Thus, their activities are most often studied in the presence of enoyl derivatives in vitro. The activity of Rv0241c was first measured spectrophotometrically (at 263 nm) in the presence of short-chain CoA derivatives, crotonoyl-CoA (C4:1) and 3-hydroxybutyryl-CoA (C4). At a 25 μM concentration of substrate, the specific activities of the protein were 16.2 ± 1.0 and 0.93 ± 0.07 μmol min−1 mg−1, respectively. The purified recombinant Rv0241c protein proved to be active and, as expected, its activity was greater in the presence of the enoyl-CoA substrate than in that of the 3-hydroxyacyl-CoA.
Substrate specificity of Rv0241c.
The specificity of Rv0241c for the chain length of the substrate was then studied with a series of longer chain derivatives, C8 to C20 trans-2-enoyl-CoAs. To minimize the solubility problems encountered with such amphipathic molecules, the experiments were performed at a fixed low substrate concentration (2.5 μM). The enzyme displayed a broad chain length specificity since it worked in the presence of short-, medium-, and long-chain molecules, yet with a predilection for the C8 to C12 substrates (Fig. 3B). For comparison, the FAS-IIMt dehydratases HadAB and HadBC are not active in the presence of C4 to C8 trans-2-enoyl-CoAs, under the same experimental conditions, but use only longer-chain molecules, C12 to C20 (28).
To the best of our knowledge, FAS-II is the only system of fatty acid biosynthesis that is ACP-dependent in mycobacteria. It has been shown that it uses the AcpM protein as ACP unit (19, 31). Thus, an investigation of the ACP dependence of Rv0241c was decisive in order to assess its involvement in FAS-IIMt. The behaviors of Rv0241c in the presence of AcpM or CoA derivatives were compared by using a fixed concentration of trans-2-octenoyl-AcpM, trans-2-octenoyl-ACPEc, or trans-2-octenoyl-CoA as substrates. The data clearly showed a strict dependence of Rv0241c on CoA as an acyl chain carrier (Fig. 3C). Indeed, the enzyme was not active in the presence of the AcpM derivative. Interestingly, however, Rv0241c was able to metabolize an enoyl chain carried by the heterologous ACPEc, although much less efficiently than the CoA derivative (Fig. 3C). For comparison, under the same experimental conditions, HadAB and HadBC work only in the presence of the ACP derivative (28).
Altogether, these experiments demonstrate that Rv0241c is a 2-trans-enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydratase, with a relatively broad chain length specificity.
In vitro essentiality study of the Rv0241c gene.
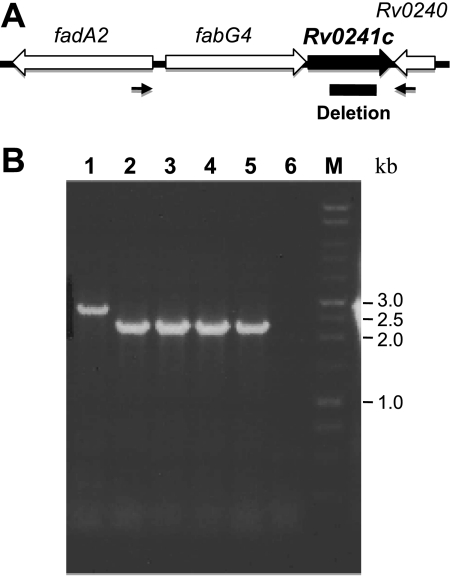
To examine the essentiality of Rv0241c enzyme, we used a two-step homologous recombination method to construct an in-frame, unmarked deletion of Rv0241 gene in M. tuberculosis H37Rv (Fig. 4). The mutant was viable, and its growth rate was similar to that of the wt strain (data not shown), showing that Rv0241c is not essential for M. tuberculosis survival in vitro, as predicted previously (30).
FIG. 4.
Construction of the Rv0241c deletion mutant. (A) Chromosomal organization of the region. The deletion zone is drawn. Arrows indicate the primers used for PCR screening. (B) PCR confirmation of deletion strains. The primers indicated in panel A were used to amplify the Rv0241c region. Wild-type strains gave a product of 2.7 kb, the deletion strains gave a product of 2.2 kb. Lane 1, wt strain; lanes 2 to 5, deletion strains; lane 6, negative control; lane M, 1-kb markers. Deletions were verified by Southern hybridization.
