Abstract
During development, gene activation is stringently regulated to restrict expression only to the correct cell type and correct developmental stage. Here, we present mechanistic evidence that suggests DNA methylation contributes to this regulation by suppressing premature gene activation. Using the mouse Myogenin promoter as an example of the weak CpG island class of promoters, we find that it is initially methylated but becomes demethylated as development proceeds. Full hypersensitive site formation of the Myogenin promoter requires both the MEF2 and SIX binding sites, but binding to only one site can trigger the partial chromatin opening of the nonmethylated promoter. DNA methylation markedly decreases hypersensitive site formation that now occurs at a detectable level only when binding to both MEF2 and SIX binding sites is possible. This suggests that the probability of activating the methylated promoter is low until two of the factors are coexpressed within the same cell. Consistent with this, the single-cell analysis of developing somites shows that the coexpression of MEF2A and SIX1, which bind the MEF2 and SIX sites, correlates with the fraction of cells that demethylate the Myogenin promoter. Taken together, these studies imply that DNA methylation helps to prevent inappropriate gene activation until sufficient activating factors are coexpressed.
The development of multicellular organisms requires a highly specific, spatially, and temporally restricted pattern of gene expression. Remarkably, this is achieved from a relatively limited set of transcription factors, many of which are expressed in more than one lineage but which nevertheless activate distinct programs of gene expression within individual lineages. One way in which these distinct expression programs are achieved is via cooperation with lineage-specific coactivators or corepressors. For example, during murine hematopoiesis, individual stem cells are primed to express genes that are typical of a number of different lineages (17), but lineage-specific corepressors then restrict this expression to cause commitment to a single lineage (21).
A second tool that could restrict inappropriate developmental gene activation is DNA methylation. The established correlations between DNA methylation and gene inactivity (11), together with the fact that DNA methylation can be stably inherited (47), gave support to an attractive model in which DNA methylation represses gene activity in nonexpressing tissues (15, 34). Despite the attractiveness of the model, direct evidence that DNA methylation regulates gene activation during development has been elusive. Evidence favoring this hypothesis includes the observation that the loss of the maintenance methyltransferase, DNMT1, in Xenopus (xDNMT1) triggered premature gene activation (39); likewise, the conditional knockout of Dnmt1 in mouse fibroblasts resulted in the upregulation of 10% of genes (18). In contrast, an analysis of Dnmt1 knockout mice showed that the expression of certain lineage-specific genes was unaltered in embryonic day 9.5 (e9.5) embryos (45).
Recent genome-wide methylation analyses have suggested why DNA methylation apparently has such variable effects; these studies propose that promoter CpG density is central to determining if and how DNA methylation affects gene expression (46). Methylated CpG dinucleotides are bound by methylated DNA binding proteins (MBDs), and the affinity of these proteins for a given promoter influences the extent to which MBDs inhibit transcription factor binding (6). CpG island promoters have a high CpG density, but normally they are unmethylated in all tissues regardless of the expression of their linked gene (7). When these promoters do become methylated, however, such as on the inactive X chromosome, then they are strongly repressed, most likely due to high-affinity binding by MBDs (6). Genes with a low promoter CpG density also do not show a strong correlation between expression and DNA methylation; in this case, the methyl-CpG density is thought to be too low to stably bind repressive methylated DNA binding proteins (46). In contrast, an inverse correlation between DNA methylation and RNA polymerase II occupancy was found for promoters with an intermediate CpG density, thus raising the possibility that DNA methylation, via MBD binding, plays a role in modulating their activity. These promoters account for about 12% of promoters, have an average ratio of observed to expected CpG of 0.5, and have been called weak CpG islands (46).
Although these studies establish a correlation, whether DNA methylation indeed contributes to the regulation of these promoters during development is unknown. DNA methylation levels undergo major changes during development. Shortly after fertilization, the male pronucleus undergoes a dramatic demethylation (30); this is followed by a more gradual demethylation of the maternal genome between the 2- and 8-cell stages to reduce the overall methylation level to about 30% of that in somatic cells. This initial demethylation is followed by a wave of de novo methylation around the time of implantation (day 5 post coitum; e5) (33). Consequently, the genes activated after the wave of de novo methylation are the ones that are more likely to be regulated by DNA methylation.
In this report, we addressed if and how DNA methylation affects developmental gene activation using the Myogenin gene as a model. The promoter of this gene is a weak CpG island, and in mice its expression is detected first in somites (that are found along the anterior-posterior axis of the embryo) after the wave of de novo methylation, at day e8.5 (35); by day e9.5, its expression follows a rostro-caudal gradient where only the most anterior somites express the gene (49). This expression pattern offers a major advantage, since both progenitor and expressing cells can be isolated from the same embryo by the dissection of spatially distinct somites (Fig. 1 A). Also, Myogenin gene expression can be recapitulated with only 133 bp of 5′ flanking DNA, and although this region additionally is bound by a Y box protein in adults (4), only three known sites are bound by transcription factors during embryogenesis: a canonical E box sequence, recognized by the muscle regulatory factors (MRFs; MYF5, MYOD, MYOGENIN, and MRF4), a MEF2 site (bound by MEF2A, MEF2B, MEF2C, or MEF2D), and a SIX binding site that is bound by the SIX family of proteins (49). These three sets of transcription factors have distinct patterns of expression. The MRFs are muscle specific, and of these, MYF5 is first detected in the epaxial domain of somites at day 8 post coitum (e8.0), while Mrf4 expression is detected, also from e8.0, in the dorsal half of the somites (9, 40, 43). MYF5 initiates and coordinates the myogenic cascade by activating the expression of the late MRF, Myogenin, at e8.5 (10), whereas MyoD expression is observed later at e10.5. In vertebrates, the Mef2 genes are widely expressed, and some reports suggest that the MEF2 proteins are ubiquitous (reviewed in reference 32). Of the Six family, Six1 and Six4 are highly expressed in muscle, with Six1 being detected in somites by e8 (14, 27).
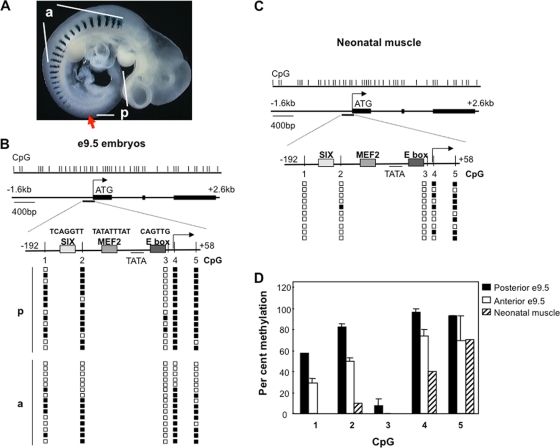
FIG. 1.
