Abstract
Cyclin-dependent kinase-associated protein 1 (Cks1) is involved in the control of the transcription of a subset of genes in addition to its role in controlling the cell cycle in the budding yeast Saccharomyces cerevisiae. By directly ligating Cks1 onto a GAL1 promoter-driven reporter, we demonstrated that Cks1 acts as a transcription activator. Using this method, we dissected the downstream events from Cks1 recruitment at the promoter. We showed that subsequent to promoter binding, Cdc28 binding is required to modulate the level of gene expression. The ubiquitin-binding domain of Cks1 is essential for implementing downstream transcription events, which appears to recruit the proteasome via ubiquitylated proteasome subunits. We propose that the selective ability of Cks1 to bind ubiquitin allows this small molecule the flexibility to bind large protein complexes with specificity and that this may represent a novel mechanism of regulating transcriptional activation.
Transcription is a complex process, which involves multiple levels of regulation. Cks proteins are evolutionarily conserved and bind cyclin-dependent kinases with high affinity (7). In the budding yeast Saccharomyces cerevisiae, Cks1 binds the cell cycle kinase Cdc28. It was previously demonstrated that Cks1 function was required for the transcription of a subset of genes in the yeast genome, which require rapid upregulation, such as CDC20 (22) and GAL1. An intact Cks1/Cdc28 complex is required to maintain efficient transcription at these loci. Nonetheless, the kinase activity of Cdc28 per se is not essential (31).
Cdc28/Cks1-mediated transcription takes place through the recruitment of the proteasome to actively transcribing promoters and open reading frames (31). The proteasome is a large molecular machine that uses ATPases (via interactions with the lid subcomplex) to unfold polyubiquitinated proteins and then uses its core barrel to proteolytically degrade the unfolded proteins. Johnston and colleagues initially described a genetic link between the proteasome and RNA polymerase II (Pol II)-regulated transcription in the early 1990s. In subsequent studies, the function of the proteasome at different stages of transcription, from promoter activation to transcript elongation to termination, was delineated (8). Some of these processes, such as transcript termination, clearly require the proteolytic activity of the proteasome (12), while for others, the role of proteolysis is less clear (10, 11, 14).
A relationship between Cks1 and the proteasome was initially revealed by genetic analysis (16). Specifically, the RPN3 gene, which encodes a proteasome subunit, was identified as enhancing the temperature-sensitive phenotypes of both cks1 and cdc28-1N mutants (cdc28-1N encodes a catalytically active Cdk1 kinase that is deficient in Cks1 binding). Subsequent studies indicated that Cdc28 interacts directly with the proteasome biochemically (16). Despite these studies, to date, it remains unknown exactly how the Cks1/Cdc28 complex recruits the proteasome to chromatin.
In this study, we aimed to investigate whether Cks1 possesses transcription-activating properties if artificially ligated onto a promoter. Furthermore, we characterized the functional domain of Cks1, which is essential for transcription-specific roles.
MATERIALS AND METHODS
Yeast strains.
Yeast one-hybrid experiments were carried out with yeast strain AH109 (Clontech) (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ MEL1). Plasmids expressing Cks1-Gal4 fusion proteins were constructed in frame into vector pGBKT7 (2μm plasmid; Clontech). Sequences of cloning primers are available upon request.
All chromatin immunoprecipitation experiments were carried out with yeast strain W303 (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100). Gal4-Cks1 or the cks1-E94Q mutant (or their non-Gal4-tagged counterparts) was C-terminally tagged with the affinity Flag tag and cloned into expression vector pCM252 (Euroscarf) (5) for tetracycline-dependent expression.
Measurements of transcriptional activation strength: yeast-one hybrid assay.
HIS3 gene expression was measured by growing yeast cells on selective medium lacking tryptophan (Trp) (−Trp) and lacking histidine (His) (−His). TRP was the selective marker on vector pGBKT7. The activity of the HIS3 reporter was quantified by increasing amounts of 3-aminotriazole (3-AT; Sigma), a competitive inhibitor of His3. The lowest concentration of 3-AT that inhibited growth was considered the activation strength of the gene construct. Each experimental data point was an average of data from triplicate experiments.
RNA analysis.
Total RNA was extracted by using an RNeasy kit (Qiagen) and the corresponding RNase-free DNase (Qiagen). Equal amounts of total RNA were used in one-step reverse transcription (RT)-quantitative PCRs (qPCRs) (Qiagen) to quantify HIS3 expression by using primers specific to the transcript. Primers specific to the ACT1 transcript were used to control for loading. The following formula was used for normalization: y = (2−CT/2−control CT) × 100. Primer sequences are available upon request.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed as described previously by Kuras and Struhl (19), with modifications. W303 transformed with Flag-tagged Gal4 DNA-binding domain (Gal4DBD)-Cks1 constructs (carrying the TRP marker) was incubated for 12 h in dextrose medium −Trp medium in the presence of doxycycline (5 μg/ml) to induce the expression of the tagged protein. Cells were washed three times and incubated with medium without doxycycline to shut off the transcription of tagged Gal4DBD-Cks1. Samples were harvested at log phase and treated with 1% formaldehyde for 15 min at room temperature at 0, 5, or 7 h after Tet shutoff. Cross-linking was halted by using a final concentration of 125 mM glycine for 5 min. Lysate preparation and immunoprecipitation of Flag-tagged proteins were carried out by using the anti-Flag M2 affinity gel (Sigma-Aldrich). Eluants after reverse cross-linking were analyzed by using qPCR. PCR oligonucleotide sequences are available upon request. Experiments were repeated in triplicates.
