Abstract
Reactive oxygen species are generated within peroxisomes during peroxisomal metabolism. However, due to technological difficulties, the intraperoxisomal redox state remain elusive, and the effect of peroxisome deficiency on the intracellular redox state is controversial. A newly developed, genetically encoded fluorescence resonance energy transfer (FRET) probe, Redoxfluor, senses the physiological redox state via its internal disulfide bonds, resulting in a change in the conformation of the protein leading to a FRET response. We made use of Redoxfluor to measure the redox states at the subcellular level in yeast and Chinese hamster ovary (CHO) cells. In wild-type peroxisomes harboring an intact fatty acid β-oxidation system, the redox state within the peroxisomes was more reductive than that in the cytosol, despite the fact that reactive oxygen species were generated within the peroxisomes. Interestingly, we observed that the redox state of the cytosol of cell mutants for peroxisome assembly, regarded as models for a neurological metabolic disorder, was more reductive than that of the wild-type cells in yeast and CHO cells. Furthermore, Redoxfluor was utilized to develop an efficient system for the screening of drugs that moderate the abnormal cytosolic redox state in the mutant CHO cell lines for peroxisome assembly without affecting the redox state of normal cells.
Peroxisomes are single membrane-bound organelles harboring at least one H2O2-generating oxidase and one H2O2-decomposing catalase, and they are present in virtually all eukaryotic cells, from yeast to mammals. The most conserved activity of peroxisomal metabolism is the β-oxidation of fatty acids (27).
Peroxisome assembly requires more than 20 PEX gene products, termed peroxins, in any given organism (5). The impairment of peroxisomal protein transport caused by mutations in PEX genes causes fatal human peroxisome biogenesis disorders (PBDs) (34). In the cells of such PBD patients, essential enzymes normally localized to peroxisomes are found mostly in the cytosol. Mammalian cell lines harboring mutations in peroxins (including fibroblasts from PBD patients) grow well in cell culture. On the other hand, pex mutants of the methylotrophic yeast Pichia pastoris can grow normally on glucose but not oleate or methanol (37).
Peroxisomal metabolic pathways can generate a high level of reactive oxygen species (ROS) (32). Therefore, peroxisomal disorders have been studied with a focus on the generation of ROS. However, the relationship between PBDs and the intracellular redox state is unclear (13, 32).
Peroxisomes have long been thought to be in a more highly oxidized state than the cytosol due to this generation of ROS. However, there is no reported experimental evidence supporting this notion. We previously identified a 20-kDa peroxisomal membrane protein, named Pmp20, in methanol-induced peroxisomes of methylotrophic yeasts. Pmp20 had a glutathione (GSH) peroxidase activity, suggesting the presence of glutathione within the peroxisomes (9). However, we and other groups of investigators have been unable to determine the levels of the reduced and oxidized forms of glutathione due to technical difficulties and therefore have been unable to assess the redox state within peroxisomes by conventional biochemical methods.
In general, the intracellular redox state is determined by the levels of redox-related metabolites that are generated by multiple metabolic pathways. (We herein refer to the “redox state” as an intracellular environment at steady state, which is distinct from oxidative stress or ROS, which functions as a signal for further intracellular events such as apoptosis.) Therefore, the redox state is considered to reflect the overall metabolic status. While the standard redox potential (E0′) is a general index used to express the redox state of a compound, it cannot be used to describe the intracellular redox state because it does not take into account various physiological considerations, such as the cytosol, where many compounds coexist in a mixture of various redox states (14). Therefore, the equilibrium redox state in living cells has been estimated from indices such as the ratio of oxidized and reduced forms of glutathione, from indirect indices of the redox state, such as the NAD(P)H ratio (12, 40), or from the level of the expression of antioxidant enzymes. However, the measurement of these indices often yields contradictory results, making it difficult to evaluate the physiological redox state using any single index. This situation might have led to misunderstanding the redox state in cells from patients with PBDs. Reductive conditions could occur during conditions of oxidative stress, when the ROS defense system is functioning normally.
With the aim of determining the intracellular redox state directly, we developed a fluorescent redox probe, Redoxfluor, with a novel sensing mechanism. Several green fluorescent protein (GFP) variants that report the in vivo redox state (roGFP [4, 7], rxYFP [18, 24, 25]) or H2O2 level (HyPer [3]) have been developed since the start of our research. However, none of these reporters have been used to visualize the redox state in mammalian cytosol, and differences in the redox potential between normal and pathological states have not been reported.
In the present work, we developed a Redoxfluor that discriminates the redox state of peroxisome assembly mutant cell lines (34) from that of the normal cell line. Our findings shed light on how to tackle problems with monitoring the spatiotemporal dynamics of the redox state within living mammalian cells and also should pave the way for the development of a screen for drugs that can affect various metabolic disorders with abnormal redox state.
MATERIALS AND METHODS
Construction of plasmids driving expression of Redoxfluor.
A recombinant gene encoding Redoxfluor (Fig. 1 A) was constructed as follows: the DNA fragment encoding Yap1 CRD (I601 to N650) DNA was amplified by PCR using either one of the two primer sets: (i) a forward primer containing an SphI site and a reverse primer containing sequences encoding a short linker (GG) and a ClaI site, or (ii) a forward primer containing a ClaI site and a reverse primer containing an SacI restriction site. The two PCR products were ligated, fused at the 5′ end to a DNA fragment encoding cerulean (29) (a version of enhanced cyan fluorescent protein, or CFP) and at the 3′ end to a DNA encoding citrine (6) (a version of enhanced yellow fluorescent protein, YFP) (Fig. 1A). The constructed Redoxfluor C probe DNA was cloned into the pRSETA-vector with N-terminal hexa-His tag (His6) (Invitrogen) for expression. The plasmid encoding Redoxfluor A probe was generated from the plasmid encoding Redoxfluor C probe using a QuikChange PCR-based mutagenesis kit (Stratagene).
FIG. 1.