Myogenin promoter becomes demethylated during skeletal muscle differentiation. (A) Expression pattern of Myogenin at e9.5. An embryo from transgenic mice in which lacZ expression is under the control of the Myogenin 5′ regulatory region was stained for β-galactosidase. Anterior and posterior somites (including presomitic mesoderm) are indicated. The red arrow indicates the most posterior somite expressing Myogenin. (B and C) Combined bisulfite analysis of the endogenous Myogenin promoter and this promoter in the transgene array (49) in (B) posterior (p) and anterior (a) somites of e9.5 embryos and (C) skeletal muscle cells isolated from neonatal mice. The upper panel shows the position of CpG dinucleotides in the Myogenin promoter and gene. Filled boxes in the second panel indicate Myogenin exons. Bisulfite products were analyzed by both direct sequencing and by cloning and PCR, with equivalent results. Unmethylated and methylated CpG dinucleotides, as determined by sequencing the clones from a representative experiment, are shown by open and filled squares, respectively. The sequences of the transcription factor binding sites are given above the names of these sites; none of these have a CpG dinucleotide, but since MBDs can indirectly repress the activation of methylated promoters (6), the presence of CpG in the transcription factor binding site is not essential for repression. (D) Graph showing the average percent methylation of each cytosine in the Myogenin promoter, calculated as (number of methylated alleles/total number of clones analyzed) × 100. Error bars show the standard errors from three independent experiments; a total of 30 clones each from anterior and posterior somites were sequenced.
Here, we examined whether DNA methylation affects the developmental activation of the Myogenin gene. We find that the Myogenin promoter initially is methylated prior to activation and subsequently becomes demethylated. We show that further DNA methylation substantially decreases hypersensitive site (HS) formation at the Myogenin promoter but does not abolish it completely. Instead, we find that binding to both MEF2 and SIX binding sites typically is required for the detectable activation of the methylated promoter. This suggests that at early stages of somite formation, methylation decreases the probability of Myogenin promoter opening until both SIX and MEF2 proteins are coexpressed. Consistent with this, single-cell analysis showed that the degree of demethylation is similar to the number of cells that coexpress Six1 and Mef2A, and knockdown studies show that SIX1 is required for demethylation. Taken together, these studies suggest an ordered mechanism of activation whereby DNA methylation prevents inappropriate chromatin opening in most cells in the population until a sufficient number of specific activators are present.
MATERIALS AND METHODS
Transgenic animals and tissues.
All experiments were performed using GZ1092 mice (49) under a license from the United Kingdom Home Office. Embryos were taken at e9.5, where the morning of the vaginal plug was considered to be e0.5. Somites and presomitic mesoderm were dissected into phosphate-buffered saline (PBS), and as many axial structures and lateral mesoderm as possible were removed. The tissue was divided into three samples: presomitic mesoderm plus somites 1 to 3 (for a 25-somite embryo), somites 4 to 12, and somites 13 to 25 (Fig. 1A). The region comprising somites 4 to 12 was stained for β-galactosidase as described previously (49). When individual cells were needed, tissues were mechanically disaggregated and passed through a 0.7-μm filter. Neonatal muscle was obtained from the tongues of 3- to 4-day-old mice.
Plasmids and in vitro methylation.
The −4.2myo wild-type (wt) construct was prepared by fusing the Myogenin genomic sequence from −4.25 to +1.43 kb (49) to a 1.2-kb XhoI fragment [containing the poly(A) tail from the rabbit β-globin gene] from PSTC (37) in pBluescript S/K (+/−). The mutation of the factor binding sites in the Myogenin promoter was performed as described previously (38, 49) using the QuikChange XL mutagenesis kit (Stratagene); pREP7 was from Invitrogen. In vitro methylation was performed using SssI (New England Biolabs); mock-methylated DNA was treated identically, except that SssI was omitted. The plasmids were checked by digestion with HpaII and by direct sequencing following bisulfite treatment to ensure they were fully methylated. To ensure that CpG density adjacent to the Myogenin promoter resembled that found in the endogenous locus as closely as possible, 4.25 kb of upstream sequence and 1.4 kb of downstream gene sequence were included, and plasmid sequences were removed prior to transfection. Following stable transfection into C2C12 cells, the methylation status of the constructs was checked again by restriction enzyme analysis and Southern blotting as well as by bisulfite sequencing.
Cell culture and transfections.
Murine C2C12 myoblasts were cultured in growth medium (GM): Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal calf serum, 4 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Differentiation was induced by incubation in differentiation medium (DM); this is identical to GM, except fetal calf serum is replaced with 2% horse serum. Cells were transfected with 4.5 μg of −4.2myo wt and 1.5 μg of pREP7 using Lipofectamine reagent (Invitrogen) and selected with 400 μg/ml of hygromycin for 10 days. Hygromycin-resistant clones were pooled and kept in growth medium supplemented with 200 μg/ml of antibiotic.
Bisulfite treatment.
Bisulfite genomic sequencing was performed as described previously (12), with minor modifications: embryonic tissues were lysed at 37°C for at least 3 h in a buffer containing 100 mM NaCl, 10 mM Tris, pH 8, 25 mM EDTA, 0.5% SDS, and 0.3 mg/ml proteinase K; genomic DNA was extracted with phenol-chloroform and ethanol precipitated. Prior to bisulfite treatment, samples were digested with KpnI for at least 3 h, followed by phenol-chloroform extraction and precipitation. Treatment was performed with a saturated sodium bisulfite solution (between 5.20 and 5.69 M HSO3−) and incubation at 55°C in the dark for 2.5 h. Desulfonated samples were precipitated in the presence of 0.8 M ammonium acetate and 20 μg of glycogen. The bisulfite-treated DNA then was amplified by nested PCR using the oligonucleotides given in Table 1; the PCR products were either directly sequenced or cloned into pBluescript S/K (+/−) via the EcoRI and XbaI sites, and individual clones were sequenced.
TABLE 1.
