Abstract
The 26S proteasome, a central enzyme for ubiquitin-dependent proteolysis, is a highly complex structure comprising 33 distinct subunits. Recent studies have revealed multiple dedicated chaperones involved in proteasome assembly both in yeast and in mammals. However, none of these chaperones is essential for yeast viability. PAC1 is a mammalian proteasome assembly chaperone that plays a role in the initial assembly of the 20S proteasome, the catalytic core of the 26S proteasome, but does not cause a complete loss of the 20S proteasome when knocked down. Thus, both chaperone-dependent and -independent assembly pathways exist in cells, but the contribution of the chaperone-dependent pathway remains unclear. To elucidate its biological significance in mammals, we generated PAC1 conditional knockout mice. PAC1-null mice exhibited early embryonic lethality, demonstrating that PAC1 is essential for mammalian development, especially for explosive cell proliferation. In quiescent adult hepatocytes, PAC1 is responsible for producing the majority of the 20S proteasome. PAC1-deficient hepatocytes contained normal amounts of the 26S proteasome, but they completely lost the free latent 20S proteasome. They also accumulated ubiquitinated proteins and exhibited premature senescence. Our results demonstrate the importance of the PAC1-dependent assembly pathway and of the latent 20S proteasomes for maintaining cellular integrity.
The 26S proteasome is a eukaryotic ATP-dependent protease responsible for the degradation of proteins tagged with polyubiquitin chains (21). The ubiquitin-dependent proteolysis by the proteasome plays a pivotal role in various cellular processes by catalyzing the selective degradation of short-lived regulatory proteins as well as damaged proteins. Thus, the proteasome is essential for the viability of all eukaryotic cells.
The 26S proteasome is a large protein complex consisting of two portions; one is the catalytic 20S proteasome of approximately 700 kDa (also called the 20S core particle), and the other is the 19S regulatory particle (RP; also called PA700) of approximately 900 kDa, both of which are composed of a set of multiple distinct subunits (70). The 20S proteasome is a cylindrically shaped stack of four heptameric rings, where the outer and inner rings each are composed of seven homologous α subunits (α1 to α7) and seven homologous β subunits (β1 to β7), respectively (5). The proteolytic active sites reside within the central chamber enclosed by the two inner β-rings, while a small channel formed by the outer α-ring, which is primarily closed, restricts the access of native proteins to the catalytic chamber. Thus, the 20S proteasome is a latent enzyme. Appending 19S RP, which consists of 19 different subunits, to the α-ring enables the 20S proteasome to degrade native proteins; 19S RP accepts ubiquitin chains of substrate proteins, removes ubiquitin chains while unfolding the substrates, and feeds the substrates into the interior proteolytic chamber of the 20S proteasome through the α-ring that is opened when the C-terminal tails of the ATPase subunits of 19S RP are inserted into the intersubunit spaces of the α-ring (24, 62, 74). However, it also has been reported that some denatured or unstructured proteins can be degraded directly by the 20S proteasome even in the absence of 19S RP and ubiquitination (37, 39).
Much attention has been focused on how such a highly elaborate structure is achieved. Recent studies have identified various proteasome-dedicated chaperones that assist in the assembly of the proteasome in eukaryotic cells (23, 40, 56, 57, 65, 66). In yeast, while most of the proteasome subunits are essential for viability, the deletion of any of these chaperones does not cause lethality. In fact, many, if not all, of the deletions exhibit subtle phenotypes. In mammalian cells, although the knockdown of the assembly chaperones reduced proteasome assembly and thus proteasome activity, leading to slow cell growth, the degree of reduction was much lower than that which occurred following the knockdown of the proteasome subunit itself (33, 35, 40). These results indicate that the assembly chaperones play an auxiliary role in proteasome biogenesis.
Proteasome assembly chaperone 1 (PAC1) is one of the assembly chaperones originally identified in mammalian cells (34). PAC1 plays a role in α-ring formation that occurs during the initial assembly of the 20S proteasome; it also prevents the aberrant dimerization of the α-ring. As is the case for most assembly chaperones, the knockdown of PAC1 in mammalian cells decreases proteasome activity but to a lesser extent than that in, for example, β2 knockdown (34, 35). Therefore, both PAC1-dependent and -independent assembly pathways exist in cells, but the importance of the PAC1-dependent pathway remains elusive. To further elucidate the biological significance of PAC1 and PAC1-dependent proteasome biogenesis, we generated conditional mouse mutants carrying an inactivating mutation in Psmg1, the gene coding for PAC1 protein, in the whole body, the nervous system, and in the liver. Our results demonstrate that PAC1 is essential for the development of a mouse, and that it plays important roles in maintaining cellular integrity in quiescent tissue. Our study revealed for the first time the importance of chaperone-mediated proteasome biogenesis in a whole-body mammalian system and may provide valuable knowledge in medical drug development targeting proteasomes.