Domain structure, schematic representation, and in vitro characterization of Redoxfluor. (A) Schematic drawing of Yap1 comprising a basic leucine-zipper DNA-binding domain (bZIP), an n-CRD (N279 to R313), and a c-CRD (N565 to N650). Redoxfluor comprises a fusion of cerulean, a tandemly repeated fragment of the Yap1 c-CRD (I601 to N650), and citrine. (B) Emission spectra of Redoxfluor before (black) and after (red) exposure to H2O2 (100 μM) at 434 nm excitation. (C) Responses of Redoxfluor C probe (red) and A probe (black) to various reagents. The purified probes were incubated with H2O2 (100 μM), ATZ (10 mM), or BSO (100 μM) until the emission intensities following 434 nm excitation reached constant values. The FRET ratios (YFP intensity/CFP intensity) are shown relative to those obtained from untreated control samples. The data represent the means ± standard deviations (n = 3).
Expression and purification of Redoxfluor.
Escherichia coli BL21(DE3) cells were transformed with the constructed expression plasmids encoding Redoxfluor A or C probe. Cultures of the transformants grown in L broth at log phase (optical density at 610 nm [OD610] of 0.5) were treated with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 12 h at 23°C to induce the expression of Redoxfluor. Cells were collected and lysed by sonication in buffer A (300 mM NaCl, 50 mM NaH2PO4, 10 mM imidazole, pH 8.0). His-tagged Redoxfluor was purified by two cycles of chromatography on a column of Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) according to the manufacturer's instructions. Proteins were quantified by Bradford's method with the use of a protein assay kit (Bio-Rad) with bovine serum albumin (BSA) used as the standard.
Redox titration of Redoxfluor in vitro.
The redox potentials of Redoxfluor were determined from its reaction with glutathione. Purified Redoxfluor (0.3 μM) was incubated in RT buffer (75 mM HEPES-KOH [pH 7.0], 140 mM NaCl, 1 mM EDTA) containing concentrations of GSH and glutathione disulfide (GSSG) that varied by 1 mM increments. After the equilibration of the protein samples for 20 h at 37°C, the fluorescence emission spectra were collected on an RF5300PC spectrofluorophotometer (Shimadzu Co. Ltd.) at 405 or 434 nm excitation. All manipulations were carried out in an anaerobic chamber (Coy Laboratory products).
Biochemical analysis of Redoxfluor with mPEG-maleimide.
For in vitro analysis, purified Redoxfluor proteins equilibrated with GSH-GSSG as described above were incubated with 10 mM (final concentration) of methoxy-poly (ethylene glycol)-maleimide (mPEG-maleimide) (Laysan Bio) at 30°C for 30 min. The sample was mixed with a 1/3 volume of SDS sample buffer (50 mM Tris-HCl, pH 6.8, 30% [vol/vol] glycerol, 3% [wt/vol] sodium dodecyl sulfate, 6% [vol/vol] 2-mercaptoethanol, and 60 mg/liter bromophenol blue) and boiled for 1 min. For in vivo assessments of redox states, CHO cells were cultured on 90-mm-diameter culture dishes to semiconfluence, washed twice with ice-cold phosphate-buffered saline (PBS), and lysed in 100 μl of ice-cold PBS containing 0.5% (vol/vol) of Tween 20 and Complete proteinase inhibitor cocktail (Roche). The protein concentration of the lysate was adjusted to 1 mg/ml in the same buffer used for cell lysis, and the samples (each 180 μl) were incubated with mPEG-maleimide and then SDS sample buffer as described above. The samples were subjected to SDS-PAGE with 0.5% (vol/vol) 2-mercaptoethanol contained in the running buffer, transferred to an Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore), and analyzed by immunoblot analysis using a 3,000-fold dilution of rabbit anti-GFP antiserum (Invitrogen).
Plasmids for expression of Redoxfluor, cells, and growth conditions.
The Saccharomyces cerevisiae strain used in this study was BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (23). For expression in S. cerevisiae, Redoxfluor was subcloned into the pRS416 CEN vector containing the GAP promoter and terminator. For expression in P. pastoris strain PPY12 (arg4 his4) (31), Redoxfluor was subcloned into the integrating pIB2 vector (33) containing the GAP promoter. The isogenic Pppex5Δ and Ppcta1Δ strains were generated by replacing the respective coding region with the Zeor gene. For mammalian expression, we chemically synthesized the Yap1 CRD region of Redoxfluor with the preferred codon usage and cloned the sequence into pIRESpuro3 (Clontech) downstream of a Kozak consensus sequence (16) (GCCGCCACCATG). For C probe-PTS1, the C probe was extended by PCR at the 3′ end with a sequence encoding the peroxisomal targeting signal (PTS1) (22). CHO cells were transfected with plasmid constructs using Lipofectamine 2000 (Invitrogen).
For the oxidative stress assay, S. cerevisiae and P. pastoris were grown at 28°C in SD medium (0.67% yeast nitrogen base and 2% glucose) supplemented with amino acids and transferred to the same medium containing either H2O2 (100 μM), 3-amino-1,2,4-aminotriazole (ATZ) (10 mM), or buthionine sulfoximine (BSO) (100 μM). For microscopic analyses of P. pastoris strains under peroxisome-inducing conditions, the cells were grown at 28°C for 16 h in SM medium (0.67% yeast nitrogen base without amino acids and 0.5% methanol) or YNO medium (0.67% yeast nitrogen base, 0.05% yeast extracts, and 0.5% oleate) supplemented with their auxotrophic amino acids (100 μg/ml). CHO cell lines were grown in Ham's F-12 medium supplemented with 10% (vol/vol) fetal calf serum (FCS) under 5% CO2 and 95% air.
Fluorescence microscopy.
We performed light microscopic imaging using a fluorescence-inverted microscope (IX70; Olympus) equipped with an Uplan Apo 100× magnification/1.35-numeric-aperture oil iris for yeast cells or LUCPlanFLN 40× magnification/0.60-numeric-aperture Ph2 dry iris objective lens for CHO cells using mirror/filter units XF88-2 (Omega Optical, Inc.) for CFP/YFP FRET and a U-MNIBA (Olympus) filter set for GFP. Images were captured with a charge-coupled device camera (Sensys; PhotoMetrics) and analyzed using the MetaMorph software package version 7.0 (Universal Imaging Corp). For the evaluation of FRET values of the cytoplasm or peroxisomes, more than 20 regions in each acquired FRET image were selected and subjected to region measurements using the above-mentioned software.
qRT-PCR.