Western blotting and antibodies.
Yeast lysates and Western blotting were carried out according to standard protocols. Myc-tagged Gal4DBD fusion proteins (Clontech) were detected by using the anti-Myc antibody 9E10 (Cancer Research UK [CRUK]). Flag-tagged proteins were detected by using the M2 anti-Flag antibody (Sigma). The anti-Cdc28 PSTAIRE domain antibody (Calbiochem) was used to detect Cdc28, which served as a loading control.
Antiubiquitin antibody was obtained from Biomol (PW8810). Tandem affinity (TAP)-tagged Pre-9 was obtained through the Openbiosystems yeast TAP-tagged collection. Whole-cell lysates were extracted in the absence or the presence of the deubiquitinase inhibitor N-ethylmaleimide (NEM) (Sigma).
Protein identification and characterization by MS.
Sample analysis was performed by liquid chromatography (LC)-tandem mass spectrometry (MS/MS) by using an Ultimate (LC-Packings; Dionex, Amsterdam, Netherlands) high-performance liquid chromatography (HPLC) system coupled online to a three-dimensional (3D) high-capacity ion trap (HCTplus; Bruker Daltonics, Bremen, Germany) mass spectrometer via a pneumatically assisted nano-electrospray source as described previously (4). To increase the range of identifiable proteins, a multidimensional protein identification technology (MudPIT) approach based on gas-phase fractionation was used. For this purpose, each sample was run four times with different mass acquisition ranges: 300 to 600, 600 to 900, 900 to 1,200, and 1,200 to 1,500 m/z. MS/MS spectra (peak lists) were searched against the SwissProt (release 54.0, July 2007; number of entries, 276,256) or trEMBL (release 37.0, July 2007) database using Mascot, version 2.2 (Matrixscience, London, United Kingdom), and the following parameters: peptide tolerance, 2.5 Da; 13C = 0; fragment tolerance, 0.8 Da; missed cleavages, 3; instrument type, electrospray ionization ion trap (ESI-TRAP). In cases where ubiquitination was examined, the Gly-Gly tag on lysine residues (+114.1 Da) was included as a variable modification as described previously (17). Alternatively, samples were subjected to separation by ultraperformance liquid chromatography tandem mass spectrometry analysis (nano-UPLC-MS) as described previously (30). In brief, separation was performed by using a 75-μm-internal-diameter (ID) by 25-cm C18 nanoAcquity UPLC column with a 1.7-m particle size (Waters, Milford, MA) and a 90-min gradient of 2% to 45% solvent B (solvent A is 99.9% H2O-0.1% formic acid; solvent B is 99.9% acetonitrile-0.1% formic acid) on a Waters nanoAcquity UPLC system (final flow rate, 250 nl/min [7,000 lb/in2]) coupled to a Waters QTOFPremier tandem mass spectrometer (Waters, Milford, MA). Data were acquired in high-definition MSE mode (low-collision energy of 4 eV; high-collision energy ramping from 15 eV to 40 eV, switching every 1.5 s) and processed with the ProteinLynx global server (PLGS, version 2.2.5; Waters, Milford, MA) to reconstruct MS/MS spectra by combining all masses with identical retention times. Note that some fragment ions may not always be assigned to the right parent ion in the case when several ions coelute. The mass accuracy of the raw data was corrected by using Glu-fibrinopeptide (GFP) (200 fmol/μl, 700-nl/min flow rate, 785.8426 Da, [M + 2H]2+) that was infused into the mass spectrometer as a lock mass during sample analysis. Low- and high-collision energy MS data were calibrated at intervals of 30 s. The raw data sets were processed, including deisotoping, deconvolution, and peak lists generated on the basis of assigning precursor ions and fragments based on similar retention times. MS/MS spectra (reconstituted peak lists) were searched against the SwissProt database (release 54.0, July 2007; number of entries, 276,256) by using Mascot, version 2.2 (Matrixscience, London, United Kingdom), and the following parameters: peptide tolerance, 0.2 Da; 13C = 2; fragment tolerance, 0.1 Da; missed cleavages, 3; variable modifications, carbamidomethylation C, oxidation M/R/K, Gly-Gly K; instrument type, electrospray ionization-quadrupole time of flight (ESI-QUAD-TOF).
The interpretation and presentation of MS/MS data were performed according to previously reported guidelines (26). In addition, individual MS/MS spectra for peptides with a Mascot Mowse score lower than 40 (Expect score of <0.015) were inspected manually and included in the statistics only if a series of at least four continuous y or b ions was observed. Protein identification is also based on the assignment of at least two peptides. In the case where proteins were identified based on one peptide sequence, the corresponding MS/MS spectra were inspected and verified manually.
RESULTS
Tethering of Cks1 to a promoter is sufficient to activate transcription.