Primers used in bisulfite and RT-PCR analysesa
| Analysis and gene | Round | Primer |
|---|---|---|
| Bisulfite: Myogenin | 1 | atagaattcTGGGTTAGGGGTAGGTTTGTAG |
| aggtctagaCCCCATCATAAAAATAAAACTCC | ||
| 2 | agggaattcGGGGAATTATATGTAATTTATTGG | |
| aggtctagaCCCCATCATAAAAATAAAACTCC | ||
| RT-PCR | ||
| Hprt | 1 | 5′-GGGGGCTATAAGTTCTTTGC |
| 5′-TCCAACACTTCGAGAGGTCC | ||
| 2 | 5′-GTTCTTTGCTGACCTGCTGG | |
| 5′-TGGGGCTGTACTGCTTAACC | ||
| Myogenin | 1 | 5′-AATGCAACTCCCACAGCGCC |
| 5′-GGCACTCATGTCTCTCAAACGG | ||
| 2 | 5′-CTGGAGTTCGGTCCCAACCC | |
| 5′-TTCTGGACATCAGGACAGCC | ||
| Myf5 | 1 | 5′-ATGCCATCCGCTACATTGAGAGCC |
| 5′-TAGATAAGTCTGGAGCTGGAGGG | ||
| 2 | 5′-GAGGGAACAGGTGGAGAACTATTA | |
| 5′-CCGGGGTAGCAGGCTGTGAGTTG | ||
| Mef2A | 1 | 5′-CAGTACGCTAGCACTGACATGG |
| 5′-TCCGACAAAGGATTAGGGCTGG | ||
| 2 | 5′-TAACGAGCCTCATGAAAGCAGG | |
| 5′-GACATTGAGAAGTTCTGAGGTGG | ||
| Six1 | 1 | 5′-GTGGAGGCCGAGAAACTTCGC |
| 5′-CAGAGGAGAGAGTTGATTCTGC | ||
| 2 | 5′-TGCGCCGAAAATTCCCGTTGCC | |
| 5′-TATTGTTTTCGGTGTTCTCC |
Nucleotides complementary to the gene sequences are given in capital letters; the sequences to introduce EcoRI and XbaI restriction sites for cloning are given in lowercase.
Multiplex RT-PCR.
Total RNA from mouse embryos was extracted using TRIzol (Invitrogen) and treated with 5 U of DNase I (Worthington) prior to RT-PCR. First-strand synthesis and nested PCR were performed according to reference 17, except that 0.5 μl of each pair of gene-specific primers (300 ng/μl) was used in place of oligo(dT). For the first round of PCR, 40 μl of a mixture containing 1× PCR buffer (Takara Bio Inc.), 1 mM deoxynucleoside triphosphates (dNTP), and 1.25 U Taq were added to the RT reaction mixture. One μl of the first-round PCR products was further amplified using nested gene-specific primers. Both reactions were extended for 35 cycles (95°C for 1 min, 60°C for 1 min, and 72°C for 2 min). Primer sequences are given in Table 1.
Single-cell RT-PCR.
Single cells were deposited in 96-well PCR plates as described previously (17) and lysed on ice for 15 min. Cell lysates were reverse transcribed using several pairs of primers (15 ng/μl of each pair) and 48 U MMLV-RT per reaction. Nested PCR was performed as described above.
DNase I and restriction endonuclease digestions.
Nuclei were prepared from C2C12 myotubes, and DNase I digestions were performed according to reference 8. For restriction enzyme digestions, 6.5 × 106 nuclei were resuspended in 300 μl of the relevant restriction enzyme buffer and 200 U DraI or 100 U AflIII and incubated for 1 h at 37°C.
Southern blotting.
Twenty-five μg of DNA was digested with the restriction enzymes indicated and resolved on a 0.8% agarose gel, and Southern blotting was performed according to reference 3. The myo1 probe was a 358-bp HinfI-EcoRI fragment from the −4.2myo wt plasmid, and the myo2 probe was generated by PCR using the primers 5′-GAGATTGTCTGTCAGGCTGGG-3′ and 5′-GGCACTCATGTCTCTCAAACGG-3′ to amplify positions 1763 to 2107 of the Myogenin gene. The transgene was detected using a 300-bp EcoRI/ScaI fragment corresponding to the rabbit β-globin poly(A) tail (37). Hybridization was at 60°C overnight.
Knockdown of Six1.
C2C12 cells (7.5 × 104) were seeded into a 35-mm dish and cultured overnight prior to transfection with 350 pmol of each small interfering RNA (siRNA) against Six1 (24, 50) and 200 pmol of siGLO green transfection indicator (Dharmacon) using Dharmafect 3 transfection agent. One day later, the cells were transferred to DM and cultured for 48 h prior to the purification of the fluorescently labeled cells by flow cytometry. RNA and DNA were prepared as described above. Six1 expression was analyzed via RT-PCR using the primers described in Table 1; demethylation was analyzed by bisulfite treatment as described above.
RESULTS
The Myogenin regulatory region becomes demethylated during myogenesis.
To investigate if CpG methylation plays a role in restricting Myogenin gene activation during development, we first asked if the mouse Myogenin promoter is methylated or nonmethylated prior to activation. We therefore examined the methylation status of the Myogenin promoter in somites from e9.5 embryos. Somites develop from the presomitic mesoderm, and a new somite is formed every 1.5 to 2 h. In embryos at e9.5, Myogenin is expressed in the myotome of somites in a rostro-caudal pattern, where the most rostral or anterior somites express Myogenin but the most caudal or posterior somites have not yet activated the gene (49 and Fig. 1A). To ensure that embryos at the same developmental stage were examined, we used e9.5 embryos from transgenic mice in which β-galactosidase expression is under the control of the Myogenin 5′ regulatory region (49). Following the dissection of the anterior and posterior somites for analysis, central somites were stained for β-galactosidase expression; only embryos in which the most posterior somites of the central section stained negatively for β-galactosidase expression (and thus still had not activated the Myogenin promoter) were used.
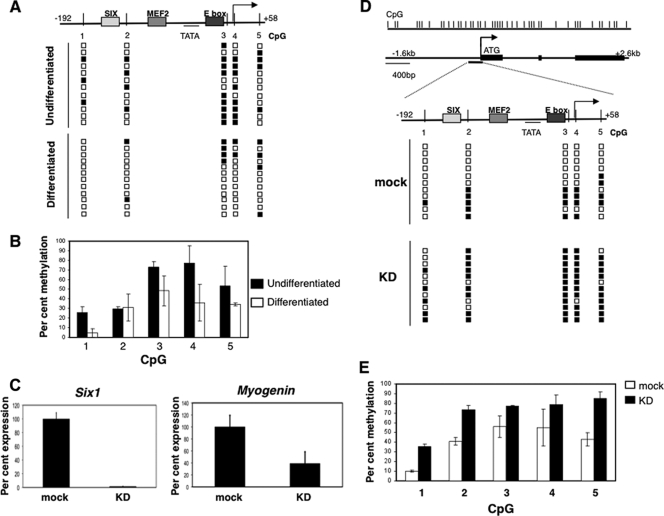
Bisulfite sequencing showed that the Myogenin minimal promoter is mostly methylated in e9.5 posterior somites (except the CpG closest to the proximal E box). However, in anterior somites where the Myogenin gene is expressed, it has become partially demethylated (Fig. 1A and B). A more detailed analysis of the data, from three independent experiments, reveals that all five CpGs in the promoter have become demethylated in some clones, whereas in others most CpG dinucleotides remain modified. Since it is difficult to isolate a totally pure population of e9.5 somites, we suggest that some of the methylated clones originated from nonsomitic tissue and thus that the level of demethylation in somites is likely to be greater than that indicated in Fig. 1.
