Abstract
In the nervous system, cell death by apoptosis plays a critical role during normal development and pathological neurodegeneration. Jun N-terminal kinases (JNKs) are essential regulators of neuronal apoptosis. The AP-1 transcription factor c-Jun is phosphorylated at multiple sites within its transactivation domain by the JNKs, and c-Jun phosphorylation is required for JNK-induced neurotoxicity. While the importance of c-Jun as a mediator of apoptotic JNK signaling in neurons is firmly established, the molecular mechanism underlying the requirement for c-Jun N-terminal phosphorylation is enigmatic. Here we identify the multifunctional protein Bag1-L as a coactivator of phosphorylated c-Jun. Bag1-L preferentially interacts with N-terminally phosphorylated c-Jun, and Bag1-L greatly augments transcriptional activation by phosphorylated c-Jun. Chromatin immunoprecipitation experiments revealed binding of Bag1-L to the promoters of proapoptotic AP-1 target genes, and overexpression of Bag1-L augmented cell death in primary neurons. Therefore, Bag1-L functions as a coactivator regulating neurotoxicity mediated by phosphorylated c-Jun.
The AP-1 transcription factor consists of a variety of dimers composed of members of the Fos (c-Fos, FosB, Fra-1, and Fra-2) and Jun (c-Jun, JunB, and JunD) families of proteins (24). The activity of the AP-1 transcription factor is strongly induced in response to numerous signals, including growth factors, cytokines, and extracellular stresses. AP-1 stimulation is mediated, in part, by the phosphorylation of c-Jun by the Jun N-terminal kinases (JNKs) (15). c-Jun N-terminal phosphorylation at serine residues 63 and 73 and threonine residues 91 and 93 within its transactivation domain is thought to increase transcription of target genes, one of which is the c-jun gene itself (2). In neurons, JNK signaling is believed to play an obligatory role in the regulation of cell death. Early work using PC12 neuron-like cells first implied the JNK pathway in caspase-dependent cell death induced by withdrawal of nerve growth factor (NGF) (54). Numerous subsequent studies in a variety of model systems have substantiated the role of JNK in neuronal apoptosis. The JNK proteins are encoded by three genes (jnk1, jnk2, and jnk3), and jnk mouse mutants have revealed roles for JNK signaling in neuronal development and disease. Mice lacking jnk1 and jnk2 display exencephaly due to deregulated apoptosis during neurogenesis (29, 41), and jnk2/jnk3 mutants show reduced neuronal loss in models of cerebral ischemia and Parkinsonian degeneration (23, 28). Small molecule inhibitors and inhibiting peptides targeting JNK have been developed and have shown therapeutic promise for treatment of neurological disorders (8, 27, 34).
Several JNK substrates have been implicated as mediators of neuronal death. JNKs were shown to phosphorylate and thereby modify the activities of several apoptosis regulators of the Bcl2 superfamily, including Bim (4, 31, 39), thereby linking JNK signaling to the mitochondrial death pathway. In addition, in response to many stimuli, JNK-mediated neuronal apoptosis is dependent on de novo transcription, and c-Jun was identified as the essential substrate in this arm of the JNK pathway (13, 51). Overexpression of a dominant-negative c-Jun mutant greatly impaired neuronal apoptosis, and c-Jun N-terminal phosphorylation plays a crucial role in JNK-dependent death (5, 17, 51). Interestingly, the dp5 and bim genes have been shown to be important targets of JNK/c-Jun-mediated transcription (40, 52), implying a convergence of the transcriptional and mitochondrial JNK death pathways.
While the importance of c-Jun as a mediator of apoptotic JNK signaling in neurons is firmly established, the molecular mechanism underlying the requirement for c-Jun N-terminal phosphorylation has proved enigmatic. We have previously described a genetic screen to discover proteins that interact with c-Jun in an N-terminal phosphorylation-dependent manner (36). This approach identified Bag1-L as a protein that preferentially interacted with the phosphorylated form of c-Jun. Bag1-L is a multifunctional protein that had previously been shown to augment transactivation by nuclear hormone receptors (47). Our analysis revealed that Bag1-L stimulated c-Jun function in a JNK-dependent manner and cooperated with c-Jun in the induction of apoptosis, suggesting that Bag1-L functions as a phosphorylation-dependent c-Jun coactivator.
MATERIALS AND METHODS
DNA constructs and transfections.
Bag1 was identified as a phosphorylation-dependent interactor of c-Jun in a previously described genetic screen (36). Full-length mouse Bag1 (accession no. NM_009736) encoding the long and short isoforms was amplified from brain mRNA by reverse transcription-PCR and cloned into pIRES2-eGFP (Clontech) for eukaryotic expression. The Bag1-L and Bag1-S expression plasmids were generated by mutating the N-terminal Leu to Met and Ala, respectively. The deletion mutants Bag1-LΔAR (amino acids [aa] 111 to 148), Bag1-LΔULD (aa 169 to 215), and Bag1-LΔBD (aa 256 to 347) were generated from Bag1-L by PCR or restriction digest (primer sequences are available on request). The coding sequence for the hemagglutinin (HA) epitope (YPYDVPDYA) was inserted by PCR at the C termini of Bag1-L, Bag1-S, Bag1-LΔAR, Bag1-LΔULD, and Bag1-LΔBD to generate tagged versions.
Optimized expression constructs for human Bag1-S (hBag1-S), Bag1-L (hBag1-L), and Bag1-S fused to a heterologous nuclear localization signal (NLS) (NLS-hBag1-S), nuclear export signal (NES) (NES-hBag1-S), hBag1-L with point mutations in the BAG domain, or hBag1-S fused to glutathione S-transferase (GST) (GST-hBag1-S) have been previously described (12, 30, 48).
Epitope-tagged wild-type c-Jun and Ala mutant c-Jun have been described previously (36).
The following luciferase reporter constructs were used: −517/−42 collagenase-luc (TRE) and −1977/−1850 urokinase-luc (CRE) were a kind gift from H. Van Dam; SW3-Ubi-Gal4-cJun (1-256) and SW7-Ubi-Gal4-cJun (Pan-Ala63,73,91,93) were a kind gift from D. Bohmann; the PathDetect c-Jun trans-reporting system containing 5× Gal4UAS-luc, Gal4-DBD-cJun (1-223), Gal4-DBD, and pFC-MEKK plasmids was obtained from Stratagene; −1568/+81 dp5-luc (CRE) has been previously described (32).
Reporter gene assays.
Cell extracts were made 32 h posttransfection, and luciferase activity was determined using the Dual-Luciferase reporter assay system (Promega). In some cases, cells were exposed to UV (100 kJ/m2) and collected 8 h after treatment. Data are expressed as fold induction after being normalized using Ubi-Renilla luciferase.
Neuronal cultures and transfections.
PC12 cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% horse serum, 5% fetal calf serum (FCS), 2 mM glutamine, and penicillin-streptomycin. Cells were grown on collagen-coated tissue culture dishes, and differentiation was induced by NGF (Promega) as previously described (46). Cells were transfected with Lipofectamine 2000 (Invitrogen).
Cerebellar granule cells (CGNs) were isolated from P7 C57BL/6J mice, and transfections of CGNs were performed at day in vitro 6 (DIV6) using calcium phosphate (Promega) as described previously (32). High levels of potassium promote survival of CGNs in vitro in the absence of neurotrophic factors. Switching CGNs into medium containing low potassium induces apoptosis, with a concomitant increase in the levels of c-Jun and N-terminally phosphorylated c-Jun. Pyknotic nuclei indicative of apoptosis were visualized after fixing and staining with 4′,6-diamidino-2-phenylindole (DAPI) as described above, with green fluorescent protein (GFP) used to mark transfected cells.
Western blot analysis.