Mycolic acid profile of the mutant.
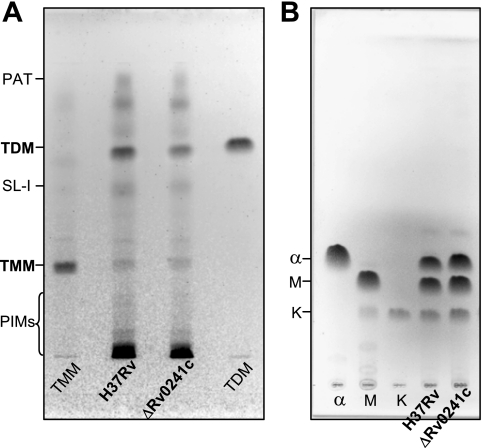
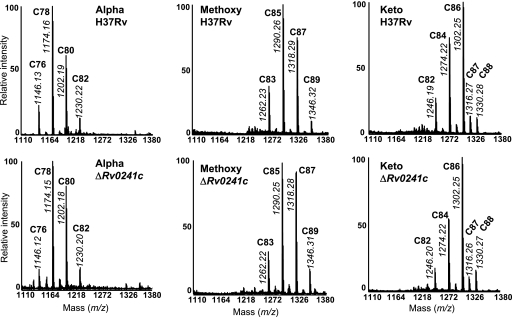
To determine the impact of the mutation on the mycolic acid content of Rv0241c deletion mutant, the complex lipids holding mycolic acids, i.e., trehalose dimycolates and trehalose monomycolates, were obtained by successive CHCl3/CH3OH extractions from the bacterial pellets and analyzed. The same types of glycolipids and other lipids were detected in M. tuberculosis ΔRv0241c mutant by comparison with the wt parent strain, M. tuberculosis H37Rv, as observed by TLC (Fig. 5). After saponification of the delipidated bacterial residues, the released mycolic acids were extracted, methylated, and analyzed by TLC. Very similar molecular profiles were observed for both the wt and the mutant strains (Fig. 5). The three types of mycolate were purified by preparative TLC and studied further by MALDI-TOF MS. Analysis of the mass spectra demonstrated that the α-, methoxy-, and keto-mycolates held the same number of carbons in M. tuberculosis ΔRv0241c as in the wt strain (Fig. 6).
FIG. 5.
Analysis of the mycolic acid content of M. tuberculosis ΔRv0241c. (A) TLC of the extractable lipids on silica gel G-60 plate using CHCl3/CH3OH/H2O (60/16/2) as the eluent. Visualization by spraying with anthrone-H2SO4 and charring. No differences between the two strains were visible using another eluent (CHCl3/CH3OH, 9/1). PAT, polyacyltrehalose; TDM, trehalose dimycolates; SL-I, sulfolipid-I; TMM, trehalose monomycolates; PIMs, phosphatidylinositol mannosides. Standards of TDM and TMM were deposited. (B) TLC of the mycolic acid methyl esters on a silica gel G-60 plate using petroleum ether-diethyl ether (9/1, five passages) as the eluent. Visualization by spraying with molybdophosphoric acid and charring. α, α-mycolates; M, methoxymycolates; K, ketomycolates. The standards α-, methoxy-, and keto-mycolates were deposited. Identical quantities of total lipids from wt and ΔRv0241c strains were deposited in panels A and B.
FIG. 6.
MALDI-TOF MS analysis of the mycolic acids of M. tuberculosis ΔRv0241c. Mass spectra of purified mycolic acid methyl esters from the mutant strain and the wt parent strain. The peaks correspond to [M+Na]+ ions. The total carbon number of each molecular species is mentioned.
In conclusion, the lack of Rv0241c enzyme does not affect the biosynthesis of all three types of mycolic acid in M. tuberculosis.