Total RNA was isolated from cells at log phase using the RNeasy minikit (Qiagen) followed by on-column DNase digestion. cDNAs were synthesized from 1 μg total RNA using random primers (Promega) and ReverTra Ace (Toyobo). After reverse transcription for 50 min at 42°C, the samples were heated for 5 min at 99°C to terminate the reaction, and 0.5 μl of RNase H was added. Quantitative reverse transcription-PCR (qRT-PCR) was performed in 20-μl mixtures in glass capillary tubes in a LightCycler (Roche Diagnostic). Primers used for the reactions are listed in Table S1 in the supplemental material. Negative control PCRs were performed without ReverTra Ace addition.
Determination of glutathione.
CHO cells were grown to confluence in the presence or absence (control culture condition) of pharmacological agents for 24 h. P. pastoris cells were cultured to mid-log phase in YNO medium at 30°C. CHO and yeast cells were harvested by centrifugation, washed once with physiological saline, and disrupted with zirconia beads in 300 μl of ice-chilled 8 mM HCl solution. Cell homogenates were centrifuged at 12,000 × g at 4°C, and the amount of GSH and GSSG in the resultant supernatant was determined as previously described (35).
RESULTS
Development and characterization of Redoxfluor as a redox indicator.
The yeast transcriptional factor Yap1 senses the intracellular redox state via its carboxy-terminal cysteine-rich domain (c-CRD) (17), and the structure of c-CRD has been reported to reflect the equilibrated and steady-state physiological redox status (1). The Redoxfluor C probe (C probe) contains a tandem repeat of the partial sequence for c-CRD (Yap1 601-650) (Fig. 1A), and it mediates fluorescence resonance energy transfer (FRET) between cerulean (29) (CFP) and citrine (6) (YFP) (Fig. 1B). The exposure of the purified C probe to H2O2 enhanced the CFP emission at the expense of the YFP emission and decreased the yellow-to-cyan emission ratio (527/476 nm), indicating an H2O2-induced loss of FRET (Fig. 1B and C).
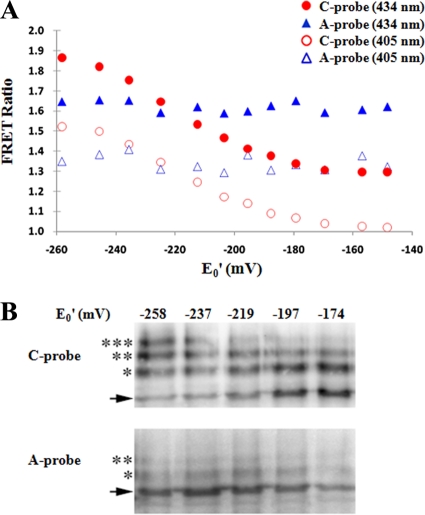
The purified C probe was used for the determination of the FRET ratio (the fluorescence intensity of YFP divided by that of CFP) in titration buffers containing various ratios of reduced (GSH) and oxidized (GSSG) glutathione (Fig. 2 A). Based on the Nernst equation with an E0 value of −240 mV for the GSH/GSSG redox couple (30), the redox midpoint potential of the C probe was determined to be −213 mV at pH 7.0 and 30°C, which was close to the value of the mammalian cytosolic redox potential (10). From the titration curve, the redox potential within the range of ∼−180 to −250 mV can be estimated from the FRET ratio (Fig. 2A). Under the conditions used for our experiment in vitro, this titration curve was unaffected by the probe concentration within a range of up to 1 μM (data not shown), which is estimated to be a much greater concentration than the level of the intracellularly expressed Redoxfluor and much lower than the intracellular GSH-plus-GSSH concentration (0.5 to 10 mM) (15). Thus, the effect of the intracellular expression of Redoxfluor on the redox state was assumed to be negligible.
FIG. 2.
Redox titration of Redoxfluor with reduced and oxidized glutathione in vitro. (A) Spectrophotometric analysis of C probe and A probe at 37°C in titration buffer containing different ratios of GSH/GSSG corresponding to the designated electropotential (E0′) gradient. The excitation wavelength was 405 or 434 nm. (B) mPEG-maleimide modification of Redoxfluor. Under the same titration conditions as those described for panel A, the purified Redoxfluor was incubated with mPEG-maleimide and subjected to immunoblot analysis. The arrows indicate the signals corresponding to the nonmodified form of the protein. The asterisks indicate signals corresponding to modified forms of the probe protein.
The redox-dependent FRET response of the C probe was supported by experiments using the control Redoxfluor A probe (A probe), in which all of the redox-sensitive cysteine residues in the C probe CRD had been replaced by alanine (Fig. 2A). The A probe did not show the C probe-like FRET response following exposure to oxidizing reagents, although we observed a slight change in FRET due to the inherent redox-responsive nature of the fluorescent protein. This CRD-independent FRET change was reduced by the use of cerulean and citrine instead of the original CFP and YFP, and the former combination of fluorescent proteins gave rise to a larger change in FRET than did the latter combination (data not shown).
The CRD-dependent oxidation of the C probe was confirmed by biochemical experiments that involved the conjugation of methoxy-poly(ethylene glycol)-maleimide (mPEG-maleimide) to free thiol residues within the probe proteins, resulting in incremental increases in the size of the probe proteins that could be detected by immunoblot analysis (Fig. 2B). Consistently with the FRET ratio analysis, the fraction of C probe protein modified with mPEG-maleimide decreased as the redox potential of the GSH-GSSG titration buffer increased. In contrast, the similar treatment of A probe protein did not show any change in the mobility of the protein as assessed by immunoblot analysis (Fig. 2B). The C probe responded to redox reagents within a pH range of 6.5 to 8.5 (see Fig. S1 in the supplemental material). Unlike the case with other fluorescent redox probes, such as roGFP, we were able to make use of the A probe as a control that enabled us to assess the true redox-dependent conformational change of the C probe.