MATERIALS AND METHODS
Gene targeting of Psmg1.
A targeting vector for Psmg1-floxed mice was constructed by inserting loxP sequences into intron 1 and intron 2 so that exon 2 was deleted upon the expression of Cre recombinase. A neomycin resistance gene cassette that was flanked by FRT sites also was inserted into intron 2. TT2 embryonic stem (ES) cells were screened as described previously (54). For Southern blot analysis, genomic DNA extracted from ES cells was digested with BamHI and hybridized with the probe indicated in Fig. 1 A. The mice carrying the mutated Psmg1 locus were crossed with Flp mice (Jackson Laboratory) to delete the neomycin cassette, generating Psmg1F/+ mice. Psmg1F/+ mice were backcrossed onto the C57BL/6J strain for at least five generations before analysis. EIIa-Cre, Nestin-Cre, and albumin-Cre transgenic mice were purchased from Jackson Laboratory. PCR primers used for mouse genotyping are listed in Table 1. Mice were housed in pathogen-free facilities. The experimental protocols were approved by the Ethics Review Committee for Animal Experimentation of the Tokyo Metropolitan Institute of Medical Science and the University of Tokyo.
FIG. 1.
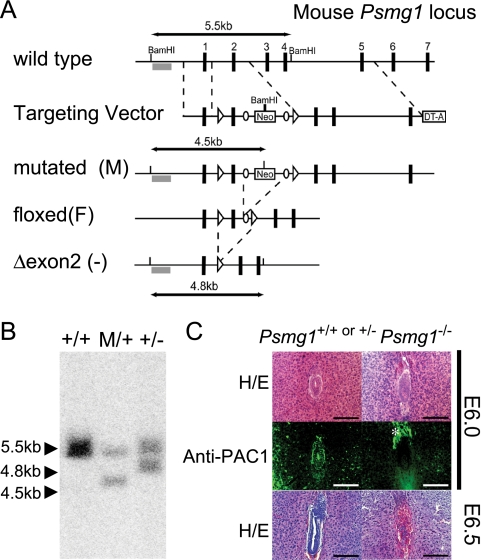
PAC1-dependent proteasome biogenesis is essential for mammalian development. (A) Strategy for the mutation of Psmg1 gene. The genomic region of the wild-type Psmg1 locus (+), the Psmg1 targeting vector, the structures of the mutated Psmg1 gene (M), the floxed gene in which the neomycin cassette (Neo) removed by flippase-mediated recombination (F), and the knockout allele in which exon 2 was deleted by Cre-mediated recombination [Δexon2 (−)] are depicted. The numbered black boxes are Psmg1 exons. The open arrowheads and circles indicate loxP and FRT sites. The probe for Southern blot analysis is shown as a gray box. DNA fragments detected by Southern blot analysis after BamHI digestion are shown. DTA, diphtheria toxin fragment A. (B) Southern blot analysis of genomic DNAs extracted from ES cells of the indicated genotypes. (C) Psmg1+/+ or Psmg1+/− (left) and Psmg1−/− (right) embryos in the uterus at E6.0 (top) and E6.5 (bottom) were morphologically identified by hematoxylin-eosin (HE) staining followed by immunofluorescent staining with anti-PAC1 antibody (middle). The signal shown with an asterisk in the immunostaining of Psmg1−/− embryo corresponds to eosinophilic structures seen outside the embryo in HE staining and probably is due to nonspecific staining.
TABLE 1.
Genotyping PCR primersa
| Detected allele | Primer designation |
Product length (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| Psmg1 wild type | a | b | 350 |
| Psmg1 mutated (M) | a | c | 640 |
| Psmg1 floxed (F) | a | d | 150 |
| Psmg1 Δexon2 (−) | e | d | 420 |
| Cre transgenes | f | g | 970 |
Primer designations and sequences are the following: a, TGTCTTCAAAAGCCACAGTCGT; b, AGGGCAAGAGCTCCTAACTAG; c, TCGTGCTTTACGGTATCGCCGCTCCCGATT; d, AGGGCAAGCGATACCGTCGAGATTAAAATA; e, GGTGATTGTGTCAACGACAGACACTTTTG; f, ATTTGCCTGCATTACCGGTCGATGCAAC; g, TGTTTCACTATCCAGGTTACGGATATAG.
Real-time PCR.