Samples were homogenized in RIPA lysis buffer (Cell Signaling) supplemented with protease inhibitor cocktail (Sigma). Western blotting was performed as described previously (36). The following antibodies were used: Bag1 (Delta Biolabs), c-Jun, JunD, JunB, phospho-c-Jun, Hsp70-agarose (Santa Cruz Biotechnology), c-Jun phosphoserine 73, JNK, phospho-JNK (Cell Signaling), phospho-JNK1/2 (Stressgen), Bim, Hsp70, FLAG, Myc epitope (9E10), HA and β-actin (Sigma), and tubulin (Abcam). The gels were transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare), and the membranes were immunoblotted with various antibodies as indicated.
Immunoprecipitations (IPs).
HEK293T and HCT116 cells were used for analysis of interactions between endogenous proteins and/or transiently expressed proteins. Cells were washed with cold phosphate-buffered saline (PBS) and subsequently lysed for 30 min at 4°C with 150 mM NaCl, 80 mM Tris-HCl (pH 7.2), 0.2% NP-40, 10% glycerol, and complete protease inhibitor cocktail (Sigma). The pellet was sonicated for nuclear extracts three times for 10 s (each) in 200 μl of buffer containing 300 mM NaCl, 0.01% SDS, 1% Triton, 2 mM EDTA, 20 mM Tris-HCl (pH 7.2), and protease inhibitor cocktail. Lysates were cleared by centrifugation for 15 min at 4°C. In some cases, cells were treated 2 h prior to collection with 50 μM JNK Inhibitor II (Merck Biosciences) or transfected with the indicated plasmids with the Lipofectamine 2000 reagent (Invitrogen) for 48 h.
Lysates were precleared for 1 h by rotation at 4°C, and proteins were immunoprecipitated overnight with the indicated antibody or antibody-agarose conjugate. Immunocomplexes were captured by rotation for 2 h with protein A- and protein G-Sepharose beads (Sigma). Immunoprecipitates were washed four times with lysis buffer, and sample proteins were separated by SDS-PAGE and subsequently transferred onto PVDF membranes (GE Healthcare) by following standard procedures.
ChIP.
Chromatin immunoprecipitation (ChIP) analysis was performed as described previously (1) using HCT116 cells. In some cases, cells were treated 2 h prior to collection with 50 μM JNK Inhibitor II (Merck Biosciences) and 25 ng/ml anisomycin (Sigma), exposed to UV (100 kJ/m2), or transfected with the indicated plasmids using the Lipofectamine 2000 reagent (Invitrogen) for 48 h.
Immunoprecipitations were carried out overnight at 4°C with rotation with the indicated antibodies. Immunocomplexes were collected by rotation for 2 h at 4°C with protein G-Sepharose beads (Sigma). The oligonucleotide sequences used to amplify the DNA fragments are available upon request. Amplified AP-1 promoter sequences were visualized by running on 3% agarose gels containing 10 μg/ml ethidium bromide (Sigma) and documented using a BioDoc-It imaging system (Ultra Violet Products Ltd.). Quantitative real-time PCR was accomplished with SYBR green incorporation (Platinum quantitative PCR supermix-uracil DNA glycosylase [UDG] with Rox reference dye; Invitrogen) using an ABI 7900HT real-time PCR system (Applied Bioscience), and the data were analyzed using the SDS 2.3 software program. Enrichment from the IP was quantified with reference to the input, and results are presented as fold enhancement over results for untreated cells.
qRT-PCR analysis.
For quantitative real-time PCR (qRT-PCR) analysis, total mRNA was isolated from HCT116 cells transfected with the indicated plasmids using the Lipofectamine 2000 reagent (Invitrogen) for 48 h using the RNeasy minikit according to the manufacturer's instructions (Qiagen). In some cases, cells were treated for 30 min with 25 ng/ml anisomycin (Sigma). For quantitative real-time PCRs, cDNA was synthesized using Invitrogen Superscript reagents according to the manufacturer's instructions. Quantitative real-time PCR was accomplished with SYBR green incorporation (Platinum quantitative PCR supermix-UDG with Rox; Invitrogen) using an ABI 7900HT real-time PCR system (Applied Bioscience), and the data were analyzed using the SDS 2.3 software program. Results were normalized to those obtained with glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and results are presented as fold induction over that for untreated cells. Primer sequences used were as previously described for c-Jun (42), dp5 (35), and Bim (7).
GST pull-down assay.
Human Bag1-S was cloned into the pGEX2TK vector (GE Healthcare). The GST-hBag1-S fusion protein was produced recombinantly in BL21 bacteria (Stratagene) and purified as described previously (12). The GST tag was cleaved with thrombin (GE Healthcare), and free hBag1-S was recovered from the supernatant.
Biotinylated c-Jun peptides encompassing amino acids 55 to 100 with residues S63, S73, T91, and T93 phosphorylated or unphosphorylated or a scrambled version were produced by the CRUK-London Research Institute Peptide Synthesis Core Facility.
Peptides were immobilized on streptavidin-coupled Dynabeads (Invitrogen) and incubated with recombinant hBag1-S in binding buffer (150 mM NaCl, 80 mM Tris-HCl [pH 7.2], 0.1% NP-40) containing phosphatase and protease inhibitors (Sigma) at 4°C for 2 h, washed, boiled, and loaded onto an SDS-PAGE gel. Western blotting was performed as described previously (36), and membranes were blotted with anti-Bag1 antibody (Delta Biolabs).
RESULTS
Bag1-L interacts with c-Jun.
Members of the JNK family of mitogen-activated protein (MAP) kinases are important regulators of neuronal death both during neurogenesis and during neurodegenerative conditions, such as Parkinson's disease and stroke. The AP-1 transcription factor c-Jun is a key substrate of neurotoxic JNK signaling, and an absence of c-Jun N-terminal phosphorylation protects from stress-induced neuronal death (5, 6). However, an explanation for the neurotoxicity of phosphorylated but not unphosphorylated c-Jun has been lacking, and the molecular mechanism of how phosphorylated c-Jun triggers cell death has been unclear.
We speculated that the JNK-mediated phosphorylation of c-Jun was modifying the interaction of the c-Jun N terminus with cofactor molecules, thereby modulating neurotoxic JNK signaling. To investigate the mechanism of JNK-mediated transcriptional regulation, we previously described a genetic screen to identify proteins that interact with c-Jun in a phosphorylation-dependent manner (phosphorylation-dependent c-Jun interactors or PDJs) (36). One of the candidate proteins (PDJ5) that preferentially interacted with N-terminally phosphorylated c-Jun encoded the known protein Bag1.
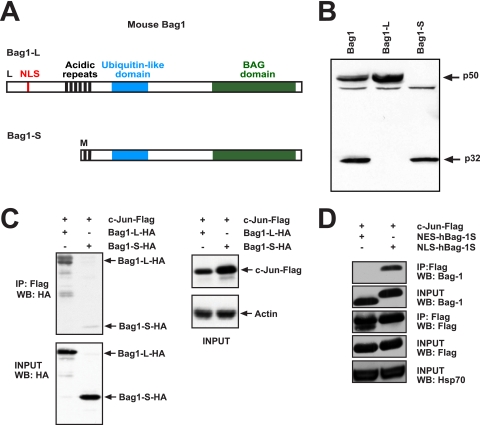
Through differential translational initiation, different Bag1 proteins are generated from a single bag1 mRNA. In human cells, isoforms of 36 kDa (called hBag1-S), 46 kDa (hBag1-M/RAP46), and 50 kDa (hBag1-L) can be detected, whereas only two Bag1 proteins, of 32 kDa (Bag1-S) and 50 kDa (Bag1-L), are found in murine cells (Fig. 1 A). Bag1-L translation initiates at an upstream CUG codon, whereas Bag1-S translation initiation takes place at an internal in-frame AUG codon (48). Therefore, Bag1 isoforms share a carboxy terminus, and the larger isoforms have unique amino-terminal sequences. A strong nuclear localization signal located in N-terminal amino acids is present in Bag1-L but not Bag1-S, which causes Bag1-L to be nuclear whereas Bag1-S is localized mainly in the cytoplasm (Fig. 1A). Mutation of the CUG initiation codon to methionine will generate a construct that gives rise solely to Bag1-L, whereas a nonsense mutation will abolish translational initiation and produce only Bag1-S (Fig. 1B).