Redoxfluor is an intracellular redox indicator.
Redoxfluor then was expressed in the yeast Saccharomyces cerevisiae and Chinese hamster ovary (CHO) cells (with optimized codon usage), which subsequently were subjected to conventional FRET imaging analysis (Fig. 3). We observed a change in the cytosolic FRET image of the C probe from green (reducing) to blue (oxidizing) in response to the treatment of both types of cell with either oxidative agents such as H2O2 or nonoxidative reagents, such as 3-amino-1,2,4-aminotriazole (ATZ), an inhibitor of catalase, or buthionine sulfoximine (BSO), an inhibitor of glutathione synthesis (Fig. 3A and B; also see Videos S1 and S2 in the supplemental material). In contrast, neither ATZ nor BSO led to a change in the FRET signal in vitro (Fig. 1C), indicating that the observed change in the FRET signal in vivo reflects an effect upon cellular metabolism rather than a direct effect of these reagents on the C probe. Similar results were obtained in cells of the methylotrophic yeast Pichia pastoris expressing the C probe (see Fig. S2 in the supplemental material).
FIG. 3.
Visualization of the redox state in yeast and CHO-K1 cells. (A) Imaging of the FRET signal generated by wild-type (C probe) or mutant (A probe) Redoxfluor in S. cerevisiae in response to H2O2 (100 μM), ATZ (10 mM), or BSO (100 μM). Following the exposure of cells to H2O2 or ATZ, the cells were incubated in fresh (drug-free) medium and were monitored at the indicated times. Bar, 2 μm. (B) The FRET ratio imaging of C probe in CHO-K1 cells in response to H2O2 (100 μM) or ATZ (10 mM) (see Videos S1 and S2 in the supplemental material). The ATZ treatment of CHO-K1 cells expressing A probe did not show a C probe-like FRET response, although a slight FRET change due to the inherent redox-responsive nature of the fluorescent protein was observed. Bar, 10 μm. (C) From the microscopic analyses shown in panel B, the relative values of the FRET signal intensity of the C probe before and after H2O2 or ATZ treatment were plotted.
The analysis of cells following the removal of H2O2 or ATZ from the culture media led to the reduction of the redox state of the cytosol (Fig. 3A and B), demonstrating that the C probe responds to the redox state in a reversible manner in yeast and CHO cells. This reversibility of the C probe response was seen even in the presence of cycloheximide (see Fig. S3 in the supplemental material), demonstrating that the reversibility of the probe response was real and not merely due to the synthesis of new probe following the washout of the drug.
Redox state within peroxisomes.
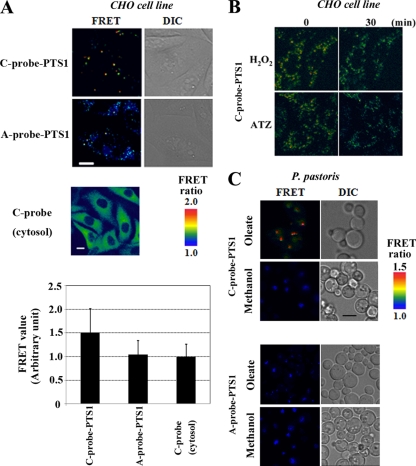
We next genetically engineered derivatives of the Redoxfluor C and A probes harboring C-terminal peroxisome targeting signal 1 (PTS1) (22). The subcellular localization of these probe proteins exhibited a punctate pattern when expressed in wild-type CHO cells (Fig. 4 A), but they exhibited a disperse pattern of distribution following expression in temperature-sensitive, pex5 mutant CHO cells (ZP105), which verifies the targeting of the probe proteins to the peroxisome.
FIG. 4.
Targeting of Redoxfluor to peroxisomes in mammalian and yeast cells. (A) The intraperoxisomal redox state assessed by C probe-PTS1 is more reductive than the cytosolic redox state in wild-type CHO cells. The FRET ratio images of A probe-PTS1 (lacking the redox-sensitive cysteine residues) and of cytosolic C probe also are shown as controls. Bar, 10 μm. The FRET values of the Redoxfluor probes are normalized to the value generated with the cytosolic C probe, which was arbitrarily set to 1.0. (B) The intraperoxisomal C probe is functional (CHO cells). The intraperoxisomal redox change represented by the FRET ratio imaging of C probe-PTS1 (also see Video S3 in the supplemental material). (C) P. pastoris wild-type strain PPY12 expressing C probe-PTS1 was transferred to oleate or methanol medium for the induction of peroxisome proliferation and was subjected to fluorescence microscopy for the FRET ratio imaging. Bar, 2 μm.
Using these peroxisome-targeted probes, we found that the intraperoxisomal redox state is more reductive than that of the cytosol in wild-type CHO cells (Fig. 4A). We observed that both the cytosolic and intraperoxisomal redox states became oxidative following the exposure of the cells to either ATZ or H2O2 (Fig. 4B; also see Video S3 in the supplemental material), thereby demonstrating that the C probe functioned normally in peroxisomes and that the peroxisomes were in a reductive state in CHO cells.
We next introduced the peroxisome-targeted Redoxfluor into the methylotrophic yeast P. pastoris. In this yeast, peroxisomes can be induced by culture on either oleate or methanol as a sole carbon source. The oleate-induced peroxisomes contain a fatty acid β-oxidation system similar to that in mammalian peroxisomes, whereas methanol-induced peroxisomes contain enzymes for methanol metabolism that are specific to the methylotrophic yeasts. We performed the FRET ratio analysis in these cells and observed that the redox state within the oleate-induced peroxisomes was more reductive than that in the methanol-induced peroxisomes (Fig. 4C). These results demonstrate that changes in the intraperoxisomal redox state can be monitored using Redoxfluor and that the redox state within peroxisomes depends on the carbon source used to induce peroxisome proliferation.
Cytosolic redox state within peroxisome assembly mutant cell lines.