The isolation of total RNA, reverse transcription, and real-time PCR analysis were performed as described previously (30). The optimal combinations of the specific PCR primers and probes (Roche), which are listed in Table 2, were designed according to the Universal Probe Assay Design Center (https://www.roche-applied-science.com/sis/rtpcr/upl/adc.jsp). β-Glucuronidase (GUSβ) was used for normalization. Real-time PCR data were analyzed by the E-method from Roche Applied Science.
TABLE 2.
PCR primers and universal probes for real-time PCR
| Gene product | Probe no. | Primer sequence (5′-3′) |
|
|---|---|---|---|
| Forward | Reverse | ||
| Gusβ | 6 | GATGTGGTCTGTGGCCAAT | TGTGGGTGATCAGCGTCTT |
| α4 | 21 | CAACAGAGCCCGGGTAGA | CGCCCATTGCTCTGTGTAT |
| β2 | 89 | TCCCGAGAGTTGTTACAGCTAA | GCACCAATGTAACCTTGATACCT |
| Rpn13 | 40 | CCCCAGACTGCAGATGAGAT | GCCGCACTGAACATACCC |
| Smp30 | 3 | CGATTCAATGATGGGAAGGT | CGTTTCCTCAGCCATGGTA |
| Cu/ZuSOD | 49 | CCATCAGTATGGGGACAATACA | GGTCTCCAACATGCCTCTCT |
| Gst-mu | 58 | CTACCTTGCCCGAAAGCAC | ATGTCTGCACGGATCCTCTC |
| Gpx2 | 2 | GTTCTCGGCTTCCCTTGC | TTCAGGATCTCCTCGTTCTGA |
| Cyp2a5 | 52 | ACCAAGGACACCAAGTTTCG | AGAGCCCAGCATAGGAAACA |
| Nqo1 | 50 | AGCGTTCGGTATTACGATCC | AGTACAATCAGGGCTCTTCTCG |
| Ercc3 | 6 | CGGGTACTCAGAGCCAAGAA | CAGGGAGTAGAAAAAGGCATTG |
| Xrcc5 | 29 | GAAGATCACATCAGCATCTCCA | CAGGATTCACACTTCCAACCT |
| Ogg1 | 20 | TTATCATGGCTTCCCAAACC | CCTCAGGTGAGTCTCTGCTTC |
| p19ARF | 106 | GGGTTTTCTTGGTGAAGTTCG | TTGCCCATCATCATCACCT |
| p53 | 60 | GTGAGGCGAGTTGTGGAAAT | TGCCACACAGCAGTGAATG |
| p21Waf1/Cip1 | 21 | TCCACAGCGATATCCAGACA | GGACATCACCAGGATTGGAC |
| p63 | 45 | AGACCTCAGTGACCCCATGT | CTGCTGGTCCATGCTGTTC |
| Noxa | 15 | CAGATGCCTGGGAAGTCG | TGAGCACACTCGTCCTTCAA |
| PUMA | 79 | TTCTCCGGAGTGTTCATGC | TACAGCGGAGGGCATCAG |
| p16INK4a | 42 | CTTCCTGGACACGCTGGT | TCTTGATGTCCCCGCTCTT |
| p15INK4b | 42 | GACACGCTTGTCGTGCTG | TGCTCTTCAGCCAAGTCTACC |
Immunoblot analysis.
Mouse brains and livers were homogenized with a Potter-Elvehjem homogenizer in a buffer containing 25 mM Tris-HCl (pH 7.5), 2 mM ATP, 5 mM MgCl2, and 1 mM dithiothreitol (DTT). The homogenates were clarified by centrifugation at 20,000 × g for 10 min at 4°C before immunoblot analysis, glycerol gradient centrifugation, and peptidase activity assay. For subcellular fractionation, the liver homogenates were clarified by passing them through a 70-μm mesh to remove debris and then were centrifuged at 100 × g for 10 min at 4°C. The precipitates were dispersed in TKM buffer (50 mM Tris-HCl [pH 7.5], 25 mM KCl, 5 mM MgCl2) containing 0.25 M sucrose and subjected to sucrose density gradient ultracentrifugation at 12,000 × g for 30 min at 4°C on a 2.3 M sucrose cushion in TKM buffer. The resultant pellets were dissolved in a buffer containing 25 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 1 mM DTT, 2 mM ATP, and 1% Triton X-100 and were centrifuged at 20,000 × g for 10 min at 4°C. The supernatant was used as the nuclear fraction. SDS-PAGE and immunoblot analysis were performed as described previously (29). Antibodies for the proteasome subunits and chaperones used in this study were described previously (30, 33-35, 40). The antibodies for polyubiquitin (FK-2; Medical & Biological Laboratories), actin (MAB1501R; Chemicon), and lamin B (M-20; Santa Cruz) were purchased.