We next examined the methylation status of the Myogenin promoter in neonatal tongues, a tissue composed almost exclusively of skeletal muscle cells of somitic origin and where high levels of Myogenin expression are observed. Bisulfite analysis showed that the promoter is unmethylated at all CpGs surrounding the transcription factor binding sites (Fig. 1C). Taken together, these data show a partial (at least 30%) demethylation of Myogenin promoter during the process of somitogenesis that is complete by the time the skeletal muscle of neonatal mice is formed (Fig. 1D).
SIX and MEF2 binding sites are required for chromatin opening.
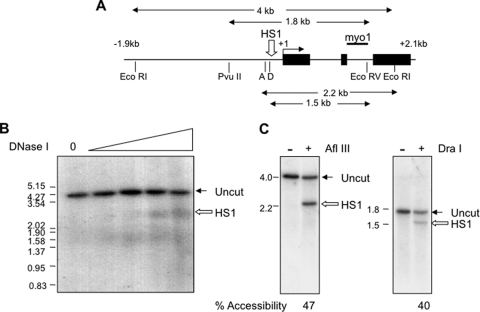
We next wished to examine whether DNA methylation influences the developmental activation of the Myogenin gene. The promoters of active genes are associated with nuclease-hypersensitive sites (HS), and we therefore asked if DNA methylation influences the formation of the HS at the Myogenin promoter. To this end, we used the myoblast cell line C2C12, which can be induced to differentiate to myotubes upon serum starvation (48). These cells faithfully recapitulate the activation of the Myogenin gene during differentiation and offer an amenable system for the subsequent mechanistic analysis of HS formation. Following the differentiation of C2C12 cells in low-concentration serum for 2 days, a hypersensitive site, HS1, is detected at the Myogenin promoter (Fig. 2 A and B). Furthermore, HS1 accessibility could be quantitatively measured by digesting C2C12 nuclei with the restriction enzymes AflIII and DraI. These cut at the 5′ end and at a central position of HS1, respectively, and, using amounts of enzyme that had been titrated to give maximal cutting, we find that HS1 is between 40 and 47% accessible to these enzymes in differentiated C2C12 cells (Fig. 2C).
FIG. 2.
Accessibility of the Myogenin promoter to DNase I and restriction enzymes in differentiated C2C12 myotubes. (A) Map of the mouse Myogenin gene from kb −1.9 to +2.3. Black boxes indicate exons and the horizontal bar, labeled myo1, indicates the probe. EcoRI, EcoRV, PvuII, AflIII (A), and DraI (D) restriction sites are shown below the gene. (B) DNase I-hypersensitive site in the Myogenin promoter. C2C12 cells were differentiated for 48 h, and genomic DNA was digested with EcoRI to map the DNase I-hypersensitive site HS1. Marker sizes (in kb) are shown to the left of the gel. (C) Accessibility of HS1 to restriction enzymes. Nuclei from differentiated C2C12 cells were harvested and digested with AflIII or DraI. Genomic DNA was analyzed as described for panel B, except that DraI accessibility was assessed using EcoRV and PvuII to generate the parental fragment.
We had previously shown that tissue-specific transcription factors additively increase the probability of HS formation when the DNA is unmethylated (8). Myogenin expression can be achieved using only 133 bp of the 5′ regulatory DNA sequence that contains binding sites for the bHLH family of MRFs and for factors of the MEF2 and SIX families of proteins (49). We therefore wanted to ask which of these factors is required to generate HS1 and, second, if DNA methylation affects its formation.
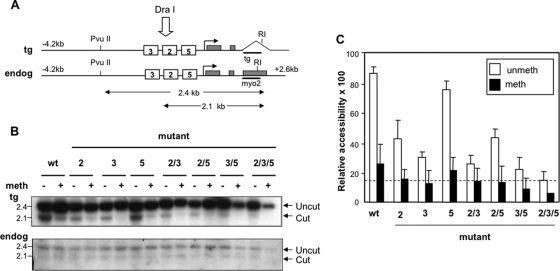
Thus, the factor binding sites within the promoter were mutated in all possible combinations; the constructs then were methylated or mock methylated in vitro with SssI methyltransferase, followed by stable transfection into C2C12 cells. Nuclei were harvested from differentiated C2C12 myotubes, and the accessibility of the promoter to DraI was determined. In all cases, the accessibility of HS1 of the endogenous gene was measured as an internal control, and the level of the accessibility of the test constructs was calculated relative to this. As can be seen in Fig. 3, when the constructs are unmethylated, the mutation of either the MEF2 (2) or SIX (3) binding site decreases HS formation by more than 50%, whereas the mutation of the proximal E box (5) alone resulted in a level of accessibility that was similar to that of the wild-type construct, suggesting that binding to this site has, at best, a minor effect on hypersensitive site formation. In contrast, the methylation of the wild-type construct (black bars) markedly decreases HS formation, and the mutation of either the MEF2 or SIX binding site together with DNA methylation causes a substantial reduction in HS1 formation. Indeed, if we consider the level of cutting when all three transcription factor binding sites are mutated to be the background (which we propose is due to restriction enzyme cleavage in the linker region between nucleosomes, denoted by the dashed line Fig. 3C), then DNA methylation decreases restriction enzyme cutting to this background level for most constructs. Accessibility above the background level was reproducibly observed only when binding sites for both MEF2 and SIX were intact. This is consistent with the idea that MEF2A and SIX1 need to cooperate to increase the probability of the hypersensitive site being formed when the promoter is methylated. However, since there is a small variation in the level of the cutting of the methylated constructs around the background level (most likely due to the difficulty in accurately quantifying the low level of accessibility), we cannot absolutely exclude the possibility that an individual factor is capable of triggering hypersensitive site formation in some cells in the population with a very low probability.
FIG. 3.
Methylation of the Myogenin promoter decreases the probability of hypersensitive site formation. (A) Diagram of the endogenous Myogenin gene (endog) and transgene (tg). White boxes represent the binding sites for bHLH (5), MEF2 (2), and SIX factors (3), and gray boxes indicate exons. The last exon in the transgene was replaced with the rabbit β-globin poly(A) tail. Cutting sites for DraI, PvuII, and EcoRI are indicated, and the sizes of the parental and DraI-cut bands are shown beneath the diagram (not to scale). The probes (tg and myo2) are indicated by horizontal bars. (B) Accessibility of the methylated and nonmethylated Myogenin promoter. C2C12 cells were stably transfected with mock methylated (−) or methylated (+) wild-type (wt) Myogenin construct or with constructs in which the factor binding sites were mutated. Following differentiation for 6 days, nuclei were digested with DraI; genomic DNA was cut with EcoRI and PvuII. Marker sizes (in kb) are given to the left of the gel. (C) Graph showing the relative accessibility of the transgene promoter for each cell line, normalized to the accessibility of the endogenous gene via the calculation [transgene cut/(transgene cut + uncut)]/[endogenous cut/(endogenous cut/(endogenous cut + uncut)] × 100. Error bars show standard errors from three independent experiments. The dotted line shows the level of accessibility when all factor binding sites are mutated and is considered to be the background level of accessibility due to restriction enzyme cutting in between nucleosomes (3).