FIG. 1.
Bag1-L interacts with c-Jun. (A) Schematic of mouse Bag1 isoforms. Translation of Bag1-L (aa 1 to 355) initiates at an upstream CUG codon, and that of Bag1-S (aa 136 to 355) initiates at an internal AUG codon. The positions of the nuclear localization sequence (NLS) (aa 70 to 76), acidic repeats (AR), (aa 111 to 148), ubiquitin-like domain (ULD), (aa 169 to 215), and BAG domain (BD) (aa 256 to 347) are indicated. (B) Western blot of cell extracts of HEK293T cells transfected with mouse Bag1, optimized mouse Bag1-L, and optimized mouse Bag1-S plasmids were immunoblotted with anti-rabbit Bag1 antibody. (C) HEK293T cells were transfected with Flag-tagged c-Jun and HA-tagged Bag1-L or Bag1-S. Cell extracts were immunoprecipitated with anti-Flag and immunoblotted with anti-HA. The interaction of Bag1-L with c-Jun is markedly enhanced versus that of Bag1-S. WB, Western blot. (D) HEK293T cells were transfected with Flag-tagged c-Jun and the human short Bag1 isoform fused to a nuclear export signal (NES-hBag1-S) or a nuclear localization signal (NLS-hBag1-S). Cell extracts were immunoprecipitated with anti-Flag and immunoblotted with anti-Bag1. Bag1-S can efficiently interact with c-Jun when targeted to the nucleus.
In order to corroborate the interaction between Bag1-L and c-Jun, which was originally observed in yeast, we performed coimmunoprecipitation (IP) experiments. Cooverexpression studies revealed interaction of c-Jun with Bag1-L, but interaction with Bag1-S was less efficient (Fig. 1C). Targeting of Bag1-S to the nucleus through the addition of a simian virus 40 (SV40)-derived NLS (NLS-hBag1-S) was sufficient to restore interaction with c-Jun, whereas Bag1-S fused to a nuclear export signal (NES-hBag1-S) did not bind (3), indicating that the Bag1 C terminus is sufficient for interaction with c-Jun but that the largely cytoplasmic localization of Bag1-S precludes c-Jun binding (Fig. 1D).
Bag1-L functions as a c-Jun coactivator.
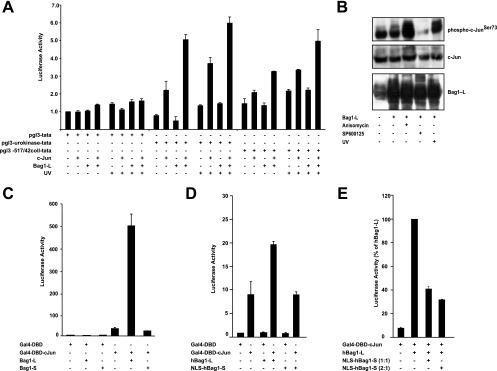
To address the functional consequences of Bag1-L binding to c-Jun, the effect of Bag1-L on c-Jun transactivation was measured. The collagenase gene contains a high-affinity AP-1 binding site, the so-called tetradecanoyl phorbol acetate (TPA) response element (TRE), which is believed to preferentially bind Jun/Fos dimers. c-Jun overexpression modestly activated a collagenase-luciferase (Coll-Luc) reporter construct, but Bag1-L expression alone had no effect. However, Bag1-L coexpression with c-Jun further augmented reporter activity, indicating that Bag1-L transactivation is c-Jun dependent (Fig. 2 A). Using a reporter construct derived from the urokinase plasminogen activator (uPA) promoter containing a CRE-like element (uPA-luc), which binds mainly Jun/Jun and Jun/ATF dimers, c-Jun/Bag1-L cooperation was more pronounced. c-Jun strongly activated uPA-luc, whereas Bag1-L expression on its own did not. However, simultaneous expression of c-Jun and Bag1-L resulted in more efficient activation. UV treatment, which induced robust c-Jun N-terminal phosphorylation (Fig. 2B), led to a further increase in reporter gene activity (Fig. 2A). Thus, Bag1-L functions as a c-Jun coactivator and appears to have a preference for CRE-like elements.
FIG. 2.
Bag1-L augments c-Jun transcriptional activity. (A) HCT116 cells were transfected with pGL3-TATA-luc, pGL3-uPA-TATA-luc, or pGL3-collagense-TATA-luc, together with c-Jun, Bag1-L, and Ubi-Rluc. Some samples were exposed to UV (100 kJ/m2) for 8 h prior to processing. Luciferase activity was normalized to that for Ubi-Rluc. Data are represented as the means of results for triplicate wells ± standard errors of the means (SEM). Data represent luciferase activity relative to that of pGL3-TATA plus empty vector-transfected cells, which was arbitrarily set to 1. (B) Western blot of cell extracts of HCT116 cells transfected with Bag1-L, which were subjected to 50 μM JNK Inhibitor II (SP600125), 25 ng/ml anisomycin, or UV (100 kJ/m2) and immunoblotted with the indicated antibodies. Note that there is no significant change in mobility of Bag1-L after JNK activation or JNK inhibitor treatment. Overexpression of Bag1-L results in an increase in hyperphosphorylated c-Jun versus that in parental HCT116 cells. Anisomycin and UV robustly increase levels of phospho-c-Jun and JNK inhibitor treatment decreases this substantially versus those in parental HCT116 cells. (C) HEK293T cells were transfected to express Gal4-DBD or Gal4-DBD-c-Jun, Bag1-L or Bag1-S, 5× Gal4UAS-luc, and the Ubi-Renilla-luc transfection control reporter gene (Ubi-Rluc). Luciferase activity was normalized to that with Ubi-Rluc. Data are represented as the means of triplicate wells ± SEM. Activity for Gal4-DBD + empty vector is arbitrarily set to 1. (D) NIH 3T3 cells were transfected to express Gal4-DBD or Gal4-DBD-c-Jun, hBag1-L or NLS-hBag1-S, 5× Gal4UAS-luc, and the Ubi-Renilla-luc transfection control reporter gene (Ubi-Rluc). Luciferase activity was normalized to that for Ubi-Rluc. Data are represented as the means of triplicate wells ± SEM. Activity for Gal4-DBD plus empty vector is arbitrarily set to 1. Targeting Bag1-S to the nucleus does not result in an increase in luciferase reporter activity. (E) NIH 3T3 cells were transfected to express Gal4-DBD-c-Jun or hBag1-L ± NLS-hBag1-S (in either a 1:1 or 2:1 ratio), 5× Gal4UAS-luc, and the Ubi-Renilla-luc transfection control reporter gene (Ubi-Rluc). Luciferase activity was normalized to that for Ubi-Rluc. Data are represented as the means of results for triplicate wells ± SEM. Targeting of Bag1-S to the nucleus and coexpression with Bag1-L result in a significant reduction of luciferase reporter activity.
To test whether the transcriptional stimulation of c-Jun by Bag1-L is independent of heterodimerization with other AP-1 family members, we used a fusion protein that replaced the basic region/leucine zipper (bZIP) domain of c-Jun with the heterologous DNA-binding domain of the yeast transcription factor GAL4 (GAL4-DBD). Whereas GAL4-DBD did not stimulate a luciferase reporter driven by Gal4 operators, GAL4-DBD-Jun expression resulted in significant activation. Bag1-L did not influence GAL4-DBD function but augmented GAL4-DBD-Jun transactivation substantially, while Bag1-S had no effect (Fig. 2C). Targeting of the Bag1-L C terminus to the nucleus (NLS-hBag1-S) was not sufficient for coactivator function (Fig. 2D). Conversely, overexpression of NLS-hBag1-S had a dominant-negative function and decreased reporter gene activation by full-length Bag1-L (Fig. 2E), possibly due to competition for c-Jun binding.
The BAG domain of Bag1-L is required for c-Jun interaction and transactivation.