Glycerol density gradient analysis.
Clarified homogenates were subjected to 8 to 32% (vol/vol) linear glycerol density gradient centrifugation (22 h; 83,000 × g) as described previously (55). For immunoblot analysis, the fractionated samples were precipitated by cold acetone and dissolved in SDS sample buffer containing β-mercaptoethanol.
Assay of proteasome activity.
The peptidase activity of the proteasome was measured using a fluorescent peptide substrate, succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (Suc-LLVY-MCA; Peptide Institute), as described previously (55). The degradation assay of the recombinant 35S-labeled ornithine decarboxylase (ODC) was performed as described previously (30).
Histological examination.
Embryos in utero and brains subjected to 5-bromo-2′-deoxyuridine (BrdU) staining were fixed by immersion in phosphate-buffered saline (PBS) containing 4% paraformaldehyde (PFA), embedded in paraffin, and sectioned. Other tissues were processed as described previously (45). Meyer's hematoxylin and eosin, toluidine blue, and oil red O stainings were performed by conventional methods. Immunofluorescent analysis was performed as described previously (45). Anti-calbindin-D-28K (EG-20; Sigma) and anti-NeuN (4G2; Abcam) were purchased. Alexa Fluor 647 goat anti-rabbit IgG and Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probes) were used as secondary antibodies to detect the primary antibodies. For senescence-associated β-galactosidase (β-gal) activity staining, cryosections of Psmg1F/F liver were washed in PBS and incubated in a freshly prepared buffer containing 40 mM citrate-phosphate (pH 6.0), 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactosidase, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2 at 37°C for 12 h.
BrdU staining.
Postnatal day 0 (P0) mice, P3 mice, and pregnant mice at embryonic day 15.5 (E15.5) were intraperitoneally injected with PBS containing BrdU (Sigma; 50 μg/g body weight) three times every hour before dissection. Paraffin sections of their brains were deparaffinized, subjected to microwave-enhanced antigen retrieval in 0.05% citraconic anhydride, and then incubated in 0.2 M HCl at 37°C for 30 min, followed by immunofluorescent staining using Alexa Fluor 488-conjugated anti-BrdU antibody (Molecular Probes).
RESULTS
Early embryonic lethality of PAC1-null mice.
To create Psmg1-flox mice, a targeting vector was designed to modify the Psmg1 gene by homologous recombination in ES cells so that exon 2 was flanked by loxP sites, leading to the deletion of the Psmg1-flox gene when expressing the DNA recombinase Cre (Fig. 1A). We included FRT sites to remove the neomycin cassette after the generation of chimeric mice by crossing these with Flp mice. Homologous recombination in an ES cell clone and the deletion of the floxed gene by the infection of the clone with Cre-expressing adenovirus were confirmed by Southern blotting (Fig. 1B). Germ line mice were crossed with Flp mice to obtain Psmg1F/F mice. Psmg1F/F mice were healthy and fertile, indicating that the presence of the loxP sites did not affect PAC1 function in vivo.
We first crossed Psmg1F/F mice with mice expressing Cre recombinase throughout the whole body under the control of the adenovirus-derived EIIa promoter to obtain PAC1-null (Psmg1−/−) mice. Psmg1 heterozygous (Psmg1+/−) mice were born healthy and fertile without any noticeable pathological phenotypes. However, the subsequent intercrossing of the heterozygous mice failed to produce any viable homozygous (Psmg1−/−) mice. To further characterize these mice, embryos in utero were analyzed at various developmental stages. Psmg1−/− mice were evident at E3.5 with the appearance of normal blastocysts (data not shown). In E6.0 to 6.5 embryos, a cylinder-like two-layered cellular structure was observed in normal embryos (Fig. 1C). In contrast, PAC1-deficient embryos, which were identified by immunostaining with anti-PAC1 antibody, exhibited disorganized structure at E6.0 (Fig. 1C). At E6.5, PAC1-deficient embryos were mostly absorbed, while wild-type or heterozygous embryos developed into the normal egg cylinder (Fig. 1C). These data indicate that PAC1 is essential for mammalian embryonic development.
Loss of PAC1 leads to poor proteasome activity in the brain.