Methylation decreases the number of cells that form the HS.
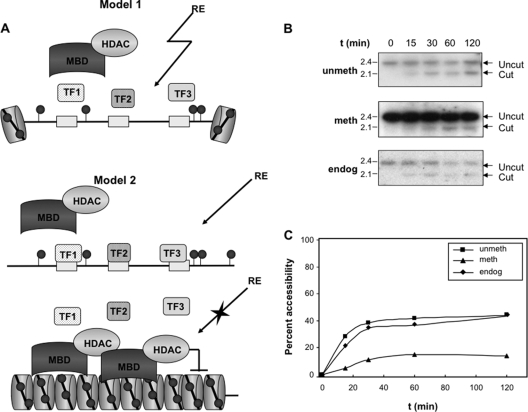
These data indicate that DNA methylation decreases the probability of HS formation, but it does not abolish it completely when the MEF2 and SIX sites both are intact. Our previous studies using nonmethylated constructs showed that when the transcription factor binding sites are mutated, the HS forms in fewer cells, but in the cells where the HS is formed, it is fully accessible (8). However, this is not necessarily the case when the promoter is methylated; the persistent binding of methylated DNA binding proteins might create a sterically less accessible HS to reduce the level of cutting by restriction enzymes (Fig. 4 A, model 1). To test this possibility, we performed a time course of restriction digestion. Using cell lines transfected with the methylated and unmethylated wild-type Myogenin promoter, we find that the methylated hypersensitive site is digested with the same kinetics as the nonmethylated site but that the overall level of digestion is lower (Fig. 4B and C). This suggests that DNA methylation decreases the probability of HS formation by decreasing the number of cells in which it is formed. However, in the cells where it is formed, it is fully accessible. Taken together with the data in Fig. 3, these findings indicate that binding to the MEF2 and SIX binding sites plays an important role in the generation of HS1, and that these factors can trigger full chromatin opening at HS1 even when promoter CpG dinucleotides are methylated. Moreover, since both MEF2 and SIX1/4 usually are required for detectable HS formation when the promoter is methylated, the coexpression of both factors may promote the activation of the methylated Myogenin promoter during development.
FIG. 4.
Methylation of the Myogenin promoter decreases the number of cells in which the hypersensitive site is formed. (A) Models of hypersensitive site formation. Model 1 proposes that the nucleosome, methylated DNA binding proteins (MBD), and transcription factors bind the promoter simultaneously to slow cutting by the restriction enzyme but do not completely prevent its accessibility. Model 2 proposes an all-or-none mechanism in which the hypersensitive site is either fully formed (upper) or completely inaccessible (lower). (B) Untransfected C2C12 cells and those transfected with either the unmethylated or the methylated wild-type constructs were differentiated for 6 days. Nuclei then were harvested and digested with DraI for the times indicated, and genomic DNA was prepared and analyzed as described for Fig. 3. Marker sizes (in kb) are given to the left of the gel. (C) Graph showing the percent accessibility of the endogenous locus (diamonds) and unmethylated (squares) and methylated (triangles) transgenes following cutting by DraI.
Posterior somites in e9.5 embryos coexpress MEF2A and SIX1 prior to Myogenin activation.
We next examined the expression profile of the MRFs, Mef2 and Six family members, during embryonic myogenesis. To this end, we performed multiplex RT-PCR analysis in single cells (17) sorted from anterior and posterior somites from e9.5 embryos. Tissues were disaggregated, and individual cells were deposited into 96-well plates by flow cytometry. The cells were lysed and reverse transcribed using several primer pairs, and the reaction then was split into new 96-well plates where nested PCR was performed using the gene-specific primers shown. Figure 5 A shows a representative single-cell RT-PCR experiment. Consistent with its broader expression pattern, we find that Mef2A is expressed in all cells from both anterior and posterior somites. The expression of Six1 is restricted mainly to the dermomyotome and myotome compartments of the somites and to the presomitic mesoderm immediately posterior to the most recently formed somite (27). In our single-cell analysis, we find that 30 to 40% of anterior and posterior somitic cells coexpress Mef2A and Six1. Therefore, the very factors that seem to be required to trigger measurable HS formation at the methylated Myogenin promoter are coexpressed in posterior somites where the Myogenin promoter is methylated, thus raising the possibility that these factors prime the Myogenin promoter for activation.
FIG. 5.
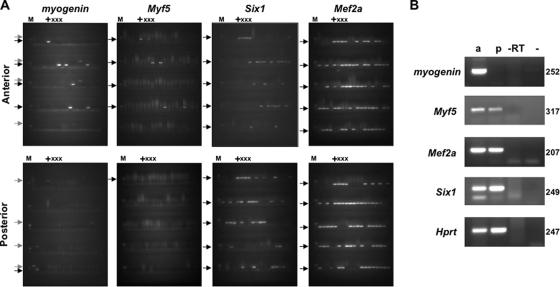
Expression of Mef2A, Six1, Myf5, and Myogenin in e9.5 anterior and posterior somites. (A) Expression in single cells. Single cells from anterior and posterior somites (upper and lower panels, respectively) were deposited into single wells of a 96-well plate by flow cytometry. Following multiplex RT-PCR using the primers for the specific genes shown in the second round, the samples from the individual wells were loaded onto the 96 lanes of the gel shown. The first lane in each panel shows the molecular weight marker (M). Lanes 2 to 5 are from wells that contain no cells, whereas lane 6 is from a well that contains 100 cells (+) and lanes 7 to 9 are from wells that contain 10 cells each (x). All other lanes are from wells that contain one cell each. Black arrows indicate RT-PCR products. The presence of a higher-molecular-weight band in the Myogenin RT-PCR product is due to contamination with genomic DNA and is indicated by lighter arrows. The fastest-migrating band in the gel of the Myf5 RT-PCR products is the primers. (B) Multiplex RT-PCR analysis in anterior (lane 1) and posterior (lane 2) somites of e9.5 embryos, using oligonucleotides to amplify Myogenin, Myf5, Mef2A, Six1, and Hprt cDNAs. Lane 3 does not contain reverse transcriptase in the RT reaction mixture, whereas lane 4 does not contain RNA (negative controls). The sizes of the products (in bp) are given to the right of each panel.
In contrast to Mef2A and Six1, the expression of Myf5 is very different between anterior and posterior somites at this developmental stage. This factor is first expressed in posterior somites (Fig. 5B) but the number of cells expressing it is very low, since we could detect it only in wells containing 100 cells in some experiments (Fig. 5A). Consistent with the requirement of MYF5 for transcription (49), Myogenin expression is not detected in posterior somites in our single-cell analysis. In anterior somites, however, Myf5 is detected in about 10% of the cells, and most of the Myf5-positive cells also express Myogenin (Fig. 5A). Taken together with the data from C2C12 cells, these studies suggest an ordered progression of Myogenin activation in which binding to the MEF2 and SIX binding sites of the methylated Myogenin promoter in posterior somites increases the probability of HS1 formation. We further propose that the subsequent expression of Myf5, primarily in the more mature anterior somites, leads to the full activation and transcription of the demethylated promoter. These studies also provide the first analysis of gene activation during embryogenesis at the single-cell level.