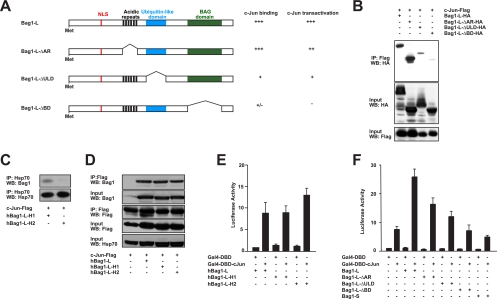
We determined which domain within the Bag1-L C terminus was required for interaction with c-Jun. The Bag1 protein contains several copies of a 6-amino-acid repeat that conform to a TR/QSEEX consensus, the function of which is unclear. Moreover, all Bag1 isoforms contain a ubiquitin-like domain (ULD) and a so-called BAG domain (BD) (Fig. 3 A). This is a 70-amino-acid-residue domain located at the very carboxy terminus, which comprises a pair of antiparallel amphipathic alpha-helices which mediate interaction with the Hsc70 and Hsp70 chaperone molecules and other proteins (11, 45). Binding to chaperones via this region is thought to play a critical role in many Bag1 functions (47).
FIG. 3.
Deletion of the BAG domain impairs Bag1-L/c-Jun interaction and c-Jun-mediated transcriptional stimulation. (A) Schematic representation of Bag1-L deletion mutants and their biological activities. (B) HEK293T cells were transfected with Flag-tagged c-Jun and HA-tagged Bag1-L, Bag1-L-ΔAR, -ΔULD, or -ΔBD. Cell extracts were immunoprecipitated with anti-Flag and immunoblotted with anti-HA. The interaction of Bag1-LΔBD with c-Jun is markedly reduced versus that of parental Bag1-L. (C) HEK293T cells were transfected with Flag-tagged c-Jun and human Bag1-L with point mutations in helix 1 or helix 2 of the BAG domain (hBag1-L-H1 or hBag1-L-H2). The mutation in helix 2 renders this version incapable of binding to Hsp70. The extracts were immunoprecipitated with anti-Hsp70 and immunoblotted with anti-Bag1 to confirm previous findings that the helix 2 mutant cannot interact with Bag1 (47). (D) HEK293T cells were transfected with Flag-tagged c-Jun and the human long Bag1 isoform or mutants with point mutations in helix 1 or helix 2 of the BAG domain (hBag1-L-H1 and hBag1-L-H2). Cell extracts were immunoprecipitated with anti-Flag and immunoblotted with anti-Bag1. hBag1-L, hBag1-L-H1, and hBag1-L-H2 can all interact with c-Jun. Thus, the interaction of Bag1-L with c-Jun is independent of Hsp70. (E) NIH 3T3 cells were transfected to express Gal4-DBD or Gal4-DBD-c-Jun; hBag1-L, hBag1-L-H1, or hBag1-L-H2; 5× Gal4UAS-luc; and the Ubi-Renilla-luc transfection control reporter gene (Ubi-Rluc). Luciferase activity was normalized to that for Ubi-Rluc. Data are represented as the means of results for triplicate wells ± SEM. Activity for Gal4-DBD + hBag1-L is arbitrarily set to 1. Helix mutants promote c-Jun-mediated transcriptional stimulation to an extent similar to that for parental hBag1-L. (F) NIH 3T3 cells were transfected to express Gal4-DBD or Gal4-DBD-c-Jun and Bag1-L, Bag1-L-ΔAR, -ΔULD, or -ΔBD, or Bag1-S, 5× Gal4UAS-luc, and the Ubi-Renilla-luc transfection control reporter gene (Ubi-Rluc). Luciferase activity was normalized to that for Ubi-Rluc. Data are represented as the means of results for triplicate wells ± SEM. Activity for Gal4-DBD + empty vector is arbitrarily set to 1. Deletion of the BAG domain abolishes the ability of Bag1-L to enhance c-Jun-mediated transcriptional stimulation.
To determine which domains would contribute to c-Jun interaction, we generated epitope-tagged Bag1-L deletion mutants lacking individual domains (Fig. 3A). ULD deletion led to a substantial reduction in c-Jun interaction, and removal of the BAG domain rendered the mutant HA-Bag1-LΔBD protein essentially incapable of c-Jun interaction (Fig. 3B). It is thus conceivable that both domains are part of the c-Jun binding interface.
The BAG domain mediates Bag1-L interaction with nuclear hormone receptors (14, 18, 21). These interactions are believed to occur indirectly via heat shock proteins, which was elegantly demonstrated for Bag1-L/androgen receptor interaction (11). Also, it was previously shown using recombinant proteins in an in vitro system that Bag1 could interact with a wide range of proteins, including c-Jun in addition to c-Fos and other transcription factors (56). Notably, these experiments were performed in the presence of high concentrations of the heat shock protein Hsc70, indicating that in these assays Bag1 might interact with denatured proteins via intermediary heat shock proteins (56). To assess whether heat shock proteins associated with Bag1-L play a role in physiological c-Jun binding, we made use of point mutations in the BAG domain that specifically impair Hsc70 and Hsp70 chaperone interaction with Bag1-L (11, 49). We confirmed previous observations that a double point mutation in helix 2 of Bag1-LE283A,K286A (hBag1-L-H2) results in a greatly reduced ability to interact with heat shock proteins (30) (Fig. 3C) while Bag1-LE226A,K230A (hBag1-L-H1) was unaffected. The Bag1-LE283A,K286A (hBag1-L-H2) mutant protein interacted with c-Jun and was able to cooperate efficiently with c-Jun in transcriptional activation (Fig. 3D and E). Thus, Bag1-L directly interacts with c-Jun through its BAG domain without the aid of heat shock proteins.
We next assessed the ability of individual Bag1-L deletion mutants to promote c-Jun-mediated transcriptional activation in NIH 3T3 cells. Deletion of the BAG domain abolished transcriptional cooperation between Bag1 and c-Jun (Fig. 3F). Overall, the ability of Bag1-L deletion mutants to stimulate c-Jun activity correlated well with the strength of binding to c-Jun (Fig. 3A).
N-terminal phosphorylation is required for c-Jun interaction with Bag1-L.
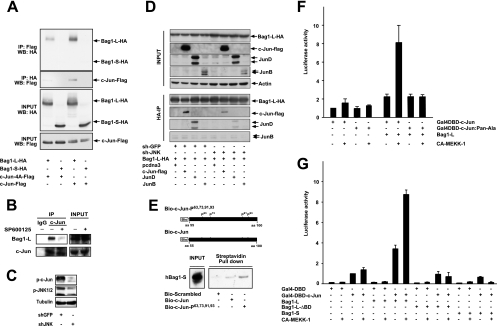
Next, we wanted to validate whether the interaction between c-Jun and Bag1-L was dependent on JNK phosphorylation, as suggested by the identification of Bag1-L as a phosphorylation-dependent interactor of c-Jun in the genetic screen in yeast. To investigate the phosphorylation dependency of c-Jun/Bag1-L interaction in mammalian cells, wild-type c-Jun or a mutant c-Jun allele lacking all N-terminal phosphorylation sites (c-Jun4A) was coexpressed with Bag1-L in HEK 293T cells. Whereas wild-type c-Jun efficiently bound to Bag1-L, c-Jun4A interacted less well (Fig. 4 A). In addition, endogenous Bag1-L bound to c-Jun, but pharmacological inhibition of JNK activity, which greatly decreased the levels of c-Jun N-terminal phosphorylation (Fig. 2B), reduced Bag1-L/c-Jun interaction (Fig. 4B). Moreover, RNA interference (RNAi)-mediated depletion of jnk1 and jnk2 (shJNK) reduced the protein levels of the active phosphorylated JNK protein and c-Jun phosphorylation (Fig. 4C) and consequently decreased the efficiency of c-Jun/Bag1-L interaction (Fig. 4D). Interestingly, Bag1-L also interacted with JunD but not with JunB, and shJNK treatment also reduced Bag1-L/JunD interaction (Fig. 4D). Phosphorylated peptides encompassing the c-Jun N terminus interacted with recombinant hBag1-S to a greater extent than unphosphorylated peptides, indicating a direct phosphorylation-dependent physical interaction between c-Jun and Bag1 (Fig. 4E). Thus, N-terminal phosphorylation of c-Jun increases interaction with Bag1-L.