Recent studies have suggested links between the impairment of the ubiquitin-dependent protein degradation system and neurological disorders such as neurodegeneration (26). Therefore, as a next step, we investigated the role of PAC1 in the central nervous system by crossing Psmg1F/F mice with mice that express Cre under the control of the Nestin promoter. The Nestin-Cre allele has been shown to result in very efficient recombination in neuronal and glial precursor cells starting at around E10.5 (78). Homozygous mutant pups (Psmg1F/F:Nes) were born at the Mendelian frequency as genotyped at P1 without gross abnormality (n = 59 for Psmg1F/F:Nes out of 224 pups from the crossing of Psmg1F/+:Nes and Psmg1F/F). However, they exhibited growth retardation and neurological disorders, including ataxia, tremor, and abnormal limb-clasping reflexes when held by the tail (Fig. 2 A). They died at around P21, with no pups surviving beyond the weaning stage.
FIG. 2.
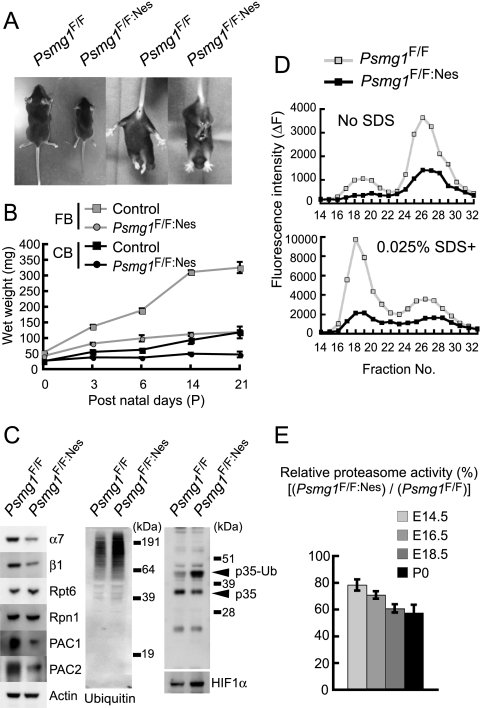
Decreased proteasome activity in Psmg1F/F:Nes mouse brains. (A) Growth retardation (left two panels) and abnormal limb-clasping reflexes (right two panels) in Psmg1F/F:Nes mice at P21. When lifted by the tail, Psmg1F/F mice behaved normally, extending their hind limbs and bodies. In contrast, Psmg1F/F:Nes mice bent their legs toward their trunk or tightened their back limbs to their bodies and anterior limbs. (B) Impairment of weight gain in Psmg1F/F:Nes brains. Control (Psmg1F/F) and Psmg1F/F:Nes cerebra and cerebella were dissected, and their wet weight was measured on the indicated days. The data are shown as the means ± standard deviations (SD) (n > 3 each). (C) Extracts from Psmg1F/F and Psmg1F/F:Nes brains at P21 were subjected to immunoblot analysis using the indicated antibodies. p35-Ub denotes monoubiquitinated p35. (D) The extracts used for panel C were fractionated by 8 to 32% glycerol gradient centrifugation. An aliquot of each fraction was used for an assay of the chymotryptic activity of the proteasomes using Suc-LLVY-MCA in the absence (upper panel) or presence (lower panel) of 0.025% SDS. (E) Extracts from Psmg1F/F and Psmg1F/F:Nes brains at E14.5, E16.5, E18.5, and P0 were used for an assay of the chymotryptic activity of the proteasomes using Suc-LLVY-AMC as a substrate. The data are shown as means ± SD (n > 3 each) relative activity.
The cerebrum and cerebellum weights of mutant mice were reduced as early as P3 and were only one-third of those of control mice at P21 (Fig. 2B). Whole-brain lysates of P21 Psmg1F/F:Nes and control (Psmg1F/F) mice were analyzed by immunoblotting for PAC1 protein expression as well as for its heterodimeric partner PAC2, several proteasome subunits, and ubiquitin. PAC1 protein contents were reduced by approximately 90% in brain lysates of Psmg1F/F:Nes mice (Fig. 2C). Considering that PAC1 is a short-lived protein that is rapidly degraded upon the biogenesis of the 20S proteasome, the residual PAC1 protein likely is due to its expression in the blood vessels and in the meninges of the brain, where Cre-mediated DNA recombination does not occur (17, 27, 34). The PAC2 protein was similarly reduced (Fig. 2C), which is consistent with the notion that PAC1 and PAC2 proteins are stable only when they form a heterodimer (34). The proteasome subunits α7 and β1, both of which are subunits of the 20S proteasome, were significantly reduced, whereas the 19S RP subunits, Rpt6 and Rpn1, were marginally increased (Fig. 2C). Immunoblotting for ubiquitin revealed that the mutant mouse brain accumulated a larger amount of ubiquitin-conjugated proteins than the control mouse brain (Fig. 2C). We also observed an increase in two known short-lived proteins, p35 and HIF1α, that are degraded by the ubiquitin-proteasome pathway (59, 68), the former accumulating mainly in a ubiquitinated form, as described previously (59) (Fig. 2C). These data indicate attenuated proteasome activity in the Psmg1F/F:Nes brain.