SIX1 is required for demethylation of the Myogenin promoter.
Notably, the number of somites that coexpress MEF2A and SIX1 (30 to 40%) is similar to the percentage of clones that are fully demethylated (33%) in anterior somites (Fig. 1B). Since these transcription factors, in combination, increase the probability of hypersensitive site formation at the methylated Myogenin promoter (Fig. 3), and since Myogenin activation likely occurs sequentially as cells mature from posterior into anterior somites, we next wished to investigate if SIX1 cooperates with MEF2A to bring about not only HS1 formation but also the observed demethylation.
Mef2A is broadly expressed, and since the Myogenin promoter remains methylated in non-muscle cells (25), it is unlikely that this factor alone is responsible for demethylation. Instead, it seemed possible that the coexpression of Six1 is required. To test this hypothesis, we made use of the fact that the Myogenin promoter becomes demethylated as C2C12 cells differentiate from myoblasts to myotubes (Fig. 6 A and B). We therefore performed RNA interference (RNAi) against Six1 using two distinct siRNAs (24, 50), followed by the differentiation of C2C12 cells to myotubes. Quantitative PCR showed that Six1 had been knocked down to <1% of the levels in mock-transfected cells (Fig. 6C), while control experiments showed that C2C12 cells differentiate to myotubes in the absence of Six1 (data not shown) and that a marker of differentiation still is expressed (Fig. 6C). Consistent with our hypothesis that SIX1 plays a pivotal role in regulating the demethylation of the Myogenin promoter, we find that following the knockdown of Six1, many of the promoter CpGs remain methylated, whereas in mock-treated cells the methylation of the Myogenin promoter is reduced by about 30% (Fig. 6D and E).
FIG. 6.
SIX1 regulates the demethylation of the Myogenin promoter. (A) The Myogenin promoter is demethylated upon the differentiation of C2C12 cells. The level of the methylation of the Myogenin promoter from differentiated and undifferentiated C2C12 cells from one representative experiment. (B) The percent methylation of promoter CpGs, calculated as described for Fig. 1. The data represent the averages from two independent experiments; error bars indicate the standard errors. (C) Knockdown of Six1. The level of Six1 expression was determined in cDNA prepared from cells mock transfected or transfected with siRNA against Six1 via quantitative PCR, normalizing to the level of Hprt expression; Myogenin expression was used as a control to check that the cells had differentiated. (D) Knockdown of Six1 prevents demethylation of the Myogenin promoter. C2C12 cells were differentiated for 48 h following the knockdown of Six1 (KD) or mock knockdown (mock), and the level of methylation was determined by the bisulfite sequencing of individual clones. The diagram shows the methylation of 11 (mock) and 12 (KD) representative clones. (E) The graph shows the percent methylation calculated from sequencing >20 clones of each type from two independent experiments. While the trend in methylation changes in the C2C12 cell line reflects those that occur in primary tissues in vivo, the methylation of CpG 3 is considerably higher, and we suggest this is due to differences in DNA methylation between cell lines and primary tissues that have been described previously (1).
DISCUSSION
Transcription activators are expressed in a range of cell types but activate only a subset of their potential target genes in any given cell. Various models have been proposed to explain this phenomenon, including the requirement for the correct combination of factors for gene activation, the presence of cell-specific coactivators, and the absence of repressors (21). Here, we have examined an alternative hypothesis, namely, that epigenetic modifications influence the access of transactivators to their binding sites and subsequent chromatin activation. Using a promoter with a weak CpG island as a model, we show that DNA methylation substantially reduces promoter-hypersensitive site formation. However, it does not abolish it. Instead, our data suggest that DNA methylation increases the number of transactivators typically required for measurable hypersensitive site formation. In turn, this implies that DNA methylation helps to prevent illegitimate gene activation and thus restrict expression to the correct developmental stage.
Indeed, our mutagenesis studies indicate that when the Myogenin promoter is nonmethylated, binding to the SIX binding site alone generates a hypersensitive site with 50% of the probability of the wild-type enhancer (the 2/5 mutant shown in Fig. 3B). This implies that in the absence of promoter methylation, the hypersensitive site would be formed in 50% of the cells that express Six1. DNA methylation restricts detectable promoter activation to cells where both MEF2 proteins and SIX1/4 are coexpressed.
Methylation not only increases the requirement for both transcription factors but also significantly reduces the level of hypersensitive site formation. In turn, this suggests that the efficient activation of the Myogenin promoter requires demethylation following binding to the MEF2 and SIX binding sites. Thus, while binding to the MEF2 and SIX binding sites can generate the hypersensitive site, there could be continued competition between these transcription factors and the methylated DNA binding proteins. In this scenario, demethylation would be required to tip the balance in favor of an active promoter. It is unlikely that this demethylation is needed to directly allow MRFs to bind to the proximal E box: DNA methylation does not affect binding to this site (data not shown), and the CpG adjacent to the proximal E box is barely methylated even in posterior somites. Instead, our knockdown studies indicate that SIX1 is required to recruit the demethylation machinery, suggesting that in the cells that coexpress MEF2A and SIX1, these factors increase the probability of an active chromatin conformation being formed for subsequent transcription activation.
Two previous studies have suggested that DNA methylation outside of promoter regions can lead to an altered chromatin structure and the repression of transcription (16, 20). However, this does not appear to be the mechanism of the developmental repression of the Myogenin promoter. First, if methylation outside the promoter region caused the formation of a repressive chromatin structure, we would predict that methylation causes an equivalent reduction in restriction enzyme accessibility for every mutated Myogenin promoter. However, this is not what we observe (Fig. 3). Second, recent genome-wide studies have demonstrated that transcription leads to a high level of gene body methylation and low CpG methylation around the transcription start sites (2, 22, 41). This high level of gene body methylation has not been found to lead to transcriptional repression, as would be expected if non-promoter methylation triggers repression. Likewise, previous studies showed that DNA methylation within 100 bp of the transcriptional start site is required to mediate repression (26); this is consistent with the idea that MBDs need to be in close proximity of the transcription start site to prevent transcription factor binding. We suggest that our data can be explained by such a model, where the binding of MBDs to the methylated promoter leads to competition with transcription factors to thus modulate hypersensitive site formation.
Demethylation before transcription.