FIG. 4.
Bag1-L preferentially interacts with phosphorylated c-Jun. (A) HEK293T cells transfected with Flag-tagged wild-type c-Jun or the c-Jun phosphoacceptor mutant (S63A S73A T91A T93A) ± Bag1-L-HA or Bag1-S-HA were immunoprecipitated with anti-HA and immunoblotted with anti-Flag and the converse. Bag1-L/c-Jun interaction is impaired with a c-Jun phosphoacceptor mutant. (B) HCT116 cell extracts ± SP600125 (JNK inhibitor) were immunoprecipitated with rabbit IgG for c-Jun or a rabbit IgG and immunoblotted with anti-Bag1. The interaction of Bag1-L/c-Jun is abrogated by the inhibition of JNK activity. (C) Western blot of cell extracts of HCT116 cells transfected with shGFP or shJNK for 48 h. Immunoblotting was performed with the indicated antibodies and shows JNK knockdown results in reduced detection of p-c-Jun and p-JNK1/2. Tubulin detection confirmed equal loading of extracts. (D) HCT116 cells were transfected with Bag1-L-HA and Flag-tagged wild-type c-Jun, JunD, or JunB ± shGFP or shJNK as indicated. Bag1-L was immunoprecipitated with anti-HA, and immunoblotting was performed for c-Jun (Flag), JunD, and JunB as indicated. The interaction of Bag1-L and c-Jun is markedly reduced by shJNK treatment. Bag1-L/JunD interaction is also JNK dependent, whereas JunB shows no interaction. (E) Purified recombinant hBag1 was incubated with c-Jun biotinylated peptides (previously immobilized on streptavidin-coupled Dynabeads) as indicated. Complexes were immunoblotted with anti-Bag1, showing that the interaction is direct and is markedly enhanced with phosphorylated peptide. (F) NIH 3T3 cells were transfected to express Gal4-DBD-c-Jun or Gal4-DBD-c-Jun:Pan-Ala, Bag1-L ± pFC-MEKK, 5× Gal4UAS-luc, and the Ubi-Renilla-luc transfection control reporter gene (Ubi-Rluc). Luciferase activity was normalized to Ubi-Rluc. Data are represented as the means of results for triplicate wells ± SEM. Activity for Gal4-DBD-c-Jun + empty vector is arbitrarily set to 1. The ability of Bag1-L to enhance c-Jun-mediated transcriptional stimulation is phosphorylation dependent. (G) NIH 3T3 cells were transfected to express Gal4-DBD or Gal4-DBD-c-Jun; Bag1-L, Bag1-L-ΔBD, or Bag1-S ± pFC-MEKK; 5× Gal4UAS-luc; and the Ubi-Renilla-luc transfection control reporter gene (Ubi-Rluc). Luciferase activity was normalized to that for Ubi-Rluc. Data are represented as the means of results for triplicate wells ± SEM. Activity for Gal4-DBD-c-Jun + empty vector is arbitrarily set to 1. Bag1-LΔBD and Bag1-S cannot augment phosphorylated c-Jun-mediated transcriptional stimulation.
To assess the role of c-Jun N-terminal phosphorylation, the cooperation of GAL4-DBD-Jun derivatives with alanine replacements of all JNK phosphoacceptor residues with Bag1-L was tested in NIH 3T3 cells. GAL4-DBD-Jun and GAL4-DBD-JunPan-Ala showed similar transcriptional activities in the absence of stimulation, and Bag1-L expression failed to efficiently increase reporter activity. In contrast, when JNK activity was stimulated by expression of a constitutively active form of mitogen-activated protein kinase kinase kinase 1 (CA-MEKK-1), a strong cooperation between Bag1-L and GAL4-DBD-Jun but not with GAL4-DBD-JunPan-Ala was observed (Fig. 4F). This cooperation required direct physical interaction between Bag1-L and c-Jun, since deletion of the BAG domain that prevented c-Jun binding was incapable of stimulating reporter activity (Fig. 4G).
Bag1-L promotes expression of proapoptotic c-Jun target genes and neuronal death.
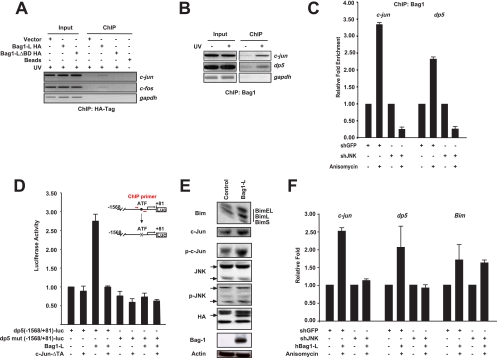
The transcriptional JNK death pathway is mediated through c-Jun-dependent transcriptional induction of the dp5 and bim genes, which encode proapoptotic members of the Bcl2 superfamily. Moreover, c-Jun increases AP-1 activity transcriptionally since c-Jun autoregulates its own transcription via 2 proximal AP-1 binding sites (37), and a functional AP-1 site is also present in the c-fos promoter (22). Chromatin immunoprecipitation (ChIP) assays of HA-tagged Bag1-L revealed efficient binding of Bag1-L to the c-jun and c-fos promoters but no binding to a control (gapdh) promoter. HA-Bag1-LΔBD, which lacks the BAG domain and thus cannot interact with c-Jun (Fig. 3A), did not bind to c-Jun-dependent promoters (Fig. 5 A). The proapoptotic dp5 gene is a direct target of the JNK-c-Jun pathway and contributes to c-Jun-mediated apoptosis (32, 46). Bag1-L bound to the CRE element in the dp5 promoter, and binding of Bag1-L to both the c-jun and dp5 promoters was strongly increased when JNK activation was triggered by UV (Fig. 5B). In addition, anisomycin treatment induced recruitment of Bag1-L to the AP-1 sites in the c-jun and dp5 promoters, but jnk1/2 knockdown prevented binding of Bag1-L to both promoters (Fig. 5C).
FIG. 5.
Bag1-L positively regulates the proapoptotic markers dp5 and Bim. (A) Chromatin immunoprecipitation was performed using HCT116 cells transfected with empty vector or HA-tagged versions of Bag1-L and Bag1-LΔBD. Cells were subsequently treated with 100 kJ/m2 UV for 2 h prior to collection and processing. ChIP was performed for the HA tag and binding to AP-1 sites in the c-jun and c-fos promoter regions determined by PCR using previously described primers (22, 37). Bag1-L exhibits in vivo occupancy of the c-Jun promoter, and deletion of the Bag domain results in loss of binding to AP-1 elements. (B) Endogenous ChIP was performed using HCT116 cells ± 100 kJ/m2 UV for 2 h prior to cell collection and processing using anti-Bag1. Binding of Bag1 to AP-1 sites in the c-Jun promoter was determined as detailed for panel A, and that to the human c-Jun/ATF-2 [TGATGTAA] site in the dp5 promoter was determined (46). Interaction with AP-1 sites in the c-Jun and dp5 promoters is augmented by JNK activation. (C) HCT116 cells were transfected with the indicated plasmids for 48 h. Cells were treated or not with 25 ng/ml anisomycin for 30 min prior to cell collection and processing. ChIP was performed using anti-Bag1. Binding of Bag1 to AP-1 sites in the c-Jun and dp5 promoters, detailed in panels A and B, was performed by real-time PCR. Enrichment from the IP of each sample was quantified with reference to the individual input, and results are presented as fold enhancement over results for untreated cells and are the means of results for triplicate wells ± SEM. Interaction of Bag1 with AP-1 sites in the c-Jun and dp5 promoters, augmented by anisomycin treatment, is blocked by JNK1/2 knockdown. (D) PC12 cells were differentiated with NGF and transfected to express Bag1-L and/or c-JunΔTA, −1568/+81 dp5-luc (CRE: TGATGTAA) or mut dp5-luc (CRE: TAACGTCT [mutated bases are underlined]), and the Ubi-Renilla-luc transfection control reporter gene (Ubi-Rluc). Luciferase activity was normalized to that for Ubi-Rluc. Data are represented as the means of results for triplicate wells ± SEM. Activity for −1568/+81 dp5-luc + empty vector is arbitrarily set to 1. The ability of Bag1-L to enhance AP-1 reporter activity is blocked by dominant-negative c-Jun. (E) PC12 cells were differentiated with NGF and transfected to express Bag1-L or empty vector. Transfection efficiency was 70%. Cells were collected, extracts were prepared, and immunoblotting was performed with the indicated antibodies. Bag1-L overexpression is accompanied by increased phosphorylated c-Jun levels and the increased expression of the proapoptotic BH3-only protein Bim. (F) HCT116 cells were transfected with the indicated plasmids for 48 h. Cells were treated or not with 25 ng/ml anisomycin for 30 min prior to collection, and mRNA was prepared for quantitative real-time-PCR. Results were normalized to those obtained with gapdh, and results are presented as fold induction over that for untreated cells and are the means of data for triplicate wells ± SEM. Primer sequences used were as previously described for c-Jun (42), dp5 (35), and Bim (7). The increase in c-Jun and dp5 transcript levels induced by anisomycin treatment is blocked by shJNK, but that of Bim is unaffected.