Indeed, the analysis of the chymotryptic activity of the proteasome following glycerol gradient centrifugation of the brain lysates revealed a decrease in the activity of the 26S proteasome and the free 20S proteasome (Fig. 2D). The time-course analysis of proteasome activity showed that the activity of the PAC1-deficient brain declined with the developmental process; while mutant brains still exhibited approximately 80% of the activity of the controls at E14.5, it gradually decreased to only 60% at birth (Fig. 2E). Immunoblot analyses of fractions separated by glycerol gradient centrifugation of E12.5, E16.5, and P0 brains showed a parallel, gradual decrease in the free 20S proteasomes (see Fig. S1A in the supplemental material), which was further confirmed by the time-course analysis of the activities of 20S and 26S proteasome fractions (see Fig. S1B in the supplemental material). Of note, when total levels of 20S proteasome gradually decreased as development proceeded, the levels of 26S proteasome were maintained until free 20S was lost, suggesting that when both 19S RP and 20S proteasomes exist in cells, they are preferentially assembled into 26S proteasomes rather than existing as free forms. These results indicate that the loss of PAC1 caused an inefficient 20S proteasome biogenesis and hence decreased proteasome activity, which led to the impaired degradation of ubiquitinated proteins.
PAC1 deficiency leads to severe defects in brain development.
We next performed histological analyses of these brains. PAC1-deficient brains exhibited malformations of the cerebrum and cerebellum (Fig. 3 A to C). The cerebrum of the mutant mice displayed cortical thinning and reduced hippocampus and dentate gyrus, although the layer formation of the pyramidal cells and granule cells was not disturbed (Fig. 3A). The morphological abnormality was more apparent in the cerebella of the mutant mice. Not only was the size markedly reduced compared to that of the controls but normal lobulation also was absent from the mutant cerebella (Fig. 3B).
FIG. 3.
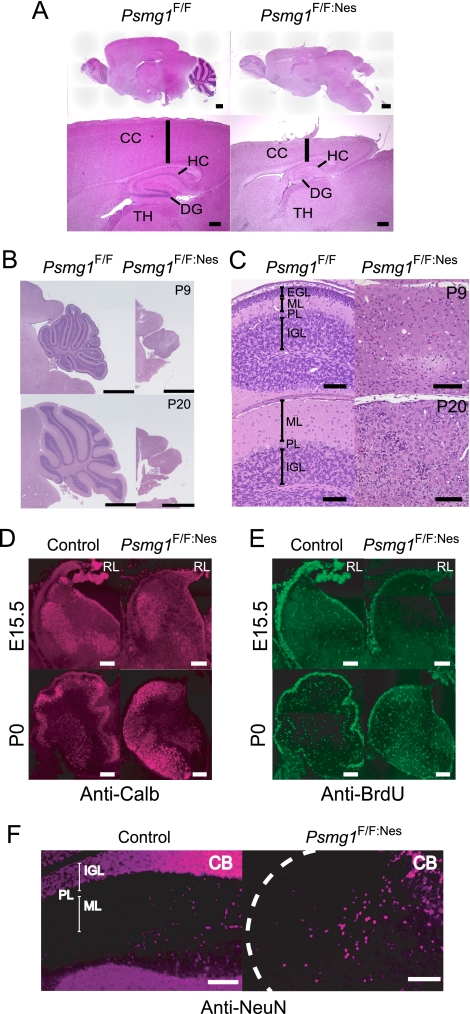
Disorganized brain structure in Psmg1F/F:Nes mice. (A) Sagittal sections of Psmg1F/F and Psmg1F/F:Nes forebrains at P21 were stained with hematoxylin-eosin. The sections were observed at low (upper panel; scale bar, 600 μm) and high (lower panel; scale bar, 300 μm) magnification. CC, cerebral cortex; HC, hippocampus; TH, thalamus; DG, dentate gyrus. (B) Histological analysis of Psmg1F/F and Psmg1F/F:Nes cerebella at P9 and P20. A section of each brain was stained with hematoxylin-eosin. Scale bar, 1 mm. (C) High-magnification images of images from panel B. Scale bar, 100 μm. EGL, external granule layer; ML, molecular layer; PL, Purkinje cell layer; IGL, internal granule layer. (D) Immunofluorescent staining of Purkinje cells in the cerebellum at E15.5 and P0 by anticalbindin antibody. (E) BrdU staining following BrdU injection into E15.5 pregnant mice and P0 mice. (F) Mature neurons in the cerebellum at P21 were visualized by immunofluorescent staining against NeuN. Abbreviations are the same as those for panel B.
