Abstract
The growth factor independence 1 (Gfi1) gene was originally discovered in the hematopoietic system, where it functions as a key regulator of stem cell homeostasis, as well as neutrophil and T-cell development. Outside the blood system, Gfi1 is essential for inner-ear hair and intestinal secretory cell differentiation. To understand the regulatory hierarchies within which Gfi1 operates to control these diverse biological functions, we used a combination of comparative genomics, locus-wide chromatin immunoprecipitation assays, functional validation in cell lines, and extensive transgenic mouse assays to identify and characterize the complete ensemble of Gfi1 regulatory elements. This concerted effort identified five distinct regulatory elements spread over 100kb each driving expression in transgenic mice to a subdomain of endogenous Gfi1. Detailed characterization of an enhancer 35 kb upstream of Gfi1 demonstrated activity in the dorsal aorta region and fetal liver in transgenic mice, which was bound by key stem cell transcription factors Scl/Tal1, PU.1/Sfpi1, Runx1, Erg, Meis1, and Gata2. Taken together, our results reveal the regulatory regions responsible for Gfi1 expression and importantly establish that Gfi1 expression at the sites of hematopoietic stem cell (HSC) emergence is controlled by key HSC regulators, thus integrating Gfi1 into the wider HSC regulatory networks.
The growth factor independence 1 gene (Gfi1) was originally identified in a retroviral screen designed to identify regulatory pathways that could initiate interleukin-2 independence in T cells (7). Gfi1 is the founding member of a small family of transcription factors and contains six C-terminal zinc fingers, as well as an N-terminal SNAG domain, which function as DNA-binding and transcriptional repressor domains, respectively (9, 41). Gfi1 is expressed in hematopoietic stem cells (HSCs) (11), specific subsets of T cells (39), granulocytes, monocytes, and activated macrophages (13). Outside the hematopoietic system, Gfi1 expression has been reported within sensory epithelia, the lungs, neuronal precursors, the inner ear, and intestinal epithelia and during mammary gland development (4, 14, 31, 37). Gfi1−/− mice are completely neutrophil deficient (12, 13). Moreover, recent studies have shown that Gfi1−/− HSCs are unable to maintain long-term hematopoiesis, because elevated levels of proliferation lead to eventual exhaustion of the stem cell pool (11, 40). Gfi1−/− mice have also been reported to suffer from hearing loss and from a high sensitivity to bacterial endotoxin (12, 13).
Key transcription factor genes such as Gfi1 function as components of wider transcriptional regulatory networks. cis-Regulatory elements form the building blocks of these networks, and their detailed functional characterization is required to reveal the regulatory hierarchies within which factors such as Gfi1 operate (6). A previous study characterized the Gfi1 promoter region, which was found to contain multiple Gfi1 binding motifs, suggesting repression via an autoregulatory feedback loop (39). However, no comprehensive locus-wide analysis of Gfi1 regulatory elements has been reported thus far. When analyzing the transcriptional control of other key hematopoietic regulators, we have shown previously that a locus-wide approach is essential since most regulatory elements are distal to the promoters (5, 16, 18, 21, 24-26, 32). Moreover, the combination of comparative genomic analysis with transgenic mouse reporter assays proved to be a particularly powerful strategy for the identification of bona fide regulatory elements. Given the function of Gfi1 as a major regulator of several developmental processes, identification of the molecular mechanisms that control Gfi1 expression would significantly expand our understanding of the underlying transcriptional networks.
In the present study we used a combination of comparative genomics, locus-wide ChIP-on-chip, transgenic mouse assays, and ChIP-Seq technologies to identify regulatory elements within the Gfi1 locus. Five distinct regions spread over a total of nearly 100 kb were shown to direct expression to subdomains of known sites of endogenous Gfi1 expression. Moreover, functional analysis of a distal upstream enhancer allowed us to identify six upstream regulators of Gfi1 in early hematopoietic cells, thus integrating Gfi1 into the wider regulatory networks controlling early blood development.
MATERIALS AND METHODS
Expression analysis.
Gfi1 and β-actin amplicons, either murine or human, were cloned into the pGEM-T Easy vector (Promega) to generate standard curves with known amounts of DNA. Reverse transcription-PCR was performed using Moloney murine leukemia virus (MMLV) RT (Invitrogen) and random hexamers. Gfi1 expression was normalized to β-actin. See Supplementary Table 1 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html for all the primers used in real-time PCR analyses.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (25). Immunoprecipitation was performed with antibodies against H3Ac (catalog no. 06-599; Millipore), PU.1 (sc-352x; Santa Cruz), Scl (sc-12954x; Santa Cruz), Runx1 (ab23980-100; Abcam), Erg (sc-354x; Santa Cruz), Meis1 (sc-10599x; Santa Cruz), and Gata2 (sc-9008x; Santa Cruz). The oligonucleotides used to generate the Gfi1 tiling array were designed by Primer3 (28) on repeat masked sequence-spanning Gfi1 and flanking genes (chr5: 106838278 to 106923132, mm7). The oligonucleotides were spotted in triplicate using a BioRobotics MicroGrid II total array system. Array design files have been submitted to ArrayExpress. The resulting data were plotted by using the Variable Width Bar Graph Drawer (http://hscl.cimr.cam.ac.uk/genomic_tools.html). Material was validated by real-time PCR prior to hybridizations/sequencing (for primers used, see Supplementary Table 1 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). All experiments have been deposited in ArrayExpress. Mouse coordinates were mapped to the human genome by using the liftover function provided by the UCSC Genome Center (http://genome.ucsc.edu/).
Transgenic analysis.
F0 transgenic mouse embryos were generated by pronuclear injection of lacZ reporter fragments as described previously (32). Whole-mount images were acquired by using a Nikon Digital Sight DS-FL1 camera attached to a Nikon SM7800 microscope (Nikon, Kingston-upon-Thames, United Kingdom). Images were processed using Adobe Photoshop and Adobe Illustrator (Adobe Systems, San Jose, CA). The numbers of transgenic mice generated for each construct are summarized in Supplementary Table 2 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html. The Gfi1 bacterial artificial chromosome (BAC) with lacZ targeted into the Gfi1 ATG was generated by using homologous recombination in Escherichia coli (details are available on request).
Cell culture, transfection, and analysis.