There has been considerable debate as to whether DNA methylation patterns are a cause or a consequence of transcription. For example, studies by the Bestor laboratory have shown that the active chromatin mark, H3K4 trimethylation, modulates de novo DNA methylation by preventing the binding of DNMT3L and the subsequent recruitment of the de novo methyltransferase DNMT3A2 (29); this suggests that in some circumstances the DNA methylation pattern is a consequence of transcription. While this could play a role in regulating the targeting of de novo methylation, this is unlikely to be the mechanism by which DNA methylation is lost from previously methylated promoters. Indeed, we find that in anterior somites at e9.5, the level of demethylation (33%) is higher than the percentage of cells that express Myogenin (10%). This suggests that in our case, demethylation precedes transcriptional activation.
Other studies also have implied that transcription is not required for demethylation. For example, the induction of estrogen receptor binding to the avian vitellogenin promoter triggered the loss of methylation prior to gene transcription (19), and the removal of methyl groups from both strands of the α-actin regulatory region was found to be a prerequisite for actin gene transcription (31). Likewise, more recent studies have shown that the binding of the glucocorticoid receptor to the rat tyrosine aminotransferase promoter is sufficient to induce demethylation, which then allows the binding of other transcription activators and chromatin remodelling (44). Similarly, the analysis of chromatin reorganization in the chicken lysozyme transgene locus during macrophage differentiation showed that the transient binding of some transcription factors and DNA demethylation occur before the onset of gene expression (23, 42). Thus, consistent with our observations, these studies suggest that the binding of specific transcription factors is the prime trigger for demethylation. The mechanism by which demethylation is achieved, however, remains controversial (28).
SIX and MEF2A establish an active chromatin state.
Our data unveil a novel and unexpected role of both MEF2 and SIX proteins in opening the Myogenin promoter. The mutation of either site decreases hypersensitive site formation by more than 50% in C2C12 cells; this, together with our single-cell analysis of developing mouse embryos, strongly suggests that the initial chromatin remodelling in posterior somites, before Myf5 expression is upregulated, depends on the coordinate action of these two families of proteins. Although the mechanism remains to be fully determined, one possibility is that MEF2 and SIX proteins directly recruit chromatin remodelling complexes for promoter opening. Consistent with this hypothesis, it has been shown previously that under certain conditions the SWI/SNF remodelling complex can be recruited to the Myogenin promoter in a complex with MEF2 and independently of the MRFs (36).
Despite being necessary for the transcription of the Myogenin gene (49), our results indicate that the proximal E box does not play an important role in hypersensitive site formation. At first this appears to contradict previous studies using MYOD-ER-transformed fibroblasts that showed that MYOD recruits chromatin remodellers for hypersensitive site formation (13). Notably, however, MYOD is recruited through its interaction with the homeodomain protein PBX, which is constitutively bound to the Myogenin promoter upstream of the SIX binding site (5). Thus, in this fibroblast system, the E box is required only after hypersensitive site formation for stable MYOD/DNA binding and transcription activation. MyoD is not expressed until e10.5, (9) and in e9.5 somites the MRF that activates Myogenin is most likely MYF5 (49). Whether MYF5 recruits chromatin remodellers in a way similar to that of MYOD in fibroblasts to trigger a further wave of chromatin remodelling remains to be determined. However, it also is possible that in this normal developmental context, the prior chromatin remodelling triggered by MEF2 and SIX proteins is sufficient to allow MYF5 binding.
Taken together, our studies uncover a further layer in the regulation of the ordered activation of the Myogenin promoter. Using e9.5 somites to take a developmental snapshot of gene activation and genetic studies of C2C12 cells, we propose that DNA methylation helps to restrict Myogenin activation until both MEF2A and SIX1 are coexpressed. The binding of SIX1 then is required for demethylation and promoter chromatin activation. Since 12% of tissue-specific genes have weak CpG island promoters, we suggest this level of control might also function to prevent the premature or inappropriate activation of a substantial number of other tissue-specific genes.
Acknowledgments
We thank Ian Titley for flow cytometry and Gill May for advice on multiplex RT-PCR.
D.S. and D.P. were supported by funds from ICR, and J.B. gratefully acknowledges support from fellowships from the Lister Institute of Preventive Medicine and the University of Leeds.
Footnotes
Published ahead of print on 24 May 2010.
REFERENCES
- 1.Antequera, F., J. Boyes, and A. Bird. 1990. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 62:503-514. [DOI] [PubMed] [Google Scholar]
- 2.Ball, M. P., J. B. Li, Y. Gao, J. H. Lee, E. M. LeProust, I. H. Park, B. Xie, G. Q. Daley, and G. M. Church. 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 27:361-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann, M., A. Mamais, F. McBlane, H. Xiao, and J. Boyes. 2003. Regulation of V(D)J. recombination by nucleosome positioning at recombination signal sequences. EMBO J. 22:5197-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berghella, L., L. De Angelis, T. De Buysscher, A. Mortazavi, S. Biressi, S. V. Forcales, D. Sirabella, G. Cossu, and B. J. Wold. 2008. A highly conserved molecular switch binds MSY-3 to regulate myogenin repression in postnatal muscle. Genes Dev. 22:2125-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkes, C. A., D. A. Bergstrom, B. H. Penn, K. J. Seaver, P. S. Knoepfler, and S. J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465-477. [DOI] [PubMed] [Google Scholar]
- 6.Bird, A. 1992. The essentials of DNA methylation. Cell 70:5-8. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. P. 1986. CpG-rich islands and the function of DNA methylation. Nature 321:209-213. [DOI] [PubMed] [Google Scholar]
- 8.Boyes, J., and G. Felsenfeld. 1996. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 15:2496-2507. [PMC free article] [PubMed] [Google Scholar]
- 9.Buckingham, M. 1992. Making muscle in mammals. Trends Genet. 8:144-148. [DOI] [PubMed] [Google Scholar]
- 10.Buckingham, M., L. Bajard, T. Chang, P. Daubas, J. Hadchouel, S. Meilhac, D. Montarras, D. Rocancourt, and F. Relaix. 2003. The formation of skeletal muscle: from somite to limb. J. Anat. 202:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eden, S., and H. Cedar. 1994. Role of DNA methylation in the regulation of transcription. Curr. Opin. Genet. Dev. 4:255-259. [DOI] [PubMed] [Google Scholar]
- 12.Frommer, M., L. E. McDonald, D. S. Millar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Molloy, and C. L. Paul. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U. S. A. 89:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerber, A. N., T. R. Klesert, D. A. Bergstrom, and S. J. Tapscott. 1997. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 11:436-450. [DOI] [PubMed] [Google Scholar]