A reporter construct derived from the dp5 promoter was robustly activated by Bag1-L, and Bag1-L-mediated dp5-luciferase reporter gene activation was dependent on the integrity of the AP-1 site and was blocked by overexpression of a dominant-negative allele of c-Jun (Fig. 5D). Bag1-L overexpression increased c-Jun protein levels and increased Bim expression (Fig. 5E). In agreement with this, overexpression of Bag1-L also induced endogenous c-jun, dp5, and Bim mRNA expression, and jnk1/2 knockdown decreased gene activation by Bag1-L (Fig. 5F). Therefore, Bag1-L augments the transcription of proapoptotic genes in a c-Jun-dependent manner.
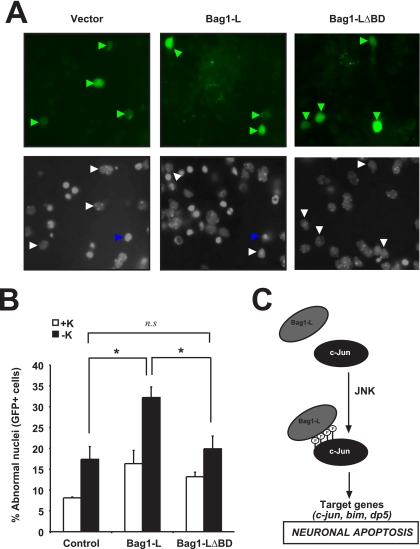
Primary cerebellar granule neurons (CGNs) are a well-characterized model system for studying neuronal death. CGNs depend on potassium depolarization for survival and undergo apoptosis when deprived of potassium. Dp5 is upregulated in a JNK/c-Jun-dependent manner during apoptosis induced by potassium deprivation, and knockdown of Dp5 by small interfering RNA rescued neurons from apoptosis (32, 46). Since Bag1-L cooperated with c-Jun in dp5 regulation (Fig. 5B and C), we examined the function of Bag1-L in neuronal death. Since bag1 mRNA knockdown would result in depletion of both the Bag1-S and Bag1-L proteins, which have different and sometimes opposing functions in cell death, we decided to investigate the consequences of Bag1-L overexpression. Ectopic Bag1-L expression exacerbated neuronal cell death induced by potassium deprivation, whereas Bag1-LΔBD had no significant effect compared to results for the control (Fig. 6 A and B). Therefore, Bag1-L augments neuronal death in primary CGNs.
FIG. 6.
Bag1-L overexpression increases cell death of CGNs. (A and B) Primary cerebellar neurons were isolated and cultured in medium containing high K+, which promotes their survival. At day in vitro 6 (DIV6), CGNs were transfected with Bag1-L, Bag1-LΔBD, or empty vector together with GFP to mark transfected cells. Cell death was measured after 48 h (including ±12 h of K+ withdrawal, which induces apoptosis) and is shown as a percentage of total cells analyzed. Representative images of each condition indicating normal nuclei (white arrows) and apoptotic nuclei (blue arrows) are shown. An asterisk (P < 0.05) indicates statistical significance; n.s, not statistically significant. (C) A model illustrating the coactivation of apoptotic c-Jun/JNK signaling by Bag1-L in neurons.
DISCUSSION
The JNK/c-Jun pathway has been implicated in the control of neuronal viability in several clinically relevant neurological disorders. JNK activation is observed in the affected areas of Alzheimer's disease (AD) patients (44), and JNK mediates Aβ neurotoxicity in animal models (43). Likewise, an absence of jnk2/jnk3 isoenzymes and JNK inhibition reduced neurotoxicity in mouse models of Parkinson's disease (23, 50). A peptide inhibitor of c-Jun N-terminal kinase was shown to protect against both acoustic trauma-induced auditory hair cell death and ischemic neuronal damage in a model of stroke (9, 38). Consequently, JNK inhibition has been proposed as a therapeutic strategy for neurodegenerative disorders (10, 27, 34).
N-terminal phosphorylation of c-Jun is an important mediator of JNK neurotoxicity. jnk3-deficient mice show reduced kainate-induced excitotoxicity, and the same phenotype is seen in junAA knock-in mice, in which the JNK phosphoacceptor serines 63 and 73 are mutated to alanines (5, 55). Similarly, trophic factor deprivation-induced death was significantly delayed in junAA homozygous primary sympathetic neurons (6). Expression of a c-Jun dominant-negative mutant or the junAA knock-in mutation diminished the induction of bim and dp5 RNA and protein levels after NGF withdrawal (6, 32, 46, 52). In addition, the mixed-lineage kinase inhibitor CEP-1347, which prevents JNK activation, also reduces the increase in bim and dp5 mRNA levels after NGF deprivation (32, 46).
While the importance of phosphorylated c-Jun as a mediator of JNK neurotoxicity is well documented, a molecular explanation for why and how c-Jun N-terminal phosphorylation contributes to neuronal cell death has been elusive. In this study, we found that Bag1-L binds preferentially to the phosphorylated form of c-Jun and augments target gene transcription by c-Jun/AP-1. Interestingly, Bag1-L also can bind JunD but not JunB in a phosphorylation-dependent manner (Fig. 4D). JNKs phosphorylate c-Jun with high efficiency and JunD less efficiently, but JunB is not a substrate for JNKs. It has been proposed that JunD, which lacks a JNK binding domain, is phosphorylated by JNK through heterodimerization with c-Jun (26). It is conceivable that c-Jun may also induce a similar transphosphorylation of Bag1-L by JNK; however, we could not detect a significant change in electrophoretic mobility in Bag1-L upon either JNK activation or inhibition (Fig. 2B). Thus, Bag1-L is capable of interacting with two JNK-phosphorylated AP-1 transcription factors.
Bag1-L but not the shorter isoforms can bind to DNA in a sequence-independent manner by making use of positively charged regions close to the amino terminus (19). Thus, while the C-terminal c-Jun interaction domain of Bag1 is present in both the short and long isoforms, the presence of the NLS and the DNA binding domain within the N terminus explains why Bag1-L but not Bag1-S functions as a c-Jun coactivator.
The two main Bag1 isoforms have very different functions in neuronal apoptosis. Bag1-S can interact with the antiapoptotic Bcl-2 protein and suppresses apoptosis, and increased neuronal death was observed in Bag1−/− mice (20). In contrast, here we show that in the context of JNK/c-Jun-mediated cell death, Bag1-L augments the proapoptotic function of c-Jun. Bag1-S and Bag1-L are generated from the same mRNA by different mechanisms. Production of Bag1-L is cap dependent, whereas synthesis of Bag1-S is dependent upon the presence of an internal ribosome entry segment (IRES). Translation from the Bag1 IRES was shown to become less efficient during some forms of apoptosis (16), indicating that translational control of Bag1 may play a role in the induction of apoptosis.