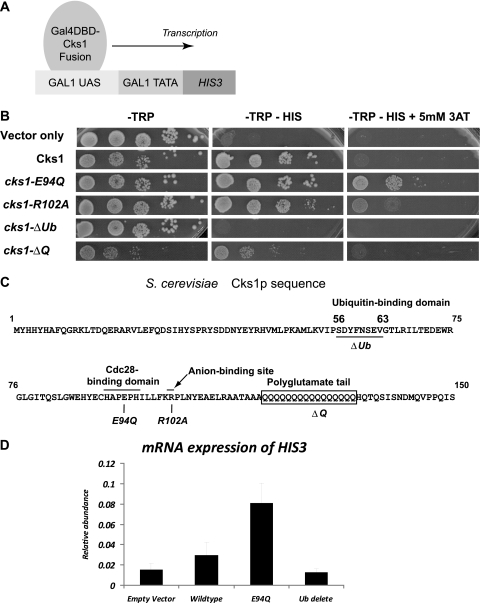
In order to investigate Cks1-dependent events at the promoter, we employed a yeast one-hybrid system as described previously (28). If Cks1 possessed properties of an activation domain (AD), its fusion to the Gal4 DNA-binding domain (Gal4DBD) should be sufficient to activate the transcription of a reporter gene with a GAL1-driven promoter (Fig. 1 A). This was indeed the case. A yeast strain expressing the Gal4DBD vector alone was unable to sustain growth on media without histidine (Fig. 1B). However, a yeast strain carrying the wild-type Cks1-Gal4DBD construct was able to express the reporter gene HIS3, thus allowing growth. By the addition of 3-aminotriazole (3-AT), which is a competitive inhibitor of the HIS3 gene product, the activity of the HIS3 reporter could be quantified (Table 1). Compared to the relative activity on yeast transcriptional activators reported previously by Titz et al. using this assay, the activity of wild-type Cks1 is equivalent to a “moderate”-strength transcription activator (28).
FIG. 1.
Tethering of Cks1 to a promoter is sufficient to activate transcription. (A and B) Schematic representation of the yeast one-hybrid system. Cks1 or mutants of Cks1 (see C) are fused with a Myc-tagged N-terminal Gal4DBD. Yeast strains that have maintained this plasmid express the TRP auxotrophic selection marker, allowing growth in −Trp medium (B, left). If Cks1 had transcription activation properties, this alone would be sufficient to drive the expression of the reporter gene HIS3 downstream of a GAL1 promoter. The expression of His3 allows yeast strains to grow in −His selective medium, which lacks the amino acid histidine (B, center). The His3 competitive inhibitor 3-AT increases the stringency of auxotrophic selection (right). Cell survival in the presence of 3-AT depends on the relative strength of the transcription activator. Each row denotes equal numbers of yeast cells spotted onto solid medium in 10-fold serial dilutions. UAS, upstream activation sequence. (C) Constructed Gal4DBD-Cks1 mutants. Positions of functional cks-1 mutants are indicated in the yeast Cks1 sequence. cks1-E94Q is a point mutant deficient in Cdc28 kinase binding activity. cks1-R102A is an anion-binding pocket mutant. cks1-ΔQ is a polyglutamate tail deletion mutant. cks1-ΔUb is a ubiquitin-binding domain deletion mutant. (D) Transcript levels of the HIS3 reporter driven by Gal4DBD-Cks1 fusion proteins. Total RNA was extracted from yeast cells carrying plasmids, which express either Gal4DBD alone (empty vector) or Gal4DBD fused with full-length Cks1 (wild-type), Gal4DBD fused with cks1-E94Q (E94Q), or Gal4DBD fused with cks1-ΔUb (Ub delete). mRNA was reverse transcribed and quantified by using real-time PCR. HIS3 transcripts were normalized against the level of ACT1 in each sample. Each data point is a representation of data from triplicate repeat experiments.
TABLE 1.
Relative strength of transcriptiona
| Gal4DBD fusion construct | Max concn of 3-AT tolerated (mM) |
|---|---|
| Vector only | 0 |
| Cks1 | 1 |
| cks1-E94Q | 50 |
| cks1-R102A | 0.8 |
| cks1-ΔQ | 1.2 |
| cks1-ΔUb | 0 |
| cks1-ΔDYFN | 0 |
| cks1-ΔDY | 0 |
| cks1-D57AY58A | 0.05 |
| cks1-Y58A | 0.05 |
| cks1-D57A | 1 |
In the presence of 3-AT, an inhibitor of the HIS3 gene product, the relative transactivating strengths of the fusion proteins can be determined. For strains that cannot grow in −His medium (i.e., no expression of the HIS3 reporter gene), the value zero is used. For known strong transactivators such as Swi5, the maximum concentration of 3-AT tolerated is typically 50 mM with this assay (28).
Cks1 has four known structural domains: a Cdk1/Cdc28-binding site (2), a conserved anion-binding pocket that is predicted to bind a phosphorylated substrate, a yeast-specific polyglutamate tail, and a recently described ubiquitin-binding domain (27). In mammalian Cks1, the anion-binding pocket binds the phosphorylated substrate p27 (15). However, no known phosphorylated substrate has been described to bind the anion pocket in yeast Cks1. Similarly, no biological function has been ascribed to the ubiquitin-binding domain or the polyglutamate tail.