DISCUSSION
The purified Rv0241c protein displays a trans-2-enoyl hydratase/3-hydroxyacyl dehydratase activity with a strict specificity toward CoA as the carrier unit of the acyl chain, as opposed to AcpM. Furthermore, it shows a broad specificity for the chain length (C4 to C20) of the substrate, which is consistent with an earlier study predicting that this enzyme would have a substrate binding tunnel that could welcome long-chain substrates (5, 15). Rv0241c behavior is clearly distinct from that of FAS-IIMt enzymes that display a marked specificity for C12 to C20 acyl chains carried by an ACP unit (21, 25, 28, 32). Therefore, the Rv0241c protein cannot belong to the FAS-IIMt system, in sharp contrast to an earlier conclusion (9). Consistently, an Rv0241c ortholog was detected in the Corynebacterium genus, which is devoid of FAS-II system.
Interestingly, we observed that the Rv0241c enzyme could metabolize an enoyl chain linked to ACPEc, although with a weak activity. This feature and the broad chain length specificity of Rv0241c may account for its ability to artificially replace the HADs of E. coli and S. cerevisiae mitochondria in vivo during complementation experiments. Indeed, the latter proteins take part into de novo fatty acid biosynthesis pathways, as classical FAS-II systems usually do. Thus, in E. coli, FabZ has a predilection for short- to medium-chain length intermediates (11). In mitochondria, Htd2 displays a broad chain length (C2 to C16) specificity for the substrate (12).
We have previously shown that the hadABC cluster was essential to M. tuberculosis survival in vitro (28), like most of the genes encoding FAS-IIMt enzymes. In contrast, the Rv0241c KO mutant is viable, meaning that either the Rv0241c enzyme has a function redundant with that of another M. tuberculosis enzyme or that it catalyzes a nonessential step of a metabolic pathway. A BLAST search against M. tuberculosis predicted proteins, using as a probe the Rv0241c sequence, revealed the absence of a paralog or closely related protein (data not shown). In particular, Rv0241c has a poor sequence identity with HadA, HadB, and HadC subunits (6 to 19% over the whole sequences). Furthermore, Rv0241c KO mutant has no detectable change in the mycolic acid profile compared to the parent strain. These data are consistent with the enzymatic study of Rv0241c and with the conclusion that this enzyme is not implicated in the mycolic acid biosynthesis pathway. Rv0241c may belong to another mycobacterial lipid metabolism such as, for example, the potential CoA-dependent fatty acid biosynthesis systems (18, 34).
The present study illustrates the fact that giving a positive result of heterologous complementation of HAD-deficient mutants of E. coli or S. cerevisiae is not a sufficient condition for a gene to encode a HAD homolog. Other M. tuberculosis proteins of the hydratase 2 family, Rv0130 and Rv3389c, were proposed to possess a HAD function and consequently to be part of FAS-IIMt system based on the complementation of S. cerevisiae Δhtd2 mutant (9, 10). However, it has previously been shown that Rv3389c is a CoA-dependent enzyme (29), like Rv0241c. Furthermore, Rv3389c is not ubiquitous among mycobacteria (no ortholog in M. smegmatis and a pseudogene in M. leprae), whereas FAS-II enzymes are very well conserved due to their essential function. Rv0130 is not conserved through the Mycobacterium genus either (absent in M. leprae). Furthermore, it has an ortholog in Corynebacterium (28), like Rv0241c. Finally, a soluble protein extract of a recombinant yeast strain overexpressing Rv0130 exhibits 5- and 16-fold lower specific activities, respectively, in the presence of decenoyl-CoA compared to crotonoyl-CoA and trans-2-hexenoyl-CoA (10), suggesting that Rv0130 has a predilection for short-chain substrates. These data together are incompatible with an involvement of either Rv0130 or Rv3389c in the FAS-IIMt system, similarly to Rv0241c.
Acknowledgments
We thank H. Wang (Illinois University) for the generous gift of strain E. coli HW7, J. E. Walker (Medical Research Council, Cambridge, United Kingdom) for E. coli C41(DE3), and the E. coli Genetic Stock Center (New Haven, CT) for E. coli CY50. We are grateful to J. Shanklin (Brookhaven National Laboratory, Upton, NY), Y.-M. Zhang (St Jude Children's Research Hospital, Memphis, TN), D. de Mendoza (Institute of Molecular and Cell Biology, Rosario, Argentina), and A. Kastaniotis (Biocenter Oulu, Oulu, Finland) for kindly providing the pAasH, pET-15b::acpM, pDM5, and pYE352::mtfabZEc plasmids, respectively, and to S. Cole (Institut Pasteur, Paris, France) for BAC-Rv329 and total M. tuberculosis DNA. We thank H. Marrakchi (IPBS, Toulouse, France) for her assistance.