The DNA binding activity of Bag1-L is not believed to be sequence specific; thus, Bag1-L may be recruited to target gene promoters by the interaction with transcription factors. Bag1-L has been shown to interact with and stimulate transcription by nuclear hormone receptors (14, 18, 19, 53). The androgen receptor is one of the nuclear hormone receptors that interact with Bag1-L. Activation of the androgen receptor is crucial for prostate cancer growth, and Bag1-L has been found to be overexpressed in primary prostate tumors (33). Expression of c-Jun has been associated with a negative clinical outcome in prostate cancer (25), and Bag1-L stimulation of c-Jun activity may contribute to tumor growth. Thus, Bag1-L may be required to regulate c-Jun function during both cancer and neurodegeneration.
Acknowledgments
We thank H. Naumann for help with primary neuron culture and transfection and P. Angel, L. Bakiri, D. Bohmann, and M. Yaniv for providing essential reagents.
R. Sancho and X. Fontana were supported by Marie Curie Intraeuropean Fellowships (for R.S., MEIF-CT-2006-041119; and for X.F., PIEF-GA-2009-236307). J. Villadiego acknowledges the support of a postdoctoral fellowship from the Spanish Ministry of Science and Innovation. The London Research Institute is funded by CR-UK.
Footnotes
Published ahead of print on 1 June 2010.
REFERENCES
- 1.Alberts, A. S., O. Geneste, and R. Treisman. 1998. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell 92:475-487. [DOI] [PubMed] [Google Scholar]
- 2.Angel, P., K. Hattori, T. Smeal, and M. Karin. 1988. The jun proto-oncogene is positively autoregulated by its product, Jun/AP1. Cell 55:875-885. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, J. D., N. J. Arhel, S. S. Lee, A. Sharp, M. Al-Okail, G. Packham, A. Hague, C. Paraskeva, and A. C. Williams. 2005. Nuclear BAG-1 expression inhibits apoptosis in colorectal adenoma-derived epithelial cells. Apoptosis 10:301-311. [DOI] [PubMed] [Google Scholar]
- 4.Becker, E. B., J. Howell, Y. Kodama, P. A. Barker, and A. Bonni. 2004. Characterization of the c-Jun N-terminal kinase-BimEL signaling pathway in neuronal apoptosis. J. Neurosci. 24:8762-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens, A., M. Sibilia, and E. F. Wagner. 1999. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat. Genet. 21:326-329. [DOI] [PubMed] [Google Scholar]
- 6.Besirli, C. G., E. F. Wagner, and E. M. Johnson, Jr. 2005. The limited role of NH2-terminal c-Jun phosphorylation in neuronal apoptosis: identification of the nuclear pore complex as a potential target of the JNK pathway. J. Cell Biol. 170:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas, S. C., Y. Shi, J. P. Vonsattel, C. L. Leung, C. M. Troy, and L. A. Greene. 2007. Bim is elevated in Alzheimer's disease neurons and is required for beta-amyloid-induced neuronal apoptosis. J. Neurosci. 27:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borsello, T., and C. Bonny. 2004. Use of cell-permeable peptides to prevent neuronal degeneration. Trends Mol. Med. 10:239-244. [DOI] [PubMed] [Google Scholar]
- 9.Borsello, T., P. G. Clarke, L. Hirt, A. Vercelli, M. Repici, D. F. Schorderet, J. Bogousslavsky, and C. Bonny. 2003. A peptide inhibitor of c-Jun N-terminal kinase protects against excitotoxicity and cerebral ischemia. Nat. Med. 9:1180-1186. [DOI] [PubMed] [Google Scholar]
- 10.Borsello, T., and G. Forloni. 2007. JNK signalling: a possible target to prevent neurodegeneration. Curr. Pharm. Des. 13:1875-1886. [DOI] [PubMed] [Google Scholar]
- 11.Briknarova, K., S. Takayama, L. Brive, M. L. Havert, D. A. Knee, J. Velasco, S. Homma, E. Cabezas, J. Stuart, D. W. Hoyt, A. C. Satterthwait, M. Llinas, J. C. Reed, and K. R. Ely. 2001. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat. Struct. Biol. 8:349-352. [DOI] [PubMed] [Google Scholar]
- 12.Brimmell, M., J. S. Burns, P. Munson, L. McDonald, M. J. O'Hare, S. R. Lakhani, and G. Packham. 1999. High level expression of differentially localized BAG-1 isoforms in some oestrogen receptor-positive human breast cancers. Br. J. Cancer 81:1042-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao, J., M. M. Semenova, V. T. Solovyan, J. Han, E. T. Coffey, and M. J. Courtney. 2004. Distinct requirements for p38alpha and c-Jun N-terminal kinase stress-activated protein kinases in different forms of apoptotic neuronal death. J. Biol. Chem. 279:35903-35913. [DOI] [PubMed] [Google Scholar]
- 14.Cutress, R. I., P. A. Townsend, A. Sharp, A. Maison, L. Wood, R. Lee, M. Brimmell, M. A. Mullee, P. W. Johnson, G. T. Royle, A. C. Bateman, and G. Packham. 2003. The nuclear BAG-1 isoform, BAG-1L, enhances oestrogen-dependent transcription. Oncogene 22:4973-4982. [DOI] [PubMed] [Google Scholar]
- 15.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 16.Dobbyn, H. C., K. Hill, T. L. Hamilton, K. A. Spriggs, B. M. Pickering, M. J. Coldwell, C. H. de Moor, M. Bushell, and A. E. Willis. 2008. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene 27:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eilers, A., J. Whitfield, C. Babij, L. L. Rubin, and J. Ham. 1998. Role of the Jun kinase pathway in the regulation of c-Jun expression and apoptosis in sympathetic neurons. J. Neurosci. 18:1713-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froesch, B. A., S. Takayama, and J. C. Reed. 1998. BAG-1L protein enhances androgen receptor function. J. Biol. Chem. 273:11660-11666. [DOI] [PubMed] [Google Scholar]
- 19.Gehring, U. 2004. Biological activities of HAP46/BAG-1. The HAP46/BAG-1 protein: regulator of HSP70 chaperones, DNA-binding protein and stimulator of transcription. EMBO Rep. 5:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotz, R., S. Wiese, S. Takayama, G. C. Camarero, W. Rossoll, U. Schweizer, J. Troppmair, S. Jablonka, B. Holtmann, J. C. Reed, U. R. Rapp, and M. Sendtner. 2005. Bag1 is essential for differentiation and survival of hematopoietic and neuronal cells. Nat. Neurosci. 8:1169-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzey, M., S. Takayama, and J. C. Reed. 2000. BAG1L enhances trans-activation function of the vitamin D receptor. J. Biol. Chem. 275:40749-40756. [DOI] [PubMed] [Google Scholar]
- 22.Hayakawa, J., S. Mittal, Y. Wang, K. S. Korkmaz, E. Adamson, C. English, M. Ohmichi, M. McClelland, and D. Mercola. 2004. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol. Cell 16:521-535. [DOI] [PubMed] [Google Scholar]
- 23.Hunot, S., M. Vila, P. Teismann, R. J. Davis, E. C. Hirsch, S. Przedborski, P. Rakic, and R. A. Flavell. 2004. JNK-mediated induction of cyclooxygenase 2 is required for neurodegeneration in a mouse model of Parkinson's disease. Proc. Natl. Acad. Sci. U. S. A. 101:665-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jochum, W., E. Passegue, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20:2401-2412. [DOI] [PubMed] [Google Scholar]
- 25.Kajanne, R., P. Miettinen, M. Tenhunen, and S. Leppa. 2009. Transcription factor AP-1 promotes growth and radioresistance in prostate cancer cells. Int. J. Oncol. 35:1175-1182. [DOI] [PubMed] [Google Scholar]