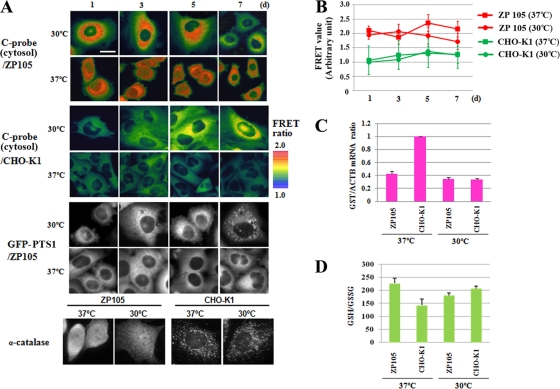
We next applied Redoxfluor to investigate the relationship between the cytosolic redox state and peroxisomal disorders. For this purpose, the cytosolic Redoxfluor C probe was introduced into the temperature-sensitive pex5 mutant cell line, ZP105. We observed a more reductive cytosolic redox state in the mutant ZP105 cells than in wild-type CHO cells at the nonpermissive temperature (37°C) (Fig. 5 A and B), and the cytosolic redox state of ZP105 cells became more oxidative following the decrease of the culture temperature to 30°C for 7 days. This change in the redox state of ZP105 cells was accompanied by the recovery of the peroxisomal import of GFP-PTS1 (Fig. 5B) and GFP-PTS2 proteins (data not shown). On the other hand, we did not observe differences in the redox state in the wild-type CHO cells between the culture temperatures at 37 and 30°C (Fig. 5A and B). These experiments demonstrate that the steady-state cytosolic redox equilibrium is more reductive when peroxisome assembly is impaired.
FIG. 5.
Cytosolic redox state is reductive in CHO cells defective in peroxisome assembly. (A) Time course of the dynamics of the cytosolic redox state and peroxisome assembly in CHO-K1 and ZP105 cells. After incubation at 37°C, cells were cultured at 37 or 30°C for 7 days. Bar, 10 μm. The import of GFP-PTS1, but not catalase, is restored in the ZP105 cell line after 7 days at 30°C, as shown in the lower panels. (B) Plotting of the relative values of the C probe FRET signal intensity under the same culture conditions as those described for panel A. (C) Relative abundance of GST mRNA, normalized to the levels of β-cytoskeletal actin (ACTB). (D) Intracellular glutathione redox ratio (GSH/GSSG).
To verify the microscopic results showing a more reductive redox state in the pex mutant cell line, cell lysates were prepared from the Redoxfluor-expressing cells and subjected to biochemical analysis using mPEG-maleimide (see Fig. S4 in the supplemental material). The incubation of cell lysate with this reagent led to a greater degree of modification of the probe proteins compared to that observed in the corresponding in vitro analyses (Fig. 2B), presumably due to a reduced level of thiol-containing substances (which could compete with the probe proteins for modification by mPEG-maleimide) upon cell breakage. The comparison of the intensities of the immunoblot signals indicated that the probe proteins in the lysate prepared from the pex5 cells were modified to a slightly greater extent by mPEG-maleimide, although the difference was very small.
We next compared the microscopic results using Redoxfluor to those obtained using conventional redox indices: the ratio between the oxidized and reduced forms of glutathione and the expression level of antioxidant enzymes (8, 28). In good agreement with the FRET imaging, we observed decreased glutathione S-transferase (GST) gene transcription at 37 and 30°C in ZP105 cells relative to the level observed in the wild-type cells (Fig. 5C). This reductive cytosolic redox state in ZP105 cells at 37°C and its shift at 30°C to a level similar to that observed in the wild-type cells also were confirmed by the measurement of the intracellular GSH/GSSG ratio (Fig. 5D). Taken together, these data demonstrate a more reductive state of the cytoplasm in the pex5 strain that was readily observed by FRET imaging using Redoxfluor.
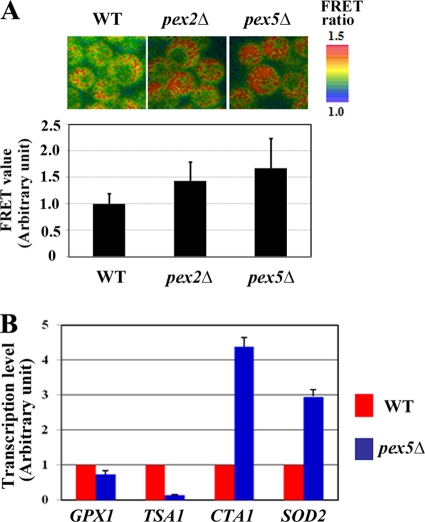
The more reductive state in the cytoplasm of peroxisome assembly mutant cells also was observed in P. pastoris pex2Δ and pex5Δ cells transferred to oleate medium (Fig. 6 A). This may be due partly to lower levels of H2O2 generation in the mutant strains. Acyl coenzyme A (acyl-CoA) oxidase is a major peroxisomal H2O2-generating enzyme that is found degraded in the cytosol of pex mutants of both mammalian and yeast cells (36). As such, the difference between the redox state observed in normal cells and peroxisome-assembly mutants might reflect differences in the efficiency of the generation and elimination of H2O2 between the wild-type and mutant cells. This notion is supported by the observation that ROS accumulation is more readily detected in CHO-K1 (wild-type) cells than in ZP105 cells using the fluorescent ROS indicator 2′,7′-dichlorodihydrofluorescein diacetate (DCF) (see Fig. S5 in the supplemental material). The import of catalase was not restored in ZP105 cells following culture at 30°C for 7 days, which is reminiscent of the phenotype of infant Refsum disease, a milder form of PBD (Fig. 5B) (26). These results suggest that mislocalization of peroxisomal catalase is not the main cause of abnormal reductive states observed in cells mutant for peroxisome assembly.
FIG. 6.