We created mutations in each of these domains (Fig. 1C) and assayed them for their respective effects on transcription (Fig. 1B). Some of the mutations did not have a major effect on changing the Cks1-mediated transcription of the reporter after the ligation of Cks1 on the promoter (Fig. 1B and Table 1). These include mutations in the putative anion-binding pocket and the extended polyglutamate tail. Of note, two mutants produced interesting and relevant findings: the deletion of the ubiquitin-binding domain completely abrogated transcription activation, and a mutation that decreases the binding of Cks1 to Cdc28 caused a hyperactivation of transcription. These results were further confirmed and quantified at the transcript level by examining mRNA by RT-PCR (Fig. 1D). These two mutants are further characterized below.
Efficient Cdc28 binding after promoter recruitment is required to modulate the level of transcription.
Cks1 interacts with its cyclin-dependent partner Cdc28, and both are required for transcription activation (31). The evolutionarily conserved Glu94 residue, which resides in the β-hinge (HXPEPH) region, interacts directly with Cdc28. The cks1-E94Q mutant was previously shown to have a reduced Cdc28-binding capacity (6). Surprisingly, this mutant increases the activator strength of the fusion protein by 50-fold (Table 1). We surmised that the apparent discrepancy between the absolute requirement for Cdc28 in transcription activation (31) and the hyperactivation observed here is due to our experimental system in artificially uncoupling events from promoter recruitment to downstream activation. Morris et al. (22) showed previously that a Cdc28 mutant that has a greatly reduced capacity to bind Cks, Cdc28-1N (6), was inefficiently removed from the CDC20 promoter (which depended on Cks1/Cdc28 for activation). We hypothesized that after promoter recruitment, Cdc28 binding to Cks1 is required to remove Cks1 from the promoter and hence act as negative feedback to tune down transcription.
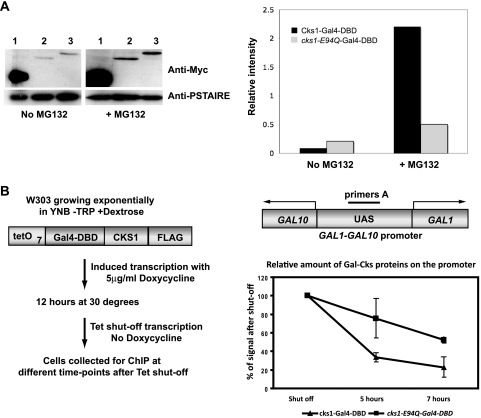
We performed Western blotting and compared the stabilities of Cks1-Gal4DBD and Cks1-E94Q-Gal4DBD in the presence and the absence of the proteasome inhibitor MG132 (Fig. 2 A). Cks1-E94Q-Gal4DBD was found to be more resistant to proteolysis endogenously. We tested this further by assaying the relative half-lives of Cks1 and the Cks1-E94Q mutant by chromatin immunoprecipitation (Fig. 2B). We introduced a tetracycline-inducible plasmid expressing either the Flag epitope-tagged Cks1-Gal4DBD or Cks1-E94Q-Gal4DBD into wild-type W303 yeast cells. The relative amount of tagged protein bound after tetracycline shutoff gave an indication of the relative half-lives of these proteins on the endogenous GAL1 promoter. In agreement with our hypothesis, we observed that Cks1-E94Q-Gal4DBD was more stable than the wild-type construct on the promoter.
FIG. 2.
The cks1-E94Q protein has a longer half-life than the wild-type protein. (A) The cks1-E94Q mutant protein is more stably expressed than the wild-type protein. (Left) Western blotting against the Myc-tagged fusion proteins was carried out. (Right) In order to quantify the amount of proteins present, the anti-Myc signal was normalized against the loading control Cdc28 by using AIDA software. The cks1-E94Q mutant fusion protein was expressed at a higher level endogenously than wild-type Cks1-Gal4DBD. The addition of the proteasome inhibitor MG132 increased the level of expression of the wild-type fusion protein, indicating that it was subjected to proteolysis in vivo. (B) The cks1-E94Q mutant remains longer on the promoter. Chromatin immunoprecipitation (ChIP) was carried out with wild-type yeast strain W303 carrying a plasmid carrying either tetracycline-inducible affinity-tagged Gal4DBD-Cks1 or Gal4DBD-Cks1-E94Q. The expression of the Gal4DBD fusion proteins was induced by incubation with doxycycline. Subsequent expression was shut off by the removal of doxycycline, and cells were harvested at log phase immediately or after 5 or 7 h. The relative amount of Gal4DBD fusion proteins on the endogenous GAL1 promoter was assayed by RT-PCR following chromatin immunoprecipitation. The signal amplified by primers spanning the upstream activation sequence of the GAL1 promoter (primers A) were normalized against primers against a transcriptionally silent control area on chromosome I. Each data point represents data from triplicate experiments. YNB, yeast nitrogen base.
The ubiquitin-binding domain of Cks1 is essential for downstream activating function.