- 26.Kallunki, T., T. Deng, M. Hibi, and M. Karin. 1996. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87:929-939. [DOI] [PubMed] [Google Scholar]
- 27.Karin, M., and E. Gallagher. 2005. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life 57:283-295. [DOI] [PubMed] [Google Scholar]
- 28.Kuan, C. Y., A. J. Whitmarsh, D. D. Yang, G. Liao, A. J. Schloemer, C. Dong, J. Bao, K. J. Banasiak, G. G. Haddad, R. A. Flavell, R. J. Davis, and P. Rakic. 2003. A critical role of neural-specific JNK3 for ischemic apoptosis. Proc. Natl. Acad. Sci. U. S. A. 100:15184-15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuan, C. Y., D. D. Yang, D. R. Samanta Roy, R. J. Davis, P. Rakic, and R. A. Flavell. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22:667-676. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. S., S. J. Crabb, N. Janghra, C. Carlberg, A. C. Williams, R. I. Cutress, G. Packham, and A. Hague. 2007. Subcellular localisation of BAG-1 and its regulation of vitamin D receptor-mediated transactivation and involucrin expression in oral keratinocytes: implications for oral carcinogenesis. Exp. Cell Res. 313:3222-3238. [DOI] [PubMed] [Google Scholar]
- 31.Lei, K., and R. J. Davis. 2003. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. U. S. A. 100:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, C., C. Ying, Z. Yuan, B. Song, D. Li, Y. Liu, B. Lai, W. Li, R. Chen, Y. P. Ching, and M. Li. 2007. dp5/HRK is a c-Jun target gene and required for apoptosis induced by potassium deprivation in cerebellar granule neurons. J. Biol. Chem. 282:30901-30909. [DOI] [PubMed] [Google Scholar]
- 33.Maki, H. E., O. R. Saramaki, L. Shatkina, P. M. Martikainen, T. L. Tammela, W. M. van Weerden, R. L. Vessella, A. C. Cato, and T. Visakorpi. 2007. Overexpression and gene amplification of BAG-1L in hormone-refractory prostate cancer. J. Pathol. 212:395-401. [DOI] [PubMed] [Google Scholar]
- 34.Manning, A. M., and R. J. Davis. 2003. Targeting JNK for therapeutic benefit: from junk to gold? Nat. Rev. Drug Discov. 2:554-565. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura, M., E. Ishida, K. Shimada, H. Nakase, T. Sakaki, and N. Konishi. 2006. Defective expression of HRK is associated with promoter methylation in primary central nervous system lymphomas. Oncology 70:212-221. [DOI] [PubMed] [Google Scholar]
- 36.Nateri, A. S., L. Riera-Sans, C. Da Costa, and A. Behrens. 2004. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303:1374-1378. [DOI] [PubMed] [Google Scholar]
- 37.Nateri, A. S., B. Spencer-Dene, and A. Behrens. 2005. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature 437:281-285. [DOI] [PubMed] [Google Scholar]
- 38.Pirvola, U., L. Xing-Qun, J. Virkkala, M. Saarma, C. Murakata, A. M. Camoratto, K. M. Walton, and J. Ylikoski. 2000. Rescue of hearing, auditory hair cells, and neurons by CEP-1347/KT7515, an inhibitor of c-Jun N-terminal kinase activation. J. Neurosci. 20:43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Putcha, G. V., S. Le, S. Frank, C. G. Besirli, K. Clark, B. Chu, S. Alix, R. J. Youle, A. LaMarche, A. C. Maroney, and E. M. Johnson, Jr. 2003. JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron 38:899-914. [DOI] [PubMed] [Google Scholar]
- 40.Putcha, G. V., K. L. Moulder, J. P. Golden, P. Bouillet, J. A. Adams, A. Strasser, and E. M. Johnson. 2001. Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron 29:615-628. [DOI] [PubMed] [Google Scholar]
- 41.Sabapathy, K., W. Jochum, K. Hochedlinger, L. Chang, M. Karin, and E. F. Wagner. 1999. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech. Dev. 89:115-124. [DOI] [PubMed] [Google Scholar]
- 42.Sancho, R., A. S. Nateri, A. Garcia de Vinuesa, C. Aguilera, E. Nye, B. Spencer-Dene, and A. Behrens. 2009. JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. EMBO J. 28:1843-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savage, M. J., Y. G. Lin, J. R. Ciallella, D. G. Flood, and R. W. Scott. 2002. Activation of c-Jun N-terminal kinase and p38 in an Alzheimer's disease model is associated with amyloid deposition. J. Neurosci. 22:3376-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shoji, M., N. Iwakami, S. Takeuchi, M. Waragai, M. Suzuki, I. Kanazawa, C. F. Lippa, S. Ono, and H. Okazawa. 2000. JNK activation is associated with intracellular beta-amyloid accumulation. Brain Res. Mol. Brain Res. 85:221-233. [DOI] [PubMed] [Google Scholar]
- 45.Takayama, S., Z. Xie, and J. C. Reed. 1999. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J. Biol. Chem. 274:781-786. [DOI] [PubMed] [Google Scholar]
- 46.Towers, E., J. Gilley, R. Randall, R. Hughes, M. Kristiansen, and J. Ham. 2009. The proapoptotic dp5 gene is a direct target of the MLK-JNK-c-Jun pathway in sympathetic neurons. Nucleic Acids Res. 37:3044-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Townsend, P. A., R. I. Cutress, A. Sharp, M. Brimmell, and G. Packham. 2003. BAG-1 prevents stress-induced long-term growth inhibition in breast cancer cells via a chaperone-dependent pathway. Cancer Res. 63:4150-4157. [PubMed] [Google Scholar]
- 48.Townsend, P. A., R. I. Cutress, A. Sharp, M. Brimmell, and G. Packham. 2003. BAG-1: a multifunctional regulator of cell growth and survival. Biochim. Biophys. Acta 1603:83-98. [DOI] [PubMed] [Google Scholar]
- 49.Townsend, P. A., A. Stephanou, G. Packham, and D. S. Latchman. 2005. BAG-1: a multi-functional pro-survival molecule. Int. J. Biochem. Cell Biol. 37:251-259. [DOI] [PubMed] [Google Scholar]
- 50.Wang, W., L. Shi, Y. Xie, C. Ma, W. Li, X. Su, S. Huang, R. Chen, Z. Zhu, Z. Mao, Y. Han, and M. Li. 2004. SP600125, a new JNK inhibitor, protects dopaminergic neurons in the MPTP model of Parkinson's disease. Neurosci. Res. 48:195-202. [DOI] [PubMed] [Google Scholar]
- 51.Watson, A., A. Eilers, D. Lallemand, J. Kyriakis, L. L. Rubin, and J. Ham. 1998. Phosphorylation of c-Jun is necessary for apoptosis induced by survival signal withdrawal in cerebellar granule neurons. J. Neurosci. 18:751-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitfield, J., S. J. Neame, L. Paquet, O. Bernard, and J. Ham. 2001. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron 29:629-643. [DOI] [PubMed] [Google Scholar]
- 53.Witcher, M., X. Yang, A. Pater, and S. C. Tang. 2001. BAG-1 p50 isoform interacts with the vitamin D receptor and its cellular overexpression inhibits the vitamin D pathway. Exp. Cell Res. 265:167-173. [DOI] [PubMed] [Google Scholar]
- 54.Xia, Z., M. Dickens, J. Raingeaud, R. J. Davis, and M. E. Greenberg. 1995. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 270:1326-1331. [DOI] [PubMed] [Google Scholar]
- 55.Yang, D. D., C. Y. Kuan, A. J. Whitmarsh, M. Rincón, T. S. Zheng, R. J. Davis, P. Rakic, and R. A. Flavell. 1997. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature 389:865-870. [DOI] [PubMed] [Google Scholar]
- 56.Zeiner, M., M. Gebauer, and U. Gehring. 1997. Mammalian protein RAP46: an interaction partner and modulator of 70 kDa heat shock proteins. EMBO J. 16:5483-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]