Redox states of P. pastoris pex mutant strains. (A) The cytosolic redox state is more reductive in the denoted P. pastoris pex mutants than in the wild-type (WT) strain during culture on oleate medium. Bar, 1 μm. The relative FRET values are normalized to the value generated from the probe in the wild-type strain, which was arbitrarily set to 1.0. (B) Transcription levels of antioxidant genes PpGPX1, PpTSA1, PpCTA1, and PpSOD2 in wild-type and Pppex5Δ strains. Values represent the means ± standard deviations (n = 3).
The findings that mitochondrial manganese-superoxide dismutase is strongly expressed in PBD cell lines has led to the conclusion that PBD cell lines suffer from oxidative stress (2). The level of transcription of genes encoding several antioxidant enzymes in P. pastoris was compared between the wild-type and pex5 mutant strains (Fig. 6B). The lower level of the expression of PpGPX (glutathione peroxidase) and PpTSA1 (cytosolic thioredoxin peroxidase) in the Pppex5Δ cells suggested that the Pppex5Δ cells are in a reductive state. Meanwhile, the transcription level of PpCTA1 (catalase) and PpSOD2 (mitochondrial manganese-superoxide dismutase) was greater in the same mutant than in the wild-type cells, suggesting a greater level of ROS generation in the Pppex5Δ cells. These results imply that the cytosol could be in a reductive state even under conditions of oxidative stress, owing to a greater level of expression of antioxidant genes induced following the generation of ROS in these cells under these conditions.
Screening for “redox modulators.”
We finally made use of Redoxfluor in the development of a screen for “redox modulators” that can restore the redox state in the patient-derived cells to a level similar to that observed in wild-type cells without affecting the redox state in normal cells. Several pharmacological agents have been suggested as potential therapeutics to treat milder forms of PBD (13, 14, 19-21, 38). We examined whether such agents could alter the intracellular redox state using Redoxfluor. The exposure of cells to trichostatin A (TSA), a histone deacetylase inhibitor, narrowed the difference between the redox states observed in CHO-K1 (wild-type) and ZP105 cells at 37°C (Fig. 7 A). TSA was more effective in doing so than either tocopherol (VE) or another histone deacetylase inhibitor, 4-phenylbutyrate (4PBA). In contrast, TSA treatment led to only a small change in the redox state of wild-type CHO-K1 cells (Fig. 7A). VE treatment led to a small change in the redox state to a more oxidative level in ZP105 cells, despite the fact that Kawada et al. speculated that VE can act as an antioxidant in PBD cell lines (13). The effects of the drugs observed by FRET ratio imaging were verified by the analyses of both the level of GST transcription and the GSH/GSSG ratio, in that TSA also was observed to narrow the differential between these indices between the wild-type and ZP105 strains more efficiently than the other compounds tested (Fig. 7C and D). The level of ROS accumulation in drug-treated ZP105 cells detected by DCF paralleled the change in redox state visualized by Redoxfluor (see Fig. S5 in the supplemental material), confirming the validity of the microscopic assessment.
FIG. 7.
Redox modulators, pharmacological agents that restore the intracellular redox state of mutant cell lines to normal levels. (A) FRET imaging of CHO cells at 37°C treated with TSA (50 nM), VE (50 M), and 4PBA (5 mM). Bar, 10 μm. (B) The values represent the relative FRET values of Redoxfluor normalized to the value from nontreated CHO-K1 (wild type) set arbitrarily to 1.0. (C) The relative abundance of GST mRNA standardized against the levels for ACTB. (D) Intracellular GSH/GSSG ratio. In panels B and C, values represent the means ± standard deviations (n = 3).
DISCUSSION
With the Redoxfluor probe developed in this study, we were able to visualize pathological redox states of a metabolic disorder in cells mutant for peroxisome assembly. The visualization of the redox state at the subcellular level has revealed that, to our surprise, the intraperoxisomal redox state is maintained in a reductive state in CHO cells, despite the generation of ROS by normal peroxisomal metabolism. This was unexpected, because peroxisomes express numerous oxidative metabolism pathways and long have been thought to exist in an oxidized state. The difference between the cytoplasmic redox potential of wild-type and pex5 CHO cells at 37°C was estimated to be approximately 9 mV (−217 mV in the wild type versus −226 mV in the pex5 cell line) when the FRET ratio monitored following excitation at 405 nm using a confocal spectrophotometric imaging system (Zeiss LSM510 Meta) was compared to the titration curve generated from the in vitro data (Fig. 2A). This small difference detected by using Redoxfluor would be difficult to detect using a biochemical assay (Fig. 5; also see Fig. S4 in the supplemental material). We are confident that the FRET signal generated by using Redoxfluor can be used directly as an index of the equilibrated redox state in a cell, shedding light on the physiological and pathological significance of the change in the steady-state redox potential caused by a metabolic defect.
In cells, the equilibrated redox state is established by the levels of various redox-related metabolites, such as GSH, NAD(P)H, thioredoxin, and ROS. Therefore, the redox state is considered to reflect the overall metabolic status. This study revealed that peroxisomes induced for oleate metabolism were more reductive than those induced for methanol metabolism in P. pastoris. This observation seems to reflect the fact that the high-energy bonds of fatty acid thioesters are stabilized in peroxisomes during β-oxidation. On the other hand, during methanol metabolism, the reduced glutathione within peroxisomes is consumed through two critical reactions for methanol metabolism, i.e., by forming hydroxymethyl glutathione nonenzymatically and by being further oxidized to S-formylglutathione to metabolize formaldehyde (39) or by Pmp20-glutathione peroxidase to detoxify intraperoxisomal ROS (9).
In many genetic disorders such as PBDs, where gene therapy is not a practical approach, drugs are used for symptomatic treatment. The screening of such drugs preferably is performed using cultured cells. The results obtained here using CHO cell pex mutants and PBD cell lines strongly suggest that an oxidative redox state is not the direct cause of PBDs. We believe that any metabolic deficiency owing to the dysfunctions of peroxisomes in the PBD cell lines, such as the impaired peroxisomal fatty acid β-oxidation, readily gives rise to an imbalance in overall cellular metabolism, resulting in a reductive cytosolic state. Therefore, the redox state in CHO cells mutant for peroxisome assembly may reflect the degree of metabolic abnormality that is correlated to the severity of PBDs. In this context, it is plausible that TSA treatment (Fig. 7) modulates metabolic defects arising from the peroxisomal abnormalities, whereas in the wild-type cell line these defects are not induced and the cells thus are not affected by the drug application.