Candidate Gfi1 cis-regulatory elements were PCR amplified from mouse genomic DNA, cloned into pGL2-Basic or pGL2 promoter vectors (Promega), and verified by sequencing (details regarding primers and region alignments are available at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html). All cloned regions vary in size but are all numbered with respect to the ATG start codon in exon 2 of the Gfi1 gene. HPC-7 cells were maintained in Iscove's modified Dulbecco medium, 10% fetal calf serum (FCS), 1.5 × 10−4 M monothioglycerol, and stem cell factor (27). 416B and MOLT4 cell lines were maintained in RPMI with 10% FCS. 293T cells were maintained in Dulbecco modified Eagle medium with 10% FCS. For MOLT4 stable transfections, 10 μg of linearized plasmid DNA and 1 μg of linearized pGK-neo were coelectroporated. After 24 h, 0.75-mg/ml G418 selection medium was added. Pools of stably transfected cells were analyzed 10 to 14 days later as previously described (1). 293T cells were transfected by using the ProFection mammalian transfection system-calcium phosphate (Promega). All assays were performed at least twice in triplicate. DNA amounts were kept constant by adding the appropriate empty control vector, and the transfection efficiency was monitored by the addition of a lacZ reporter plasmid. The pGL2 promoter Gfi1 −35kb was transfected with the following expression plasmids: pEFBOS Lmo2, pEFBOS Scl, pEFBOS Ldb1, and pCDNA3 E47 (17).
In situ hybridization.
The Gfi1 riboprobe was generated from a plasmid containing the Gfi1 full-length cDNA. Riboprobe synthesis and in situ hybridization experiments were carried out as previously described (23).
RESULTS
Comparative genomic analysis reveals proximal as well as distal conserved noncoding regions within mammalian Gfi1 loci.
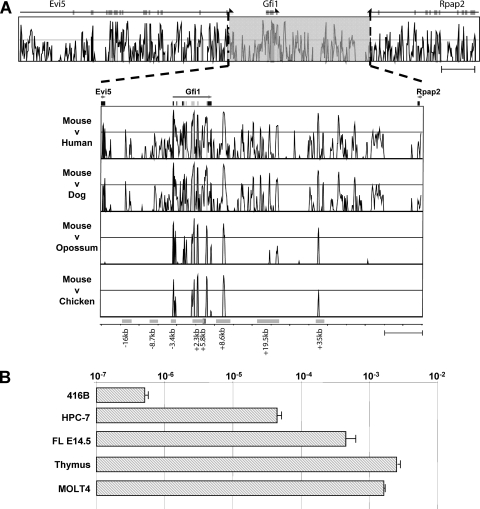
Sequence conservation of noncoding regions has long been recognized as a potential feature allowing the identification of gene regulatory sequences, and we have shown this to be an effective strategy in gene loci such as Endoglin, Lyl1, and Lmo2 (2, 8, 16, 18, 24, 25). When we compared the genomic sequences of the Gfi1 loci from mouse, human, dog, opossum, and chicken sources, we noticed that 5′ of the Gfi1 gene, noncoding sequence conservation was only apparent across eutherian mammals, whereas 3′ of Gfi1, several regions were conserved between eutherian mammals and opossums with two regions also found conserved in chicken. Taken together, this analysis highlighted a total of eight regions as illustrated in Fig. 1A: (i) two upstream regions and two intragenic regions conserved throughout the eutherians (at kb −16, −8.7, +2.3, and +5.8, respectively; numbering is assigned with respect to the ATG start codon in exon 2 of the Gfi1 gene), (ii) the −3.4kb promoter, the regions at kb +8.6 and the +35 conserved from mouse down to chicken, and (iii) the region at kb +19.5 conserved down to opossum. The conserved sequences therefore appeared to highlight a potential promoter region (at kb −3.4) and multiple candidate enhancer elements (at kb −16, −8.7, +2.3, +5.8, +8.6, +19.5, and +35). Within the hematopoietic system, Gfi1 is known to be expressed in hematopoietic precursors, hematopoietic stem cells, granulocytes, monocytes, and specific subsets of T cells (11, 13, 39). To identify cellular systems for functional validation of Gfi1 candidate regulatory elements, we performed expression analysis in a number of cell lines, as well as in primary cells (Fig. 1B). Gfi1 expression levels varied greatly between the different cell types, ranging from practically undetectable levels in 416B (myeloid progenitor), low levels in the multipotent hematopoietic progenitor cell line HPC-7, slightly higher levels in embryonic day 14.5 (E14.5) fetal liver (FL), and very high levels in whole mouse thymus and the human T-cell line MOLT4.
FIG. 1.
Comparative genomic analysis of the Gfi1 locus identifies eight potential regulatory elements. Extended Vista plot of sequence conservation between mice and humans of the loci of Evi5, Gfi1, and Rpap2. The expanded region of the Vista plot shows the sequence conservation across 85 kb of the mouse Gfi1 locus. The conservation plots correspond to (from top to bottom) mouse/human, mouse/dog, mouse/opossum, and mouse/chicken comparisons. The plots show conservation from 50 to 100% between the species based on a 100-bp window size. The exons and orientation of transcription are shown at the top of the figure. Potential cis-regulatory elements are highlighted by gray boxes, and the relevant names are displayed underneath. (B) Expression analysis of Gfi1 showing the mouse myeloid progenitor cell line (416B), the mouse multipotent hematopoietic progenitor cell line (HPC-7), mouse fetal liver (FL) on E14.5, an adult mouse thymus, and a human T-cell line (MOLT4). Gfi1 expression levels are displayed relative to β-actin.
Locus-wide ChIP-on-chip analysis identifies candidate regulatory regions.
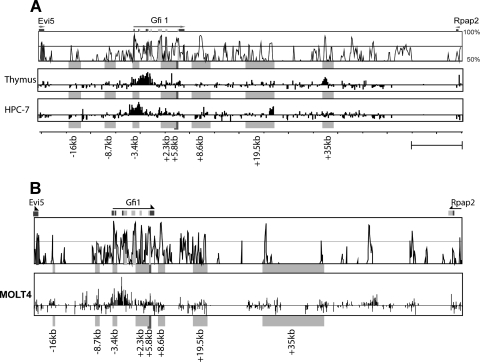
Integration of cross-species sequence conservation with long-range mapping of accessible chromatin provides a potentially powerful approach for the identification of gene regulatory elements. To identify the accessible regions within the Gfi1 locus, histone H3 acetylation ChIP-on-chip assays were performed on mouse thymus, as well as the HPC-7 progenitor cell line (Fig. 2A). The mouse array platform used covers more than 85 kb, spanning the entire Gfi1 locus and extending just into both flanking genes. In view of the high Gfi1 expression levels in the human MOLT4 T-cell line, histone acetylation was also analyzed by using an equivalent human tiling path array (Fig. 2B). Neither of the upstream regions (at kb −16 and −8.7) were acetylated in any of the mouse cell types tested, and yet low levels of acetylation can be detected in MOLT4 at both of these elements. Histone H3 acetylation levels were found to be high at the potential promoter region (kb −3.4) in HPC-7 and thymus, as well as the MOLT4 cell line, although a wider region is acetylated in the thymus material. A low level of acetylation could be detected at the kb +2.3 and +5.8 regions in all three cell types tested, potentially indicating intragenic cis-regulatory elements. The kb +8.6 region lacks any acetylation marks. In contrast, the kb +19.5 region was only acetylated in the HPC-7 and MOLT4 cells. The kb +35 element is acetylated in the thymus and MOLT4 T cells but is devoid of acetylated histone marks in HPC-7.