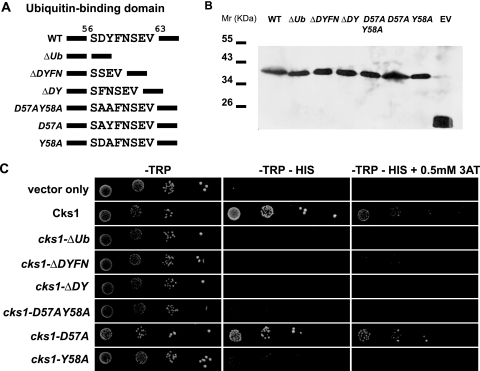
Next, we were interested in searching for domains within Cks1 that mediate downstream events of promoter activation, such as the recruitment of the proteasome. Tempe et al. (27) previously described an unusual interaction between Cks1 and ubiquitin in vitro. Through its alpha-helix, ubiquitin was shown to bind the N-terminal SDYFNSEV moiety of Cks1. Although provoking, no functional role has been ascribed to this interaction. We applied this mutant to the yeast one-hybrid assay (Fig. 1C and D and Table 1). Although stably expressed at the protein level (Fig. 3 B), the cks1-ΔUb mutant completely lacked all transcriptional activating activity despite being ligated to the promoter, representing a genuine lack of function. To exclude the possibility that the deletion of eight residues might affect the structure of the protein, we made sequential deletions (Fig. 3A). We identified the “DY” motif as two key residues important for this function and mapped the minimal functional motif to the Y58 residue (Fig. 3C and Table 1). We showed that instead of deleting the entire SDYFNSEV moiety, the deletion of the “DY” residues was sufficient to completely abrogate transcription activation. Interestingly, the replacement of these two residues with alanines (cks1-D57AY58A) was sufficient to greatly reduce transcription, confirming that it was not a mere shortening of the protein that caused our observation. Notably, changing the D57 residue alone to alanine was insufficient, as this mutant still exhibited wild-type activity (Fig. 3C and Table 1). On the other hand, although there was minimal residual activity compared to that of the ΔDY mutant (Table 1), the Y58A mutation was able to greatly reduce transcription activity by 20-fold, to the same extent as that of the cks1-D57AY58A mutant (Fig. 3C). We hypothesize that subsequent to promoter recruitment, Cks1, via binding to unknown ubiquitylated substrates via the Y58 residue, mediates downstream transcription events.
FIG. 3.
Mapping of the minimal ubiquitin-binding domain required for transcription activation in Cks1. (A) Schematic representation of mutants within the ubiquitin-binding domain of Cks1. A stretch of eight residues (positions 56 to 63) was previously demonstrated to bind ubiquitin in wild-type Cks1 (WT) (27). The deletion of these residues (ΔUb) abrogated transcription activation (Fig. 1B). The following mutants were designed to dissect the minimal function domain within the eight residues: ΔDYFN (deletion of residues 57 to 60), ΔDY (deletion of residues 57 and 58), D57AY58A (replacement of residues 57 and 58 with alanines), D57A (replacement of residue 57 with alanine), and Y58A (replacement of residue 58 with alanine). (B) All mutants are stably expressed in yeast. Western blotting using the anti-Myc antibody demonstrated that all Gal4DBD fusion mutants in the ubiquitin-binding domain were stably expressed in yeast strains carrying the plasmids (EV, Gal4DBD alone; WT, Gal4DBD-Cks1). (C) Yeast one-hybrid assay on ubiquitin domain mutants. Yeast cells harboring the above-described constructs were spotted onto selective medium in 10-fold dilutions. −Trp medium selects for yeast cells carrying the expression plasmids (left), and −Trp −His medium assays for the ability of the Gal4DBD fusion constructs to activate the transcription of the reporter gene HIS3 (middle). An example of the relative strengths of transcription activation by the mutants was demonstrated by the addition of 0.5 mM 3-AT (right). Detailed relative strengths of transcription of these mutants from triplicate experiments are shown in Table 1.
Cks1 recruits the proteasome through its ubiquitin-binding domain.
In order to explore Cks1-mediated events downstream of the promoter, we were interested in identifying other protein complexes that bound Cks1 at the promoter. To this end, we employed the Myc epitope-tagged Cks1-Gal4 fusion protein to immunoprecipitate interacting complexes. Immunoprecipitated complexes were subjected to in-solution trypsin digestion and analysis by tandem mass spectrometry (LC-MS/MS). Compared to the Gal4 vector alone, the presence of Cks1 increased the proportion of identified proteins that function in transcription and protein degradation.