- 14.Giordani, J., L. Bajard, J. Demignon, P. Daubas, M. Buckingham, and P. Maire. 2007. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc. Natl. Acad. Sci. U. S. A. 104:11310-11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holliday, R., and J. E. Pugh. 1975. DNA modification mechanisms and gene activity during development. Science 187:226-232. [PubMed] [Google Scholar]
- 16.Hsieh, C. L. 1997. Stability of patch methylation and its impact in regions of transcriptional initiation and elongation. Mol. Cell. Biol. 17:5897-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, M., D. Krause, M. Greaves, S. Sharkis, M. Dexter, C. Heyworth, and T. Enver. 1997. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11:774-785. [DOI] [PubMed] [Google Scholar]
- 18.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 19.Jost, J. P., H. P. Saluz, and A. Pawlak. 1991. Estradiol down regulates the binding activity of an avian vitellogenin gene repressor (MDBP-2) and triggers a gradual demethylation of the mCpG pair of its DNA binding site. Nucleic Acids Res. 19:5771-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kass, S. U., N. Landsberger, and A. P. Wolffe. 1997. DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol. 7:157-165. [DOI] [PubMed] [Google Scholar]
- 21.Laslo, P., C. J. Spooner, A. Warmflash, D. W. Lancki, H. J. Lee, R. Sciammas, B. N. Gantner, A. R. Dinner, and H. Singh. 2006. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126:755-766. [DOI] [PubMed] [Google Scholar]
- 22.Laurent, L., E. Wong, G. Li, T. Huynh, A. Tsirigos, C. T. Ong, H. M. Low, K. W. Kin Sung, I. Rigoutsos, J. Loring, and C. L. Wei. 2010. Dynamic changes in the human methylome during differentiation. Genome Res. 20:320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefevre, P., S. Melnik, N. Wilson, A. D. Riggs, and C. Bonifer. 2003. Developmentally regulated recruitment of transcription factors and chromatin modification activities to chicken lysozyme cis-regulatory elements in vivo. Mol. Cell. Biol. 23:4386-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, X., K. A. Oghi, J. Zhang, A. Krones, K. T. Bush, C. K. Glass, S. K. Nigam, A. K. Aggarwal, R. Maas, D. W. Rose, and M. G. Rosenfeld. 2003. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426:247-254. [DOI] [PubMed] [Google Scholar]
- 25.Lucarelli, M., A. Fuso, R. Strom, and S. Scarpa. 2001. The dynamics of myogenin site-specific demethylation is strongly correlated with its expression and with muscle differentiation. J. Biol. Chem. 276:7500-7506. [DOI] [PubMed] [Google Scholar]
- 26.Murray, E. J., and F. Grosveld. 1987. Site specific demethylation in the promoter of human gamma-globin gene does not alleviate methylation mediated suppression. EMBO J. 6:2329-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver, G., R. Wehr, N. A. Jenkins, N. G. Copeland, B. N. Cheyette, V. Hartenstein, S. L. Zipursky, and P. Gruss. 1995. Homeobox genes and connective tissue patterning. Development 121:693-705. [DOI] [PubMed] [Google Scholar]
- 28.Ooi, S. K., and T. H. Bestor. 2008. The colorful history of active DNA demethylation. Cell 133:1145-1148. [DOI] [PubMed] [Google Scholar]
- 29.Ooi, S. K., C. Qiu, E. Bernstein, K. Li, D. Jia, Z. Yang, H. Erdjument-Bromage, P. Tempst, S. P. Lin, C. D. Allis, X. Cheng, and T. H. Bestor. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oswald, J., S. Engemann, N. Lane, W. Mayer, A. Olek, R. Fundele, W. Dean, W. Reik, and J. Walter. 2000. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10:475-478. [DOI] [PubMed] [Google Scholar]
- 31.Paroush, Z., I. Keshet, J. Yisraeli, and H. Cedar. 1990. Dynamics of demethylation and activation of the alpha-actin gene in myoblasts. Cell 63:1229-1237. [DOI] [PubMed] [Google Scholar]
- 32.Potthoff, M. J., and E. N. Olson. 2007. MEF2: a central regulator of diverse developmental programs. Development 134:4131-4140. [DOI] [PubMed] [Google Scholar]
- 33.Reik, W., W. Dean, and J. Walter. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089-1093. [DOI] [PubMed] [Google Scholar]
- 34.Riggs, A. D. 1975. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 14:9-25. [DOI] [PubMed] [Google Scholar]
- 35.Sassoon, D., G. Lyons, W. E. Wright, V. Lin, A. Lassar, H. Weintraub, and M. Buckingham. 1989. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature 341:303-307. [DOI] [PubMed] [Google Scholar]
- 36.Serra, C., D. Palacios, C. Mozzetta, S. V. Forcales, I. Morantte, M. Ripani, D. R. Jones, K. Du, U. S. Jhala, C. Simone, and P. L. Puri. 2007. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell 28:200-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Severne, Y., S. Wieland, W. Schaffner, and S. Rusconi. 1988. Metal binding ‘finger’ structures in the glucocorticoid receptor defined by site-directed mutagenesis. EMBO J. 7:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spitz, F., J. Demignon, A. Porteu, A. Kahn, J. P. Concordet, D. Daegelen, and P. Maire. 1998. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl. Acad. Sci. U. S. A. 95:14220-14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stancheva, I., and R. R. Meehan. 2000. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 14:313-327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Summerbell, D., C. Halai, and P. W. Rigby. 2002. Expression of the myogenic regulatory factor Mrf4 precedes or is contemporaneous with that of Myf5 in the somitic bud. Mech. Dev. 117:331-335. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki, M. M., and A. Bird. 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9:465-476. [DOI] [PubMed] [Google Scholar]
- 42.Tagoh, H., S. Melnik, P. Lefevre, S. Chong, A. D. Riggs, and C. Bonifer. 2004. Dynamic reorganization of chromatin structure and selective DNA demethylation prior to stable enhancer complex formation during differentiation of primary hematopoietic cells in vitro. Blood 103:2950-2955. [DOI] [PubMed] [Google Scholar]
- 43.Tajbakhsh, S., E. Bober, C. Babinet, S. Pournin, H. Arnold, and M. Buckingham. 1996. Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev. Dyn. 206:291-300. [DOI] [PubMed] [Google Scholar]
- 44.Thomassin, H., M. Flavin, M. L. Espinas, and T. Grange. 2001. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 20:1974-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh, C. P., and T. H. Bestor. 1999. Cytosine methylation and mammalian development. Genes Dev. 13:26-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber, M., I. Hellmann, M. B. Stadler, L. Ramos, S. Paabo, M. Rebhan, and D. Schubeler. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 39:457-466. [DOI] [PubMed] [Google Scholar]
- 47.Wigler, M. H. 1981. The inheritance of methylation patterns in vertebrates. Cell 24:285-286. [DOI] [PubMed] [Google Scholar]
- 48.Yaffe, D., and O. Saxel. 1977. A myogenic cell line with altered serum requirements for differentiation. Differentiation 7:159-166. [DOI] [PubMed] [Google Scholar]
- 49.Yee, S. P., and P. W. Rigby. 1993. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 7:1277-1289. [DOI] [PubMed] [Google Scholar]
- 50.Yu, Y., E. Davicioni, T. J. Triche, and G. Merlino. 2006. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res. 66:1982-1989. [DOI] [PubMed] [Google Scholar]