FIG. 2.
ChIP-on-chip assays for histone H3 acetylation identify candidate regulatory regions across the Gfi1 locus. ChIP assays were performed using the hematopoietic cell lines used for expression analysis in Fig. 1B. A Vista plot of mouse/human conservation is shown at the top of the figure, with candidate cis-regulatory elements highlighted by gray boxes. (A) Histone H3 AcK9 ChIP across the mouse Gfi1 locus highlights the promoter region, as well as the kb +2.3, +5.8, +8.6, and +19.5 elements as candidate regulatory elements in hematopoietic cells. (B) Histone H3 AcK9 ChIP across the human Gfi1 locus in T cells highlights a wide region of elevated histone acetylation around the Gfi1 promoter, as well as smaller peaks at the kb −16, −8.7, +2.3, and +5.8 elements. ChIP-on-chip results are shown as the fold enrichment over the median, with a scale ranging from −1 to +5.
Taken together, the ChIP-on-chip assays using murine cells identified elevated histone acetylation at five of the eight potential regulatory elements identified by the comparative genomics approach (the −3.4kb promoter and elements at kb +2.3, +5.8, +19.5, and + 35). Importantly, no other distal acetylated regions were identified in the mouse arrays, although there were two additional acetylated regions within the human cell line which were, however, not situated in conserved regions.
Transcriptional analysis of candidate cis-regulatory elements identifies promoter and enhancer regions.
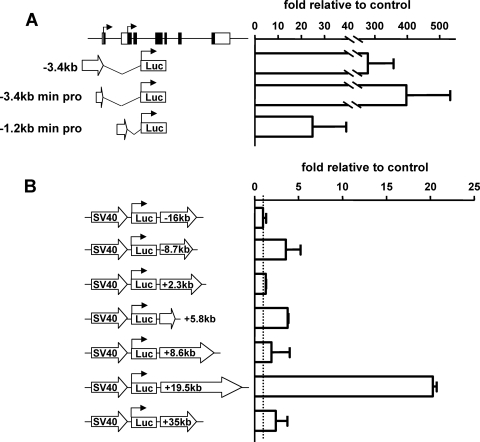
To further investigate candidate cis-regulatory regions within the Gfi1 locus, the potential regulatory elements highlighted by the comparative genomics and ChIP-on-chip assays were cloned into luciferase vectors and tested in stable transfection assays in MOLT4 T cells (Fig. 3). A closer inspection of the large acetylated region spanning 4.5 kb, including the first two exons of Gfi1, showed two independent regions of conservation and the presence of two possible ATGs, in exons 1 and 2, respectively. As previously indicated for the mouse Gfi1 gene (30), inspection of the Database of Transcriptional Start Sites (DBTSS) (see Supplementary Fig. 1 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html) confirmed two independent regions of transcriptional initiation, upstream of the exon 1 and exon 2 ATGs. These two regions of conservation were cloned to generate the luciferase reporter constructs −3.4kb min pro and −1.2kb min pro, respectively (Fig. 3A). Previously, a Gfi1 promoter was characterized (39) that corresponds to the −3.4kb promoter region described here. The regions at both kb −3.4 and kb −1.2 contained independent promoter activity, but activity of the distal −3.4kb promoter was significantly stronger (Fig. 3A).
FIG. 3.
In vitro assays identify two promoter and three enhancer elements. Stable transfections in MOLT4 cells highlight two promoter and three enhancer elements active within T cells. (A) Potential promoter elements were tested and normalized relative to cell numbers and pGL2-Basic luciferase values. (B) Potential enhancer elements were tested and normalized relative to the cell number and the pGL2 promoter.
To validate potential enhancer elements within the Gfi1 locus, all upstream and downstream regions highlighted by comparative genomics and/or ChIP-on-chip analysis were inserted into luciferase reporter constructs containing the heterologous simian virus 40 minimal promoter (Fig. 3B). The regions at kb −16, +2.3, +8.6, and +35 displayed no enhancer activity, in contrast to the regions at kb −8.7 and +5.8, which functioned as weak enhancers (Fig. 3B). The construct with the highest level of enhancer activity was the region at kb +19.5 (Fig. 3B). In summary, three of the seven conserved distal elements displayed enhancer activity when tested by stable transfection in MOLT4 cells (at kb −8.7, +5.8, and +19.5). Moreover, through detailed examination of the Gfi1 promoter region, two core regions of less than ∼300 bp were found to display promoter activity (Fig. 3A).
In vivo validation identifies cis-regulatory elements active in subdomains of endogenous Gfi1 expression.