A genetically encoded redox probe, roGFP (7), recently was used to visualize the redox status in plant leaf cytosol (around −310 mV) (11) and also in yeast and mammalian cells that were exposed to oxidoreductive reagents, including H2O2 and dithiothreitol (DTT) (4). However, the roGFP probe did not respond to cell treatment with nonoxidative reagents such as ATZ, a catalase inhibitor. Moreover, roGFP showed only a small change in the fluorescence spectrum following the exposure of cells to buthionine sulfoximine (BSO), an inhibitor of glutathione synthesis. These results imply that roGFP is neither sufficiently sensitive to visualize the redox dynamics at the physiological level in the cytosol nor sufficiently sensitive to distinguish the differences in the redox state between normal cells and patient-derived cell lines. Even the spectrum of the original GFP and other fluorescent proteins (not developed as redox probes) are known to be affected by some intracellular environment, e.g., pH, temperature, or redox state. Unlike the case with roGFP, the control Redoxfluor A probe made it possible to confirm that the change in the conformation of the Redoxfluor C probe in living cells is CRD dependent.
We currently are attempting to identify and characterize metabolites that contribute to the reductive state in mutant cells for peroxisome assembly by several approaches, including metabolome analysis. Since Redoxfluor can discriminate between physiological and pathological redox states, it may be possible to screen for compounds that can modulate the intracellular redox state in cells of patients with abnormal redox potential.
Supplementary Material
Acknowledgments
This paper is dedicated to the late Koichi Suzuki, research supervisor, CREST, Research Area Basic Technologies for Controlling Cell Functions Based on Metabolic Regulation Mechanism Analysis, who passed away on 20 April 2010. We acknowledge him for his continuous encouragement and support.
This work was supported in part by a CREST grant (to Y.F. and Y.S.) from the Science and Technology Agency of Japan, Grants-in-Aid for Scientific Research on Priority Areas 18076002 (Y.S.) and 19058011 (Y.F.), Grants-in-Aid for Scientific Research (B) 22380052 (Y.S.) and (B) 20370039 (Y.F.), Cooperative Grant of the National Project on Protein Structural and Functional Analyses (to Y.F.), and The 21st Century COE and Global COE Programs from the Ministry of Education, Culture, Sports, Science and Technology of Japan, grant from Japan Foundation for Applied Enzymology (to Y.F.). A.I. is a CREST Research Assistant Follow.
Footnotes
Published ahead of print on 24 May 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Azevedo, D., F. Tacnet, A. Delaunay, C. Rodrigues-Pousada, and M. B. Toledano. 2003. Two redox centers within Yap1 to H2O2 and thiol-reactive chemicals signaling. Free Radic. Biol. Med. 35:889-900. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart, E., I. Vanhorebeek, M. Grabenbauer, M. Borgers, P. E. Declercq, H. D. Fahimi, and M. Baes. 2001. Mitochondrial alterations caused by defective peroxisomal biogenesis in a mouse model for Zellweger syndrome (PEX5 knockout mouse). Am. J. Pathol. 159:1477-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belousov, V. V., A. F. Fradkov, K. A. Lukyanov, D. B. Staroverov, K. S. Shakhbazov, A. V. Terskikh, and S. Lukyanov. 2006. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 3:281-286. [DOI] [PubMed] [Google Scholar]
- 4.Dooley, C. T., T. M. Dore, G. T. Hanson, W. C. Jackson, S. J. Remington, and R. Y. Tsien. 2004. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 279:22284-22293. [DOI] [PubMed] [Google Scholar]
- 5.Fujiki, Y., K. Okumoto, N. Kinoshita, and K. Ghaedi. 2006. Lessons from peroxisome-deficient Chinese hamster ovary (CHO) cell mutants. Biochim. Biophys. Acta 1763:1374-1381. [DOI] [PubMed] [Google Scholar]
- 6.Griesbeck, O., G. Baird, R. E. Campbell, D. A. Zacharias, and R. Y. Tsien. 2001. Reducing the environmental sensitivity of yellow fluorescent protein. J. Biol. Chem. 276:29188-29194. [DOI] [PubMed] [Google Scholar]
- 7.Hanson, G. T., R. Aggeler, D. Oglesbee, M. Cannon, R. A. Capaldi, R. Y. Tsien, and S. J. Remington. 2004. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279:13044-13053. [DOI] [PubMed] [Google Scholar]
- 8.Hayes, J. D., and D. J. Pulford. 1995. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isozymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 30:445-600. [DOI] [PubMed] [Google Scholar]
- 9.Horiguchi, H., H. Yurimoto, N. Kato, and Y. Sakai. 2001. Antioxidant system within yeast peroxisome: biochemical and physiological characterization of CbPmp20 in the methylotrophic yeast Candida boidinii. J. Biol. Chem. 276:14279-14288. [DOI] [PubMed] [Google Scholar]
- 10.Hwang, C., A. J. Sinskey, and H. F. Lodish. 1992. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257:1496-1502. [DOI] [PubMed] [Google Scholar]
- 11.Jubany-Mari, T., L. Alegre-Batlle, K. Jiang, and L. J. Feldman. 2010. Use of a redox-sensing GFP (c-roGFP1) for real-time monitoring of cytosol redox status in Arabidopsis thaliana water-stressed plants. FEBS Lett. 584:889-897. [DOI] [PubMed] [Google Scholar]
- 12.Kasischke, K. A., H. D. Vishwasrao, P. J. Fischer, W. R. Zipfel, and W. W. Webb. 2004. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science 305:99-103. [DOI] [PubMed] [Google Scholar]