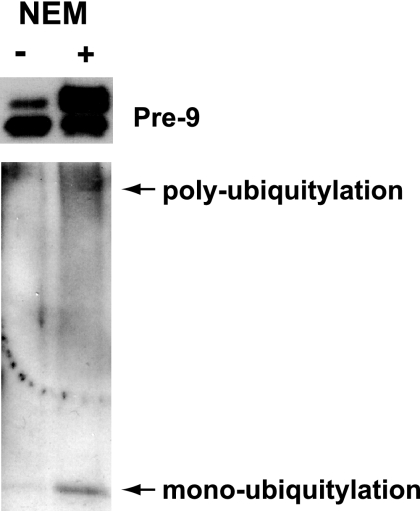
We established earlier on that the ubiquitin-binding domain was central to the Cks1-mediated transcriptional response. We therefore surmised that protein complexes downstream of this response were ubiquitylated and bound to Cks1 via their ubiquitylated subunits. We addressed this by comparing immunoprecipitated complexes from Gal4DBD-Cks1-Gal4 to those obtained from the Cks1-ubiquitin domain deletion mutant (cks1-ΔUb). As expected, only wild-type Cks1 enriched for ubiquitin (Table 2). Notably, components of the proteasome complex were identified in the wild-type Cks1-Gal4 sample but not in the cks1-ΔUb mutant. This is in accordance with what we already know for downstream of Cks1-mediated transcription (13). Other chromatin remodelers specifically enriched by Cks1-Gal4 included members of the SAGA complex and the nucleosome remodeler SWR1 (Table 2). The SAGA complex was previously shown to be activated directly by the proteasome (29). Interestingly, previously reported data that support the ubiquitylation of the proteasome already exist. A ubiquitylation site has been mapped to K199 of the Pre-9 subunit of the proteasome (3, 25). Since we specifically detected Pre-9 in wild-type Gal4DBD-Cks1 and not in cks1-ΔUb, we would like to confirm the ubiquitylation of this subunit in vivo. We purified the proteasome using a tandem affinity-tagged Pre-9 subunit (Fig. 4). Pre-9 was found to be monoubiquitylated and possibly polyubiquitylated.
TABLE 2.
The ubiquitin-binding domain of Cks1 binds the proteasome and other chromatin modifiers
| Complex | Protein identifiedb | SGD accession no.c | No. of peptidesd | Peptide sequenced |
|---|---|---|---|---|
| Proteasome | RPT3e | YDR394W | 2 | GVLLYGPPGTGKTMLVK |
| YLGEGPRMVRDVFRLAR | ||||
| PRE4 | YFR050C | 4 | GYGTQKI | |
| SSRNFSLAIIDKNTGLTFK | ||||
| KNLQVENMKWDFAKDIK | ||||
| KNLQVENMKWDFAKDIKGYGTQK | ||||
| PRE9e | YOR362C | 1 | AISVGANTSAAQTLLQMDYKDDMK | |
| SAGA | SUS1 | YBR111W-A | 1 | TMDTAQLKSQIQQYLVESGNYELISNELK |
| SGF73 | YGL066W | 3 | RSGDAEIKGIKPK | |
| QRNVNGGKSAKNGGK | ||||
| MREMFASSFSVKPGYTSPGYGAIHSRVGCLDLDR | ||||
| SWR | SWR1 | YDR334W | 2 | ALLLKVEKK |
| MSGKAHGGKGKSGAK | ||||
| SWC3 | YAL011W | 10 | IKAKLK | |
| KNDAEAK | ||||
| KFIEIAKK | ||||
| EAKTTAESTQVDVKK | ||||
| QQMQKKIAKEQK | ||||
| DKMQKMCDCVMSGGPHTFK | ||||
| FELNEWEHAMRSRRHKR | ||||
| DKMQKMCDCVMSGGPHTFK | ||||
| DKMQKMCDCVMSGGPHTFKVR | ||||
| MPAVLRTRSKESSIEQKPASRTR | ||||
| ARP6 | YLR085C | 2 | DKFGTSYLSNHIKNIK | |
| HADQVIFEEYEFDSLFKSPVAVFVPFTKSYKGEMR | ||||
| YAF9 | YNL107W | 1 | DAEVSSVYFDEIVFNEPNEEFFKILMSRPGNLLPSNK | |
| Ubiquitin | UBI4e | YLL039C | 1 | GGMQIFVK |
aImmunoprecipitated complexes purified from yeast cells expressing the Myc-tagged fusion proteins were subjected to mass spectrometry analysis (LC-MS/MS). Peptides uniquely present in the wild-type Cks1-Gal4DBD sample but absent from the cks1-ΔUb-Gal4DBD sample are listed.
Protein names are abbreviated.
Accession numbers are given for the SGD yeast database.
Number of peptides and peptide sequences obtained from MS data searches using Mascot as indicated in Materials and Methods.
Known to be ubiquitination substrates based on the SCUD database (http://scud.kaist.kr).
FIG. 4.
Western blot confirming the in vivo ubiquitylation of the proteasome subunit pre-9. Affinity-tagged Pre-9 was immunoprecipitated in the presence or absence of the deubiquitinase inhibitor N-ethylmaleimide (NEM). (Top) Western blot using the anti-TAP antibody to indicate the enrichment of tagged Pre-9. In the presence of NEM, a slow-migrating form of Pre-9 was enriched (right lane). (Bottom) Western blotting using an antiubiquitin antibody. Pre-9-enriched eluants demonstrate both monoubiquitylation and polyubiquitylation, which were preserved in the presence of NEM. The size of the monoubiquitylated band equals the size of the slow-migrating form of Pre-9 detected at the top.
DISCUSSION
In this study, we demonstrate that Cks1 acts as a transcription activator when ligated into a promoter using the yeast one-hybrid system. This is consistent with previous work showing that the recruitment of Cks1/Cdc28 to promoters directly activates the expression of certain genes, including CDC20 (22) and GAL1 (31).