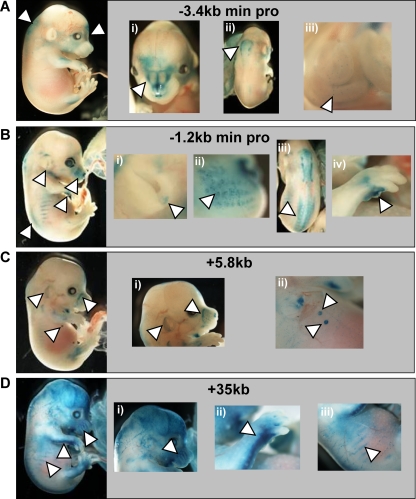
To assess the in vivo function of Gfi1 candidate cis-regulatory elements, F0 transgenic embryos were generated by pronuclear injection. Candidate elements were inserted into a lacZ reporter plasmid to generate a series of reporter constructs equivalent to the luciferase vectors tested above. Multiple transgenic embryos were generated for each construct to identify consistent staining patterns mediated by Gfi1 candidate elements. F0 embryos were assayed at E14.5, a time point that would allow detection of expression in fetal liver, as well as other tissues known to express Gfi1 within the developing ears and mammary glands. Ten regulatory regions were tested in the transgenic assays, resulting in the generation of a total of 99 transgenic embryos. Four regions displayed consistent staining patterns (called −3.4kb min pro, −1.2kb min pro, +5.8kb, and +35kb in Fig. 4): the −3.4kb min pro fragment induced lacZ expression in the developing nose, the central nervous system (CNS), and the developing gut (Fig. 4Ai, ii, and iii). The −1.2kb min promoter gave consistent lacZ expression within the ear, the Merkel cell region of the snout, the neural tube, and the limbs (Fig. 4Bi, ii, iii, and iv). The enhancer at kb +5.8 (Fig. 4C) was also active within the ear and the Merkel cell region of the snout (Fig. 4Ci) but, in addition, showed staining in the developing mammary glands of the mouse embryo (Fig. 4Cii). Finally, the region at kb +35 targeted expression to the ribs, the limbs, and the snout (Fig. 4Di, ii, and iii). Importantly, all of these expression domains have been previously reported as sites of Gfi1 expression. Transgenic assays therefore allowed us to identify four cis-regulatory elements which contribute to the endogenous expression pattern of Gfi1 (the results are summarized in Table 1). However, none of the above elements displayed consistent hematopoietic activity.
FIG. 4.
LacZ staining in transgenic embryos recapitulates the expression domains of endogenous Gfi1 expression. Transgenic mouse embryos carrying Gfi1 regulatory elements are shown. (A) −3.4kb min pro, showing staining within the nose (i), CNS (ii), and gut (iii). (B) −1.2kb min pro, showing staining of the ear (i), Merkel cells (ii), neural tube (iii), and limbs (iv). (C) +5.8kb, showing staining of the ear and Merkel cells (i) and mammary glands (ii). (D) +35kb enhancer element, showing staining in Merkel cells (i), limbs (ii), and ribs (iii). All highlighted staining patterns were found reproducibly in multiple independent founders.
TABLE 1.
Summary of LacZ staining in transgenic embryos
| Construct | Staining resulta |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nose | NT | Gut | Ear | Limb | MG | Ribs | Heart | FL | VV | |
| pGL2 Gfi1 −3.4kb min pro | + | + | + | - | - | - | - | - | - | - |
| pGL2 Gfi1 −1.2kb min pro | + | + | - | + | + | - | - | - | - | - |
| pGL2 pro Gfi1 +5.8kb | + | - | - | + | - | + | - | - | - | - |
| pGL2 pro Gfi1 +35kb | + | - | - | - | + | - | + | - | - | - |
| pGL2 pro Gfi1 −35kb | - | + | - | - | - | - | - | + | + | + |
These data represent a summary of the transgenic mouse embryos shown in Fig. 4. NT, neural tube; MG, mammary gland; FL, fetal liver; VV, vitelline vessels.
ChIP-Seq analysis identifies an additional element within the Gfi1 upstream flanking gene.
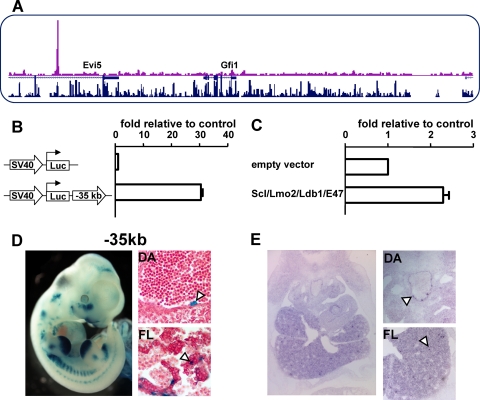
We recently published a genome-wide ChIP-Seq study reporting a genome-wide map of binding events for the HSC master regulator Scl in the progenitor cell line HPC-7 (38). This analysis highlighted the presence of a binding event of Scl at a conserved region upstream of Gfi1, within the last intron of Evi5 (Fig. 5A) and therefore not present on our tiling path arrays used for the ChIP-on-chip approach. The kb −35 region is more similar to the Gfi1 than to the Evi5 promoter and therefore represents a potential Gfi1 regulatory element. To further corroborate its potential role as a Gfi1 enhancer, we interrogated publicly available genome-wide ChIP-Seq data sets for the boundary factor CTCF, based on the previous demonstration that ubiquitous nonpromoter CTCF binding events indicate the location of boundary elements demarcating regulatory domains (31, 32, 36, 37). Of note, a ubiquitous CTCF site was found between the kb −35 region and the Evi5 promoter but not the Gfi1 promoter (see Supplementary Fig. 2 at http://hscl.cimr.cam.ac.uk/genomic_supplementary.html), thus corroborating its potential function as a Gfi1 regulatory element. Of note, the Gfi1 locus is a preferential target of the Moloney murine leukemia virus, with 85 common insert sites known to date (Retroviral Tagged Cancer Gene Database). Interestingly, 13 of the 85 integration sites reside within the same intron of Evi5 as the Scl-bound region at kb −35, a finding consistent with the notion that sequence elements in this intron of Evi5 control expression of Gfi1. This new element, Gfi1 −35kb, was cloned into a luciferase reporter construct and stably transfected into MOLT4 cells to test for enhancer activity (Fig. 5B), which demonstrated that this element clearly functions as a strong enhancer. To confirm that Scl binding was promoting activation of this element, transactivation assays were performed (Fig. 5C), which showed that the kb −35 region can be activated upon the addition of Scl.
FIG. 5.
ChIP-Seq analysis for Scl/Tal1 identifies a further distal regulatory element. (A) ChIP-Seq using a Scl antibody showed a distal region 35 kb upstream of Gfi1 that was strongly bound by Scl. (B) Stable transfections in MOLT4 show that the kb −35 construct has a high enhancer activity within T cells. (C) The Scl complex (Scl, Lmo2, Ldb1, and E47) can transactivate the kb −35 construct. (D) Transgenic analysis of E11.5 embryos highlights that the kb −35 element targets lacZ expression to tissues containing hematopoietic progenitor cells. E11.5 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-stained embryo and paraffin sections of dorsal aorta (DA) and fetal liver (FL). White arrowheads highlight reproducible staining patterns. (E) In situ hybridization using a Gfi1 probe show endogenous Gfi1 staining within an E11.5 embryo. Magnified views of the DA and FL are shown.