This study was supported in part by a doctoral fellowship (to E.S.) from the Fondation pour la Recherche Médicale and by grants from the European Community (LSHP-CT-2005-018923) and the Carl Trygger Foundation.
Footnotes
Published ahead of print on 28 May 2010.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatt, A., N. Fujiwara, K. Bhatt, S. S. Gurcha, L. Kremer, B. Chen, J. Chan, S. A. Porcelli, K. Kobayashi, G. S. Besra, and W. R. Jacobs, Jr. 2007. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. U. S. A. 104:5157-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloch, K. 1977. Control mechanisms for fatty acid synthesis in Mycobacterium smegmatis. Adv. Enzymol. Relat Areas Mol. Biol. 45:1-84. [DOI] [PubMed] [Google Scholar]
- 4.Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito, B. G. Barrell, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castell, A., P. Johansson, T. Unge, T. A. Jones, and K. Bäckbro. 2005. Rv0216, a conserved hypothetical protein from Mycobacterium tuberculosis that is essential for bacterial survival during infection, has a double hotdog fold. Protein Sci. 14:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Cronan, J. E. J., D. F. Silbert, and D. L. Wulff. 1972. Mapping of the fabA locus for unsaturated fatty acid biosynthesis in Escherichia coli. J. Bacteriol. 112:206-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubnau, E., J. Chan, C. Raynaud, V. P. Mohan, M. A. Laneelle, K. Yu, A. Quemard, I. Smith, and M. Daffe. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36:630-637. [DOI] [PubMed] [Google Scholar]
- 9.Gurvitz, A., J. K. Hiltunen, and A. J. Kastaniotis. 2009. Heterologous expression of mycobacterial proteins in Saccharomyces cerevisiae reveals two physiologically functional 3-hydroxyacyl-thioester dehydratases, HtdX and HtdY, in addition to HadABC and HtdZ. J. Bacteriol. 191:2683-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurvitz, A., J. K. Hiltunen, and A. J. Kastaniotis. 2008. Identification of a novel mycobacterial 3-hydroxyacyl-thioester dehydratase, HtdZ (Rv0130), by functional complementation in yeast. J. Bacteriol. 190:4088-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heath, R. J., and C. O. Rock. 1996. Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271:27795-27801. [DOI] [PubMed] [Google Scholar]
- 12.Hiltunen, J. K., M. S. Schonauer, K. J. Autio, T. M. Mittelmeier, A. J. Kastaniotis, and C. L. Dieckmann. 2009. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 284:9011-9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hisano, T., T. Tsuge, T. Fukui, T. Iwata, K. Miki, and Y. Doi. 2003. Crystal structure of the (R)-specific enoyl-CoA hydratase from Aeromonas caviae involved in polyhydroxyalkanoate biosynthesis. J. Biol. Chem. 278:617-624. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, C., A. Leis, M. Niederweis, J. M. Plitzko, and H. Engelhardt. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U. S. A. 105:3963-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson, P., A. Castell, T. A. Jones, and K. Backbro. 2006. Structure and function of Rv0130, a conserved hypothetical protein from Mycobacterium tuberculosis. Protein Sci. 15:2300-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi, A. K., and S. Smith. 1993. Construction, expression, and characterization of a mutated animal fatty acid synthase deficient in the dehydrase function. J. Biol. Chem. 268:22508-22513. [PubMed] [Google Scholar]
- 17.Kastaniotis, A. J., K. J. Autio, R. T. Sormunen, and J. K. Hiltunen. 2004. Htd2p/Yhr067p is a yeast 3-hydroxyacyl-ACP dehydratase essential for mitochondrial function and morphology. Mol. Microbiol. 53:1407-1421. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi, S., and T. Kusaka. 1982. New malonyl-CoA-dependent fatty acid elongation system in Mycobacterium smegmatis. J. Biochem. 92:839-844. [DOI] [PubMed] [Google Scholar]
- 19.Kremer, L., K. M. Nampoothiri, S. Lesjean, L. G. Dover, S. Graham, J. Betts, P. J. Brennan, D. E. Minnikin, C. Locht, and G. S. Besra. 2001. Biochemical characterization of acyl carrier protein (AcpM) and malonyl-CoA:AcpM transacylase (mtFabD), two major components of Mycobacterium tuberculosis fatty acid synthase II. J. Biol. Chem. 276:27967-27974. [DOI] [PubMed] [Google Scholar]
- 20.Marrakchi, H., F. Bardou, M.-A. Lanéelle, and M. Daffé. 2008. A comprehensive overview of mycolic acid structure and biosynthesis, p. 41-62. In M. D. J.-M. Reyrat (ed.), The mycobacterial cell envelope. ASM Press, Washington, DC.