In employing the yeast one-hybrid system, one has inadvertently uncoupled events at promoter recruitment from downstream transcription-activating events. We noted that Cdc28 binding is no longer necessary for transcription activation once Cks1 is on the promoter. In fact, Cdc28 binding is required to tune down transcription, as the cks1-E94Q mutant has 50 times the transactivating activity of Cks1. We showed in this study that the efficient binding of Cks1 to Cdc28 is required to destabilize the protein complex subsequent to promoter binding. This effect was observed when Cks1/Cdc28 was first studied at the CDC20 promoter (22). Morris et al. (22) showed that a Cdc28 mutant that has a greatly reduced capacity to bind Cks, Cdc28-1N (6), was inefficiently removed from the CDC20 promoter. Our finding here suggests that once Cks1 is on the promoter, Cdc28 becomes dispensable in terms of transcription activation. However, Cdc28 binding is subsequently important for the fine-tuning of the level of transcription activity. The prolonged presence of the Cks1/Cdc28 complex on the promoter leads to hyperactivation.
Cks1 has been shown to recruit the proteasome to active transcriptomes. However, it is not clear how this recruitment occurs. Analysis by tandem mass spectrometry is supportive of the presence of ubiquitylated subunits of the proteasome bound to Gal4-Cks1. We confirmed this finding by Western blotting, which showed that monoubiquitylation, as well as possible polyubiquitylation, was present.
Tempe et al. showed that Cks1 binds monoubiquitin with a very high affinity (Kd [dissociation constant] of 91 nM) via an unusual site, and unlike conventional ubiquitin-binding domains, monoubiquitin binding is favored over tetraubiquitin chains (27). We mapped the minimal functional domain to the D57Y58 residues of Cks1. The observation that the Y58A mutant was lacking in transcriptional activities suggests that possible phosphorylation at Y58 may be important for the ability of Cks1 to bind ubiquitin.
Although we cannot presently exclude the possibility that Cks1 binds free ubiquitin, we favor the model that Cks1 recruits the proteasome via a monoubiquitylated subunit (immunoprecipitation of the Cks1-bound proteasome showed ubiquitin modifications, and Western blotting of the Pre-9 subunit supports this notion. Pre-9 has also been previously shown by Tagwerker et al. to undergo ubiquitylation [25]). Activating monoubiquitylation has been proposed in models to explain the involvement of ubiquitin in transcription activation (1, 18, 23). It is yet unclear how exactly monoubiquitylation caused transcription downstream. Cks1 may act as a hub for controlling when monoubiquitylated transcription factors bind to chromatin. Further experiments are required to verify this hypothesis.
The proteasome was previously linked with the activation of the SAGA complex at the promoter (20), which is consistent with our observations. We postulate that the presence of the SAGA complex denotes downstream activating events secondary to proteasome recruitment. We were intrigued to note the presence of members of the SWR complex in Gal4-Cks1-bound extracts. The interaction between the SWR complex and Cks1 may be indirect, via the proteasome. Nonetheless, we cannot exclude the possibility that the SWR complex could potentially undergo ubiquitylation and become a direct target of Cks1.
The SWR complex is responsible for the deposition of the histone H2A variant Htz1 (or H2A.Z). The Htz1-containing nucleosome was proposed to “destabilize” nucleosomes (32). Furthermore, the deletion of Htz1 was shown previously to increase the requirement for SAGA genetically (24). As the proteasome is also implicated in activating SAGA, this could potentially be relevant mechanistically. Htz1 deposition also determines silencing boundaries (21) and may explain some of the silencing defects observed for proteasomal mutants (9). Current investigations are under way to examine the potential relationship between Cks1/Cdc28, the proteasome, and the SWR complex.
Acknowledgments
The Yu and the Kessler laboratories are both funded by the Medical Research Council (MRC). We acknowledge the Wellcome Trust for a traveling fellowship to V.P.C.C.Y. The Mascot “in-house” server is supported by the Computational Biology Research Group at the University of Oxford. We thank the DREAM program of the Chinese University of Hong Kong for scholarships to C.W.H.Y. and I.L.
Footnotes
Published ahead of print on 1 June 2010.