To assess the in vivo function of the kb −35 element, we performed F0 transgenic assays. Because the kb −35 element was identified as a Scl target region in the hematopoietic progenitor cell line HPC-7, embryos were analyzed at E11.5 since we were specifically interested in the hematopoietic stem/progenitor activity of this element. The kb −35 element showed LacZ staining of the heart, vitelline vessels, fetal liver, and dorsal aorta (Fig. 5D), the latter two of which contain hematopoietic stem/progenitor cell (HSPC) activity at this stage of mouse development. Other LacZ staining seen within the embryo shown in Fig. 5D was not consistent between multiple embryos and therefore considered ectopic. To confirm the endogenous staining pattern of Gfi1 at this stage in development, in situ hybridizations were performed on sections of E11.5 embryos. Gfi1 expression can clearly be detected within the fetal liver and dorsal aorta (Fig. 5E), which is consistent with enhancer activity of the kb −35 element.
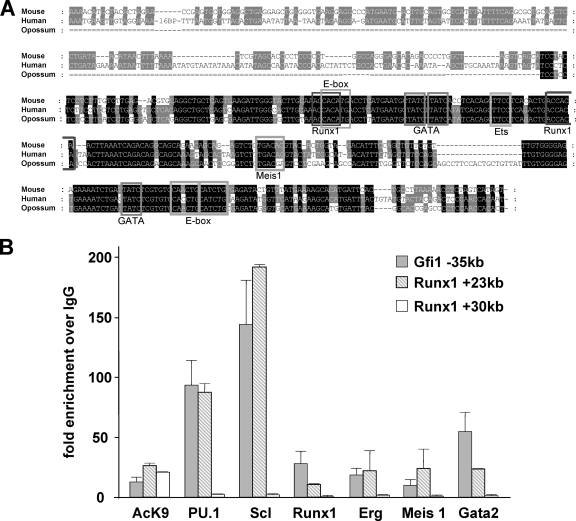
To identify additional potential candidate transcription factors that could be cooperating with Scl in driving the activity of the kb −35 element in early progenitor cells, we assessed the multispecies alignment of this element and found an array of consensus binding motifs. In addition to the E-box consensus binding site for Scl, binding sites for the key hematopoietic regulators Runx1, Gata2, PU.1, and Meis1 were detected (Fig. 6A). To confirm binding of these transcription factors to the motifs present, ChIP assays were performed within the HPC-7 cell line (Fig. 6B), which showed specific binding for all of the above transcription factors at levels comparable to those of the positive control region. Taken together, therefore, the kb −35 region exhibits consistent hematopoietic activity when tested in transgenic embryos and is controlled by a cohort of important hematopoietic transcription factors thus establishing a potential hierarchy of factors upstream of Gfi1 in early hematopoietic cells.
FIG. 6.
Characterization of the Gfi1 kb −35 element identifies several key stem cell transcription factors as upstream regulators. (A) Multispecies alignment of the kb −35 construct highlights the presence of highly conserved transcription factor motifs. (The transcription factor name is identified below the DNA motif.) (B) ChIP assay in HPC-7 cells shows clear binding of Scl, PU.1, Runx1, Erg, Meis1, and Gata2 to the kb −35 construct. All ChIP samples were normalized to IgG and compared to both a positive region (Runx1 +23kb) and a negative region (Runx1 +30kb). The Runx1 kb +23 construct has previously been shown to bind and be activated by the Scl complex of transcription factors, whereas the Runx1 kb +30 construct is known to be acetylated and within a region of open chromatin but is not bound by any transcription factors known to cooperate with Scl (17, 20). Hence, these elements are used as positive and negative regions, respectively.
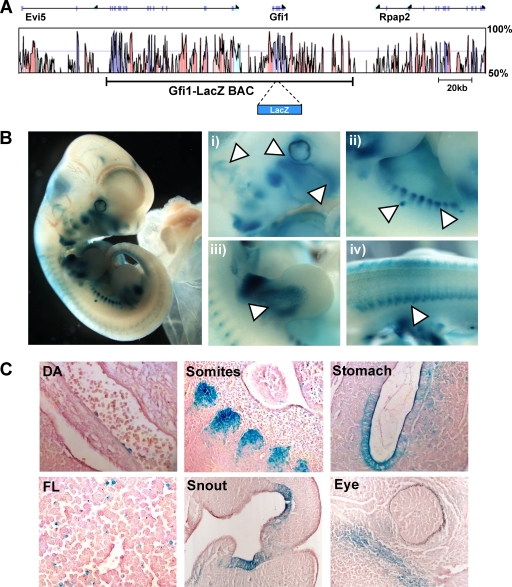
A 148-kb BAC recapitulates the activity of individual Gfi1 regulatory elements when tested in transgenic mice.
Finally, we wanted to confirm that the transcriptional activity seen in transgenic mice for the five individual regulatory elements corresponds to the gene regulatory activity of the wider Gfi1 locus. To examine this, we modified a BAC clone containing ∼148 kb of the Gfi1 locus (Fig. 7A) so that it would express the lacZ reporter gene instead of mouse Gfi1 (see Materials and Methods). When assayed in transgenic mice at midgestation, the Gfi1-lacZ BAC revealed lacZ expression at multiple sites, including the snout, ear, mammary glands, limbs, somites, and neural tube (Fig. 7B). Further analysis by histological sectioning not only confirmed specific expression in these tissues but also revealed rare lacZ-positive cells on the ventral wall of the dorsal aorta and in the fetal liver (Fig. 7C). Collectively, these expression domains closely correspond to the tissues expressing lacZ with single-element reporter constructs when tested in F0 transgenic assays. Altogether, therefore, these data suggest that we have identified five bona fide Gfi1 cis-regulatory elements that together recapitulate the endogenous expression pattern of Gfi1 at midgestation. Moreover, highly similar lacZ expression in the aorta-gonad-mesonephros (AGM) region and fetal liver hematopoietic cells with both the Gfi1-lacZ BAC and the kb −35 enhancer construct provides further evidence that while the kb −35 element may also work with the Evi5 promoter, it very likely acts on the Gfi1 promoter since the Gfi1-lacZ BAC does not contain the Evi5 promoter.
FIG. 7.
LacZ staining in mice embryos carrying the Gfi1-lacZ BAC identifies the sites of endogenous Gfi1 expression. (A) Vista plot of sequence conservation between mouse and human across the Gfi1 locus. The 148-kb region contained in the Gfi1-lacZ BAC, in which the lacZ gene has been inserted at the translational initiation ATG codon of Gfi1 by homologous recombination, is indicated below. (B) X-Gal-stained E11.5 embryos carrying the Gfi1-lacZ BAC showing lacZ expression in the eye, nose and ear (i), somites and mammary glands (ii), limbs (iii), and neural tube (iv). (C) Paraffin tissue sections from an E11.5 embryo showing lacZ expression in the dorsal aorta (DA), fetal liver (FL), somites, snout, stomach, and around the eye.