During cerebellar development, Purkinje cells emerge from the ventricular zone and stop dividing between E11 and E13. Granular cell precursors within the external granular layer are mitotically active between E15 and P14. Some of these cells stop dividing after birth, differentiate into mature granular cells, and migrate inward past the molecular layer, which is filled with dendritic trees of the Purkinje cells and the Purkinje cell layer, thus forming lobules and the inner granule cell layer that occupy the majority of a mature cerebellar cortex (73). In Psmg1F/F:Nes mice, however, this three-layered structure was completely lost (Fig. 3C).
The immunostaining of cerebellar sections at E15.5 and P0 for calbindin 1, a marker for Purkinje cells, indicated that the number of Purkinje cells in Psmg1F/F:Nes mice was comparable to that in control mice, although their distribution within the cerebellum was severely affected at P0 (Fig. 3D). This suggests that the ventricular zone cells had sufficient proteasome activity to give rise to Purkinje cells at E11 to 13, which can be inferred from the data shown in Fig. 2E.
We then analyzed the developmental processes of cerebellar granule cells that occur between E15.5 and P21. The proliferation of granular cell precursors was monitored by BrdU incorporation at E15.5 and P0. While the granular cell precursors in the external granular layer of the control cerebellum had strong signals for BrdU, those in the mutant cerebellum showed much weaker BrdU incorporation (Fig. 3E). We also stained the cerebellar sections for cleaved caspase 3, a marker for apoptosis, but we did not observe differences between Psmg1F/F:Nes and control cerebellum (see Fig. S2 in the supplemental material). These results indicate that the proliferation of granule progenitor cells in Psmg1F/F:Nes mice was severely inhibited (Fig. 3E), even though proteasome activity remained at 60 to 80% of that of the control brain (Fig. 2E), and a reduction in granular cells in Psmg1F/F:Nes cerebellum was due to a proliferation defect rather than increased cell deaths. Furthermore, immunofluorescent staining against NeuN, a marker for mature neuronal cells, the majority of which are mature granular cells, revealed that very few mature neuronal cells were present in the PAC1-deficient cerebellum at P21 (Fig. 3F). These data indicate that brain development is strongly dependent on the integral proteasome activity that is supported by PAC1-assisted proteasome formation in mice.
Important role of latent 20S proteasome in the degradation of ubiquitinated proteins.
To better determine the biochemical basis of the significance of PAC1, we generated mice lacking PAC1 in postnatal hepatocytes by crossing Psmg1F/F mice with transgenic mice that expressed Cre recombinase under the albumin promoter, Psmg1F/F:Alb (61). Psmg1F/F:Alb mice were born without any abnormal appearance or developmental defects, were fertile, and survived as long as the control mice with a nearly equal mortality rate at 18 months of age (data not shown). Both the PAC1 protein and the PAC2 protein were largely lost in the Psmg1F/F:Alb mouse liver at P14 (Fig. 4 A). In the mutant liver, the subunits of the 20S proteasome decreased, while those of 19S RP increased compared to those in the control liver (Fig. 4A). These results are similar to the observations in the mutant brains (Fig. 2D).
FIG. 4.
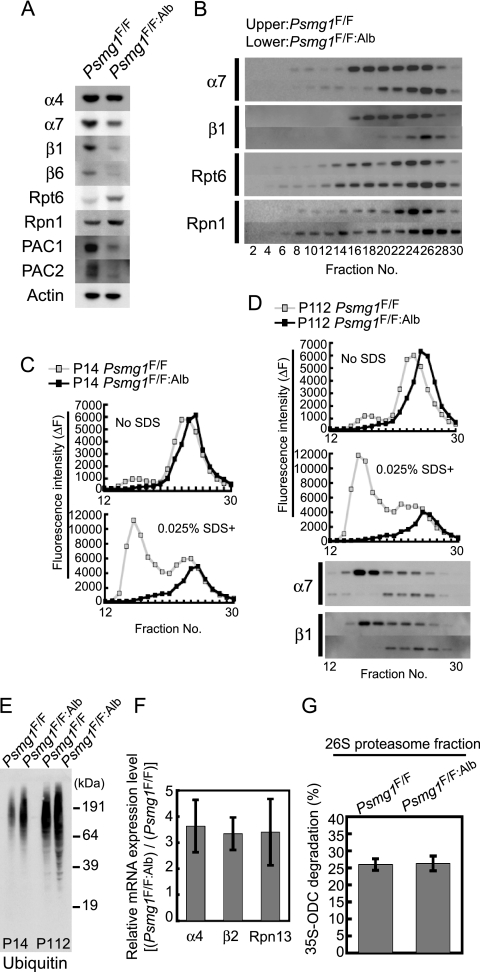