REFERENCES
- 1.Archer, C. T., L. Burdine, and T. Kodadek. 2005. Identification of Gal4 activation domain-binding proteins in the 26S proteasome by periodate-triggered cross-linking. Mol. Biosyst. 1:366-372. [DOI] [PubMed] [Google Scholar]
- 2.Arvai, A. S., Y. Bourne, M. J. Hickey, and J. A. Tainer. 1995. Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain. J. Mol. Biol. 249:835-842. [DOI] [PubMed] [Google Scholar]
- 3.Auld, K. L., C. R. Brown, J. M. Casolari, S. Komili, and P. A. Silver. 2006. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol. Cell 21:861-871. [DOI] [PubMed] [Google Scholar]
- 4.Batycka, M., N. F. Inglis, K. Cook, A. Adam, D. Fraser-Pitt, D. G. Smith, L. Main, A. Lubben, and B. M. Kessler. 2006. Ultra-fast tandem mass spectrometry scanning combined with monolithic column liquid chromatography increases throughput in proteomic analysis. Rapid Commun. Mass Spectrom. 20:2074-2080. [DOI] [PubMed] [Google Scholar]
- 5.Belli, G., E. Gari, L. Piedrafita, M. Aldea, and E. Herrero. 1998. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 26:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourne, Y., M. H. Watson, A. S. Arvai, S. L. Bernstein, S. I. Reed, and J. A. Tainer. 2000. Crystal structure and mutational analysis of the Saccharomyces cerevisiae cell cycle regulatory protein Cks1: implications for domain swapping, anion binding and protein interactions. Structure Fold Des. 8:841-850. [DOI] [PubMed] [Google Scholar]
- 7.Bourne, Y., M. H. Watson, M. J. Hickey, W. Holmes, W. Rocque, S. I. Reed, and J. A. Tainer. 1996. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell 84:863-874. [DOI] [PubMed] [Google Scholar]
- 8.Collins, G. A., and W. P. Tansey. 2006. The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 16:197-202. [DOI] [PubMed] [Google Scholar]
- 9.Ezhkova, E., and W. P. Tansey. 2004. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell 13:435-442. [DOI] [PubMed] [Google Scholar]
- 10.Ferdous, A., F. Gonzalez, L. Sun, T. Kodadek, and S. A. Johnston. 2001. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell 7:981-991. [DOI] [PubMed] [Google Scholar]
- 11.Ferdous, A., T. Kodadek, and S. A. Johnston. 2002. A nonproteolytic function of the 19S regulatory subunit of the 26S proteasome is required for efficient activated transcription by human RNA polymerase II. Biochemistry 41:12798-12805. [DOI] [PubMed] [Google Scholar]
- 12.Gillette, T. G., F. Gonzalez, A. Delahodde, S. A. Johnston, and T. Kodadek. 2004. Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl. Acad. Sci. U. S. A. 101:5904-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillette, T. G., S. Yu, Z. Zhou, R. Waters, S. A. Johnston, and S. H. Reed. 2006. Distinct functions of the ubiquitin-proteasome pathway influence nucleotide excision repair. EMBO J. 25:2529-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, F., A. Delahodde, T. Kodadek, and S. A. Johnston. 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296:548-550. [DOI] [PubMed] [Google Scholar]
- 15.Hao, B., N. Zheng, B. A. Schulman, G. Wu, J. J. Miller, M. Pagano, and N. P. Pavletich. 2005. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol. Cell 20:9-19. [DOI] [PubMed] [Google Scholar]
- 16.Kaiser, P., V. Moncollin, D. J. Clarke, M. H. Watson, B. L. Bertolaet, S. I. Reed, and E. Bailly. 1999. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 13:1190-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkpatrick, D. S., N. A. Hathaway, J. Hanna, S. Elsasser, J. Rush, D. Finley, R. W. King, and S. P. Gygi. 2006. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8:700-710. [DOI] [PubMed] [Google Scholar]
- 18.Kodadek, T. 2010. No splicing, no dicing: non-proteolytic roles of the ubiquitin-proteasome system in transcription. J. Biol. Chem. 285:2221-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 20.Lee, D., E. Ezhkova, B. Li, S. G. Pattenden, W. P. Tansey, and J. L. Workman. 2005. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123:423-436. [DOI] [PubMed] [Google Scholar]
- 21.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 22.Morris, M. C., P. Kaiser, S. Rudyak, C. Baskerville, M. H. Watson, and S. I. Reed. 2003. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 424:1009-1013. [DOI] [PubMed] [Google Scholar]
- 23.Nalley, K., S. A. Johnston, and T. Kodadek. 2006. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature 442:1054-1057. [DOI] [PubMed] [Google Scholar]
- 24.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411-422. [DOI] [PubMed] [Google Scholar]
- 25.Tagwerker, C., K. Flick, M. Cui, C. Guerrero, Y. Dou, B. Auer, P. Baldi, L. Huang, and P. Kaiser. 2006. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics 5:737-748. [DOI] [PubMed] [Google Scholar]
- 26.Taylor, G. K., and D. R. Goodlett. 2005. Rules governing protein identification by mass spectrometry. Rapid Commun. Mass Spectrom. 19:3420. [DOI] [PubMed] [Google Scholar]
- 27.Tempe, D., M. Brengues, P. Mayonove, H. Bensaad, C. Lacrouts, and M. C. Morris. 2007. The alpha helix of ubiquitin interacts with yeast cyclin-dependent kinase subunit CKS1. Biochemistry 46:45-54. [DOI] [PubMed] [Google Scholar]
- 28.Titz, B., S. Thomas, S. V. Rajagopala, T. Chiba, T. Ito, and P. Uetz. 2006. Transcriptional activators in yeast. Nucleic Acids Res. 34:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Workman, J. L. 2006. Nucleosome displacement in transcription. Genes Dev. 20:2009-2017. [DOI] [PubMed] [Google Scholar]
- 30.Xu, D., N. Suenaga, M. J. Edelmann, R. Fridman, R. J. Muschel, and B. M. Kessler. 2008. Novel MMP-9 substrates in cancer cells revealed by a label-free quantitative proteomics approach. Mol. Cell. Proteomics 7:2215-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu, V. P., C. Baskerville, B. Grunenfelder, and S. I. Reed. 2005. A kinase-independent function of Cks1 and Cdk1 in regulation of transcription. Mol. Cell 17:145-151. [DOI] [PubMed] [Google Scholar]
- 32.Zlatanova, J., and A. Thakar. 2008. H2A.Z: view from the top. Structure 16:166-179. [DOI] [PubMed] [Google Scholar]