DISCUSSION
Gfi1 encodes a transcriptional repressor with critical functions in multiple tissues, including blood stem cells, T cells, granulocytes, monocytes, and activated macrophages (13). However, the regulatory hierarchies within which Gfi1 operates to perform its pleiotropic functions have remained largely obscure. We have identified and characterized here five regulatory elements spread over nearly 100 kb of the Gfi1 locus. These five elements drive expression to subdomains of the expression pattern of endogenous Gfi1 in transgenic mice. Specifically, we show that (i) an enhancer at kb −8.7 is active in stably transfected T cells but does not direct expression to a particular tissue in transgenic embryos; (ii) an enhancer inside the gene at kb +5.8 is active in stably transfected T cells and directs expression to the ears, snout, and mammary glands in transgenic mice; (iii) an enhancer located downstream of the gene at kb +19.5 is active in stably transfected T cells but does not direct expression to a particular tissue in transgenic embryos; (iv) a phylogenetically conserved enhancer at kb +35 displays a broader staining pattern with strong expression in snout, limbs, and ribs (but with weak activity in stably transfected T cells); (v) a hematopoietic enhancer is located in an intron of the adjacent Evi5 gene and displays activity in the midgestation AGM region and fetal liver; and (vi) the previously described promoters at kb −3.4 and −1.2 are active in cells of the embryonic nose, CNS, gut, snout, ears, ribs, neural tube, and limbs. In addition to comprehensive identification and in vivo validation of regulatory elements within the Gfi1 gene locus, functional analysis of the distal upstream enhancer allowed us to identify six upstream regulators of Gfi1 in early hematopoietic cells, thus integrating Gfi1 into the wider regulatory networks controlling early blood development.
Our results illustrate that identification of the whole complement of regulatory regions for complex gene loci is becoming less of a daunting task, largely thanks to recent technological advances in screening chromatin structure and the availability of multiple complete genome sequences for comparative genomic analysis. Locus-wide comparative genomics and ChIP-on-chip analysis coupled with functional validation of candidate elements revealed that the promoter and three distal regulatory regions all drive expression to subdomains of the Gfi1 expression pattern. However, none of the elements possessed hematopoietic activity. There are two possible explanations for these findings. (i) Promoter-specific enhancer interactions or combinations of elements might be required to produce expression patterns over and above a mere additive effect, with the latter in particular having been observed previously by us and others when studying gene loci such as Lmo2 and endoglin (16, 25). (ii) An alternative explanation could be that the definition of a gene locus as the domain between the two flanking genes is too simplistic as, for example, holds true for the α-globin locus (36). We show here that for the Gfi1 locus the latter explanation is true. Locus-wide ChIP-on-chip studies failed to discover the kb −35 enhancer because it is located in the last intron of Evi5. However, after its identification by ChIP-Seq, detailed characterization of this element allowed us to establish for the first time regulatory links between Gfi1 and other known key regulators of HSCs. Based on our analysis of the regulation of Gfi1 expression, neither comparative genomics nor ChIP-on-chip analysis alone would have allowed us to readily identify all of the elements tested. However, given that the lacZ expression pattern seen with the BAC closely mirrors the sum total of all patterns seen with individual elements, it would appear that the combination of comparative genomics and ChIP-on-chip provides a successful strategy. The major remaining caveat here would be that the definition of locus size is arbitrary as illustrated by our finding of the kb −35 enhancer being situated within an intron of the neighboring gene.
One previous study had investigated the promoter sequence of Gfi1 and suggested negative cross talk between Gfi1 and Gfi1b. Importantly, however, no upstream activators linking Gfi1 into the wider stem cell networks had been identified. In contrast, our analysis of the upstream enhancer demonstrated that Gfi1 is under the control of several key players that drive HSC development. This finding is interesting for several reasons. (i) Transcription factors such as Scl and Runx1 are drivers of HSC formation, whereas Gfi1 has been predominantly associated with HSC maintenance and/or homeostasis. This suggests that either Gfi1 performs an as-yet-unknown function in early embryonic HSPCs or regulatory programs required for homeostasis may be laid down very early. (ii) Although Gfi1 is unlikely to be a key target of Scl, Runx1, Gata2, Erg/PU.1, and Meis1 in their capacity to specify HSC development in the embryo, at least two of the upstream regulators have loss-of-function phenotypes in adult HSCs that are consistent with Gfi1 being an important mediator of stem cell homeostasis. Deletion of Runx1 in adult bone marrow causes expansion of immature progenitors, together with a myeloproliferation phenotype, with a concomitant reduction of long-term HSC (LT-HSC) activity (10). Similarly, compared to wild-type mice, Runx1+/− mice have increased numbers of progenitors and yet reduced numbers of LT-HSCs (33). Moreover, levels of Gata2 critically influence the balance between HSC quiescence and proliferation, with increased levels of Gata2 inducing quiescence within HSCs (35). Therefore, both phenotypes are consistent with the Gfi1 phenotype of HSC exhaustion through excessive proliferation. (iii) Although the kb −35 distal enhancer had strong activity in the MOLT4 T-cell line, different subsets of factors may be driving this activity because Scl and Gata2 are not expressed in the T-lymphoid lineage. This raises the possibility that the closely related lymphoid transcription factor Gata3 may control Gfi1 in T cells. Of note, Gata3 recently has also been shown to be expressed in mammary glands, whiskers, and the developing CNS (22), suggesting that it may also function upstream of Gfi1 in these tissues. Interestingly, we observed conserved GATA sites in the kb −16, +2.3, +5.8, +8.6, +19.5, and +35 elements identified in the present study. Most interesting of these is the GATA site within the kb +5.8 element, since this element drives Gfi1 expression within the developing mammary gland (Fig. 4Cii). This raises the possibility that Gata3 is upstream of Gfi1 in multiple tissues but acting through different subsets of regulatory elements. Given that both Runx1 and Runx3 have been shown to be expressed within developing bones of the mouse embryo this scenario may extend to Runx factors (19). Runx factors are known to be important for the CD4/CD8 lineage choice (3, 15, 29, 34), which is also subtly altered in Gfi1−/− animals. Runx factors may therefore control Gfi1 expression in T cells in concert with Gata3 through the kb −35 enhancer but may also act on the bone-specific −1.2kb min pro and kb +35 region of Gfi1 during early bone development.