- 13.Kawada, Y., M. Khan, A. K. Sharma, D. B. Ratnayake, K. Dobashi, K. Asayama, H. W. Moser, M. A. Contreras, and I. Singh. 2004. Inhibition of peroxisomal functions due to oxidative imbalance induced by mistargeting of catalase to cytoplasm is restored by vitamin E treatment in skin fibroblasts from Zellweger syndrome-like patients. Mol. Genet. Metab. 83:297-305. [DOI] [PubMed] [Google Scholar]
- 14.Kemp, M., Y. M. Go, and D. P. Jones. 2008. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic. Biol. Med. 44:921-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosower, N. S., and E. M. Kosower. 1978. The glutathione status of cells. Int. Rev. Cytol. 54:109-160. [DOI] [PubMed] [Google Scholar]
- 16.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuge, S., N. Jones, N. Iizuka, and A. Nomoto. 1998. Crm1 (XpoI)-dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells 3:521-532. [DOI] [PubMed] [Google Scholar]
- 18.Lohman, J. R., and S. J. Remington. 2008. Development of a family of redox-sensitive green fluorescent protein indicators for use in relatively oxidizing subcellular environments. Biochemistry 47:8678-8688. [DOI] [PubMed] [Google Scholar]
- 19.McGuinness, M. C., J. F. Lu, H. P. Zhang, G. X. Dong, A. K. Heinzer, P. A. Watkins, J. Powers, and K. D. Smith. 2003. Role of ALDP (ABCD1) and mitochondria in X-linked adrenoleukodystrophy. Mol. Cell. Biol. 23:744-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGuinness, M. C., H. Wei, and K. D. Smith. 2000. Therapeutic developments in peroxisome biogenesis disorders. Expert Opin. Investig. Drugs. 9:1985-1992. [DOI] [PubMed] [Google Scholar]
- 21.McGuinness, M. C., H. P. Zhang, and K. D. Smith. 2001. Evaluation of pharmacological induction of fatty acid β-oxidation in X-linked adrenoleukodystrophy. Mol. Genet. Metab. 74:256-263. [DOI] [PubMed] [Google Scholar]
- 22.Monosov, E. D., T. J. Wenzel, G. H. Lüers, J. A. Heyman, and S. Subramani. 1996. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J. Histochem. Cytochem. 44:581-589. [DOI] [PubMed] [Google Scholar]
- 23.Okazaki, S., A. Naganuma, and S. Kuge. 2005. Peroxiredoxin-mediated redox regulation of the nuclear localization of Yap1, a transcription factor in budding yeast. Antioxid. Redox Signal. 7:327-334. [DOI] [PubMed] [Google Scholar]
- 24.Ostergaard, H., A. Henriksen, F. G. Hansen, and J. R. Winther. 2001. Shedding light on disulphide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 20:5853-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Østergaard, H., C. Tachibana, and J. R. Winther. 2004. Monitoring disulphide bond formation in the eukaryotic cytosol. J. Cell Biol. 166:337-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otera, H., T. Harano, M. Honsho, K. Ghaedi, S. Mukai, A. Tanaka, A. Kawai, N. Shimizu, and Y. Fujiki. 2000. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p·PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 275:21703-21704. [DOI] [PubMed] [Google Scholar]
- 27.Poirier, Y., V. D. Antonenkov, T. Glumoff, and J. K. Hiltunen. 2006. Peroxisomal beta-oxidation-a metabolic pathway with multiple functions. Biochim. Biophys. Acta 1763:1413-1426. [DOI] [PubMed] [Google Scholar]
- 28.Powis, G., J. R. Gasdaska, and A. Baker. 1997. Redox signaling and the control of cell growth and death. Adv. Pharmacol. 38:329-359. [DOI] [PubMed] [Google Scholar]
- 29.Rizzo, M. A., G. H. Springer, B. Granada, and D. W. Piston. 2004. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22:445-449. [DOI] [PubMed] [Google Scholar]
- 30.Rost, J., and S. Rapoport. 1964. Reduction-potential of glutathione. Nature 201:185. [DOI] [PubMed] [Google Scholar]
- 31.Sakai, Y., A. Koller, L. K. Rangell, G. A. Keller, and S. Subramani. 1998. Peroxisome degradation by micropexophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J. Cell Biol. 141:625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrader, M., and H. D. Fahimi. 2006. Peroxisomes and oxidative stress. Biochim. Biophys. Acta 1763:1755-1766. [DOI] [PubMed] [Google Scholar]
- 33.Sears, I. B., J. O'Connor, O. W. Rossanese, and B. S. Glick. 1998. A versatile set of vectors for constitutive and regulated gene expression in Pichia pastoris. Yeast 14:783-790. [DOI] [PubMed] [Google Scholar]
- 34.Steinberg, S. J., G. Dodt, G. V. Raymond, N. E. Braverman, A. B. Moser, and H. W. Moser. 2006. Peroxisome biogenesis disorders. Biochim. Biophys. Acta 1763:1733-1748. [DOI] [PubMed] [Google Scholar]
- 35.Tietze, F. 1969. Enzymatic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal. Biochem. 27:502-506. [DOI] [PubMed] [Google Scholar]
- 36.Tsukamoto, T., S. Yokota, and Y. Fujiki. 1990. Isolation and characterization of Chinese hamster ovary cell mutants defective in assembly of peroxisomes. J. Cell Biol. 110:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Klei, I. J., and M. Veenhuis. 2006. Yeast and filamentous fungi as model organisms in microbody research. Biochim. Biophys. Acta 1763:1364-1373. [DOI] [PubMed] [Google Scholar]
- 38.Wei, H., S. Kemp, M. C. McGuinness, A. B. Moser, and K. D. Smith. 2000. Pharmacological induction of peroxisomes in peroxisome biogenesis disorders. Ann. Neurol. 47:281-283. [PubMed] [Google Scholar]
- 39.Yurimoto, H., B. Lee, T. Yano, Y. Sakai, and N. Kato. 2003. Physiological role of S-formylglutathione hydrolase in C(1) metabolism of the methylotrophic yeast Candida boidinii. Microbiology 149:1971-1979. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, Q., D. W. Piston, and R. H. Goodman. 2002. Regulation of corepressor function by nuclear NADH. Science 295:1895-1897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.