- 21.Marrakchi, H., S. Ducasse, G. Labesse, H. Montrozier, E. Margeat, L. Emorine, X. Charpentier, M. Daffe, and A. Quemard. 2002. MabA (FabG1), a Mycobacterium tuberculosis protein involved in the long-chain fatty acid elongation system FAS-II. Microbiology 148:951-960. [DOI] [PubMed] [Google Scholar]
- 22.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 23.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 24.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29:123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quemard, A., J. C. Sacchettini, A. Dessen, C. Vilcheze, R. Bittman, W. R. Jacobs, Jr., and J. S. Blanchard. 1995. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry 34:8235-8241. [DOI] [PubMed] [Google Scholar]
- 26.Rock, C. O., and J. E. Cronan. 1996. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta 1302:1-16. [DOI] [PubMed] [Google Scholar]
- 27.Rock, C. O., and J. L. Garwin. 1979. Preparative enzymatic synthesis and hydrophobic chromatography of acyl-acyl carrier protein. J. Biol. Chem. 254:7123-7128. [PubMed] [Google Scholar]
- 28.Sacco, E., A. S. Covarrubias, H. M. O'Hare, P. Carroll, N. Eynard, T. A. Jones, T. Parish, M. Daffe, K. Backbro, and A. Quemard. 2007. The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 104:14628-14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sacco, E., V. Legendre, F. Laval, D. Zerbib, H. Montrozier, N. Eynard, C. Guilhot, M. Daffe, and A. Quemard. 2007. Rv3389C from Mycobacterium tuberculosis, a member of the (R)-specific hydratase/dehydratase family. Biochim. Biophys. Acta 1774:303-311. [DOI] [PubMed] [Google Scholar]
- 30.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 31.Schaeffer, M. L., G. Agnihotri, H. Kallender, P. J. Brennan, and J. T. Lonsdale. 2001. Expression, purification, and characterization of the Mycobacterium tuberculosis acyl carrier protein, AcpM. Biochim. Biophys. Acta 1532:67-78. [DOI] [PubMed] [Google Scholar]
- 32.Schaeffer, M. L., G. Agnihotri, C. Volker, H. Kallender, P. J. Brennan, and J. T. Lonsdale. 2001. Purification and biochemical characterization of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthases KasA and KasB. J. Biol. Chem. 276:47029-47037. [DOI] [PubMed] [Google Scholar]
- 33.Shanklin, J. 2000. Overexpression and purification of the Escherichia coli inner membrane enzyme acyl-acyl carrier protein synthase in an active form. Protein Expr. Purif 18:355-360. [DOI] [PubMed] [Google Scholar]
- 34.Shimakata, T., Y. Fujita, and T. Kusaka. 1977. Acetyl-CoA-dependent elongation of fatty acids in Mycobacterium smegmatis. J. Biochem. 82:725-732. [DOI] [PubMed] [Google Scholar]
- 35.Wang, H., and J. E. Cronan. 2004. Functional replacement of the FabA and FabB proteins of Escherichia coli fatty acid synthesis by Enterococcus faecalis FabZ and FabF homologues. J. Biol. Chem. 279:34489-34495. [DOI] [PubMed] [Google Scholar]
- 36.White, S. W., J. Zheng, Y. M. Zhang, and Rock. 2005. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74:791-831. [DOI] [PubMed] [Google Scholar]
- 37.World Health Organization. 2009. Global tuberculosis control: a short update to the 2009 report. World Health Organization, Geneva, Switzerland.
- 38.Zuber, B., M. Chami, C. Houssin, J. Dubochet, G. Griffiths, and M. Daffe. 2008. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190:5672-5680. [DOI] [PMC free article] [PubMed] [Google Scholar]