In conclusion, we have shown that the Gfi1 locus displays a modular arrangement of gene regulatory elements responsible for specific aspects of Gfi1 expression and spread over nearly 100 kb. Detailed characterization of a Gfi1 hematopoietic enhancer established for the first time direct regulatory links between Gfi1 and other major regulators of HSCs. Finally, identification of nonhematopoietic Gfi1 enhancers lays the foundation for future studies to establish the regulatory hierarchies within which this important regulator operates in diverse organ systems.
Acknowledgments
We are grateful to Richard Auburn at FlyChip for printing the custom arrays, to Andrew D. Wood for providing material, to Tina Hamilton for performing BAC injections, and to Nicolas Bonadies for sections.
Work in our laboratories is supported by Leukemia and Lymphoma Research UK, the Leukemia and Lymphoma Foundation, Cancer Research UK, the United Kingdom Medical Research Council, and the Kay Kendall Leukemia Fund.
N.K.W. performed research, analyzed data, and helped write the paper. S.K., Y.H.C., S.H.O., J.R.L., and J.M. performed research. K.O. performed research and helped write the paper. B.G. designed research, analyzed data, and helped write the paper.
Footnotes
Published ahead of print on 1 June 2010.
REFERENCES
- 1.Bockamp, E. O., F. McLaughlin, A. M. Murrell, B. Gottgens, L. Robb, C. G. Begley, and A. R. Green. 1995. Lineage-restricted regulation of the murine SCL/TAL-1 promoter. Blood 86:1502-1514. [PubMed] [Google Scholar]
- 2.Chan, W. Y., G. A. Follows, G. Lacaud, J. E. Pimanda, J. R. Landry, S. Kinston, K. Knezevic, S. Piltz, I. J. Donaldson, L. Gambardella, F. Sablitzky, A. R. Green, V. Kouskoff, and B. Gottgens. 2007. The paralogous hematopoietic regulators Lyl1 and Scl are coregulated by Ets and GATA factors, but Lyl1 cannot rescue the early Scl−/− phenotype. Blood 109:1908-1916. [DOI] [PubMed] [Google Scholar]
- 3.Chi, T. H., M. Wan, K. Zhao, I. Taniuchi, L. Chen, D. R. Littman, and G. R. Crabtree. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418:195-199. [DOI] [PubMed] [Google Scholar]
- 4.Clarkson, R. W., M. T. Wayland, J. Lee, T. Freeman, and C. J. Watson. 2004. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res. 6:R92-R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donaldson, I. J., M. Chapman, S. Kinston, J. R. Landry, K. Knezevic, S. Piltz, N. Buckley, A. R. Green, and B. Gottgens. 2005. Genome-wide identification of cis-regulatory sequences controlling blood and endothelial development. Hum. Mol. Genet. 14:595-601. [DOI] [PubMed] [Google Scholar]
- 6.Foster, S. D., S. H. Oram, N. K. Wilson, and B. Gottgens. 2009. From genes to cells to tissues: modeling the hematopoietic system. Mol. Biosyst. 5:1413-1420. [DOI] [PubMed] [Google Scholar]
- 7.Gilks, C. B., S. E. Bear, H. L. Grimes, and P. N. Tsichlis. 1993. Progression of interleukin-2 (IL-2)-dependent rat T-cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol. Cell. Biol. 13:1759-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottgens, B., L. M. Barton, J. G. Gilbert, A. J. Bench, M. J. Sanchez, S. Bahn, S. Mistry, D. Grafham, A. McMurray, M. Vaudin, E. Amaya, D. R. Bentley, A. R. Green, and A. M. Sinclair. 2000. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 18:181-186. [DOI] [PubMed] [Google Scholar]
- 9.Grimes, H. L., T. O. Chan, P. A. Zweidler-McKay, B. Tong, and P. N. Tsichlis. 1996. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 16:6263-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Growney, J. D., H. Shigematsu, Z. Li, B. H. Lee, J. Adelsperger, R. Rowan, D. P. Curley, J. L. Kutok, K. Akashi, I. R. Williams, N. A. Speck, and D. G. Gilliland. 2005. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hock, H., M. J. Hamblen, H. M. Rooke, J. W. Schindler, S. Saleque, Y. Fujiwara, and S. H. Orkin. 2004. Gfi-1 restricts proliferation and preserves functional integrity of hematopoietic stem cells. Nature 431:1002-1007. [DOI] [PubMed] [Google Scholar]
- 12.Hock, H., M. J. Hamblen, H. M. Rooke, D. Traver, R. T. Bronson, S. Cameron, and S. H. Orkin. 2003. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18:109-120. [DOI] [PubMed] [Google Scholar]
- 13.Karsunky, H., H. Zeng, T. Schmidt, B. Zevnik, R. Kluge, K. W. Schmid, U. Duhrsen, and T. Moroy. 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30:295-300. [DOI] [PubMed] [Google Scholar]
- 14.Kazanjian, A., D. Wallis, N. Au, R. Nigam, K. J. Venken, P. T. Cagle, B. F. Dickey, H. J. Bellen, C. B. Gilks, and H. L. Grimes. 2004. Growth factor independence-1 is expressed in primary human neuroendocrine lung carcinomas and mediates the differentiation of murine pulmonary neuroendocrine cells. Cancer Res. 64:6874-6882. [DOI] [PubMed] [Google Scholar]
- 15.Kohu, K., T. Sato, S. Ohno, K. Hayashi, R. Uchino, N. Abe, M. Nakazato, N. Yoshida, T. Kikuchi, Y. Iwakura, Y. Inoue, T. Watanabe, S. Habu, and M. Satake. 2005. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage. J. Immunol. 174:2627-2636. [DOI] [PubMed] [Google Scholar]
- 16.Landry, J. R., N. Bonadies, S. Kinston, K. Knezevic, N. K. Wilson, S. H. Oram, M. Janes, S. Piltz, M. Hammett, J. Carter, T. Hamilton, I. J. Donaldson, G. Lacaud, J. Frampton, G. Follows, V. Kouskoff, and B. Gottgens. 2009. Expression of the leukaemia oncogene Lmo2 is controlled by an array of tissue specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood 113:5783-5792. [DOI] [PubMed] [Google Scholar]
- 17.Landry, J. R., S. Kinston, K. Knezevic, M. F. de Bruijn, N. Wilson, W. T. Nottingham, M. Peitz, F. Edenhofer, J. E. Pimanda, K. Ottersbach, and B. Gottgens. 2008. Runx genes are direct targets of Scl/Tal1 in the yolk sac and fetal liver. Blood 111:3005-3014. [DOI] [PubMed] [Google Scholar]
- 18.Landry, J. R., S. Kinston, K. Knezevic, I. J. Donaldson, A. R. Green, and B. Gottgens. 2005. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood 106:2680-2687. [DOI] [PubMed] [Google Scholar]
- 19.Levanon, D., O. Brenner, V. Negreanu, D. Bettoun, E. Woolf, R. Eilam, J. Lotem, U. Gat, F. Otto, N. Speck, and Y. Groner. 2001. Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech. Dev. 109:413-417. [DOI] [PubMed] [Google Scholar]
- 20.Nottingham, W. T., A. Jarratt, M. Burgess, C. L. Speck, J. F. Cheng, S. Prabhakar, E. M. Rubin, P. S. Li, J. Sloane-Stanley, A. S. J. Kong, and M. F. de Bruijn. 2007. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 110:4188-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogilvy, S., R. Ferreira, S. G. Piltz, J. M. Bowen, B. Gottgens, and A. R. Green. 2007. The SCL +40 enhancer targets the midbrain together with primitive and definitive hematopoiesis and is regulated by SCL and GATA proteins. Mol. Cell. Biol. 27:7206-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oosterwegel, M., J. Timmerman, J. Leiden, and H. Clevers. 1992. Expression of GATA-3 during lymphocyte differentiation and mouse embryogenesis. Dev. Immunol. 3:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ottersbach, K., and E. Dzierzak. 2005. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev. Cell 8:377-387. [DOI] [PubMed] [Google Scholar]
- 24.Pimanda, J. E., W. Y. Chan, I. J. Donaldson, M. Bowen, A. R. Green, and B. Gottgens. 2006. Endoglin expression in the endothelium is regulated by Fli-1, Erg, and Elf-1 acting on the promoter and a −8-kb enhancer. Blood 107:4737-4745. [DOI] [PubMed] [Google Scholar]
- 25.Pimanda, J. E., W. Y. Chan, N. K. Wilson, A. M. Smith, S. Kinston, K. Knezevic, M. E. Janes, J. R. Landry, A. Kolb-Kokocinski, J. Frampton, D. Tannahill, K. Ottersbach, G. A. Follows, G. Lacaud, V. Kouskoff, and B. Gottgens. 2008. Endoglin expression in blood and endothelium is differentially regulated by modular assembly of the Ets/Gata hemangioblast code. Blood 112:4512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pimanda, J. E., K. Ottersbach, K. Knezevic, S. Kinston, W. Y. Chan, N. K. Wilson, J. R. Landry, A. D. Wood, A. Kolb-Kokocinski, A. R. Green, D. Tannahill, G. Lacaud, V. Kouskoff, and B. Gottgens. 2007. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc. Natl. Acad. Sci. U. S. A. 104:17692-17697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto do Ó, P., A. Kolterud, and L. Carlsson. 1998. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 17:5744-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 29.Sato, T., S. Ohno, T. Hayashi, C. Sato, K. Kohu, M. Satake, and S. Habu. 2005. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity 22:317-328. [DOI] [PubMed] [Google Scholar]
- 30.Scheijen, B., J. Jonkers, D. Acton, and A. Berns. 1997. Characterization of pal-1, a common proviral insertion site in murine leukemia virus-induced lymphomas of c-myc and Pim-1 transgenic mice. J. Virol. 71:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shroyer, N. F., D. Wallis, K. J. Venken, H. J. Bellen, and H. Y. Zoghbi. 2005. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 19:2412-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinclair, A. M., B. Gottgens, L. M. Barton, M. L. Stanley, L. Pardanaud, M. Klaine, M. Gering, S. Bahn, M. Sanchez, A. J. Bench, J. L. Fordham, E. Bockamp, and A. R. Green. 1999. Distinct 5′ SCL enhancers direct transcription to developing brain, spinal cord, and endothelium: neural expression is mediated by GATA factor binding sites. Dev. Biol. 209:128-142. [DOI] [PubMed] [Google Scholar]
- 33.Sun, W., and J. R. Downing. 2004. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood 104:3565-3572. [DOI] [PubMed] [Google Scholar]
- 34.Taniuchi, I., M. Osato, T. Egawa, M. J. Sunshine, S. C. Bae, T. Komori, Y. Ito, and D. R. Littman. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111:621-633. [DOI] [PubMed] [Google Scholar]
- 35.Tipping, A. J., C. Pina, A. Castor, D. Hong, N. P. Rodrigues, L. Lazzari, G. E. May, S. E. Jacobsen, and T. Enver. 2009. High GATA-2 expression inhibits human hematopoietic stem and progenitor cell function by effects on cell cycle. Blood 113:2661-2672. [DOI] [PubMed] [Google Scholar]
- 36.Vyas, P., M. A. Vickers, D. L. Simmons, H. Ayyub, C. F. Craddock, and D. R. Higgs. 1992. Cis-acting sequences regulating expression of the human alpha-globin cluster lie within constitutively open chromatin. Cell 69:781-793. [DOI] [PubMed] [Google Scholar]
- 37.Wallis, D., M. Hamblen, Y. Zhou, K. J. Venken, A. Schumacher, H. L. Grimes, H. Y. Zoghbi, S. H. Orkin, and H. J. Bellen. 2003. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130:221-232. [DOI] [PubMed] [Google Scholar]
- 38.Wilson, N. K., D. Miranda-Saavedra, S. Kinston, N. Bonadies, S. D. Foster, F. Calero-Nieto, M. A. Dawson, I. J. Donaldson, S. Dumon, J. Frampton, R. Janky, X. H. Sun, S. A. Teichmann, A. J. Bannister, and B. Gottgens. 2009. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood 113:5456-5465. [DOI] [PubMed] [Google Scholar]
- 39.Yucel, R., C. Kosan, F. Heyd, and T. Moroy. 2004. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J. Biol. Chem. 279:40906-40917. [DOI] [PubMed] [Google Scholar]
- 40.Zeng, H., R. Yucel, C. Kosan, L. Klein-Hitpass, and T. Moroy. 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23:4116-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zweidler-Mckay, P. A., H. L. Grimes, M. M. Flubacher, and P. N. Tsichlis. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 16:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]