Loss of latent 20S proteasomes causes inefficient degradation of ubiquitinated proteins. (A) Immunoblot analysis of liver extracts from Psmg1F/F and Psmg1F/F:Alb mice at P14 using the indicated antibodies. (B) The extracts shown in panel A were fractionated by 8 to 32% glycerol gradient centrifugation and subjected to immunoblot analysis using the indicated antibodies. (C) The fractions shown in panel B were subjected to an assay of the chymotryptic activity of the proteasome using Suc-LLVY-MCA in the absence (upper panel) or presence (lower panel) of 0.025% SDS. (D) Liver extracts from Psmg1F/F and Psmg1F/F:Alb mice at P112 were analyzed as described for panels A and B. (E) Immunoblot analysis of liver extracts from Psmg1F/F and Psmg1F/F:Alb mice at P14 and P112 using antiubiquitin antibody. (F) Transcription levels of proteasome subunits in P14 liver analyzed by real-time PCR. The relative ratios of the expression levels in Psmg1F/F:Alb liver compared to those in Psmg1F/F liver are shown. The data represent means ± standard deviations (SD) from three independent experiments. (G) Fraction 26 shown in panel B was subjected to an S35-ODC degradation assay. The data represent means ± SD from five independent experiments.
Fractionation of the liver lysates of P14 mice by glycerol gradient centrifugation followed by immunoblot analysis revealed that the amount of the 26S proteasome was comparable between the mutant and control livers, although the 26S proteasome in the mutant liver sedimented at heavier fractions than that in the control liver, probably due to the association of additional proteins, as reported previously (49) (Fig. 4B, fractions 24 to 30). However, the PAC1-deficient liver almost lost all the free 20S proteasomes that were not associated with 19S RP and thus were latent with respect to protein degradation (Fig. 4B, fractions 16 to 18). Instead, the amount of free 19S RP, which is similar in size to the 20S proteasome but is not associated with it around fraction 16, was increased (Fig. 4B; also see Fig. S3A in the supplemental material). We also observed complexes that presumably correspond to α-rings in the control liver (see Fig. S3B, fraction 14) and α-ring dimers in the PAC1-deficient liver (see Fig. S3B, fractions 16 to 18). Neither of these contained β-subunits (see Fig. S3C), which is consistent with the observation found in PAC1 knockdown cells (34). The subunit composition of the 20S proteasome in PAC1-deficient livers was almost normal, containing all of the 14 subunits at levels similar to those in wild-type livers (see Fig. S3D).
Consistently with the immunoblot analysis, the mutant liver of P14 mice exhibited a peptide-hydrolyzing activity of the 26S proteasome comparable to that of the control liver, as judged by the areas surrounded by the lines representing Suc-LLVY-hydrolyzing activities in the absence of SDS (Fig. 4C, upper panel, fractions 22 to 28). In contrast, the activity of the latent 20S proteasome, which was artificially activated by the addition of a low concentration of SDS, was completely abolished in the mutant liver (Fig. 4C, lower panel, fractions 15 to 18). The small activity peak observed in fractions 16 to 18 of the control liver is likely to represent the complex formed between the 20S proteasome and the proteasome activator PA28 complex, which also was lost in the mutant liver (Fig. 4C, upper). Surprisingly, even at P112, profiles of proteasome peptidase activities and distributions of proteasome subunits were almost the same as that at P14, i.e., mutant livers exhibited the normal peptidase activity of the 26S proteasome and a disappearance of the latent 20S proteasome (Fig. 4D).
Intriguingly, the accumulation of polyubiquitin-conjugated proteins was noted in the PAC1-deficient livers both at P14 and P112 (Fig. 4E). Consistently with this finding, transcriptions of subunits of both the 20S proteasome and 19S RP were increased by 3- to 4-fold in the mutant livers, which accounts for the increased protein content of the 19S RP subunits, as shown in Fig. 4A, and indicates the presence of proteotoxic stress due to insufficient proteasome activity (53, 63) (Fig. 4F). Since 20S assembly is impaired and unassembled subunits are rather unstable, the protein levels of the 20S subunits were reduced in PAC1-deficient livers (Fig. 4A).
Because the amount of the 20S proteasome in PAC1-deficient livers was approximately only 20 to 30% of that of the control liver, despite the increased mRNA of the proteasome subunits (Fig. 4A, B, and F), we can speculate that more than 70 to 80% of the 20S proteasome was generated in a PAC1-dependent manner, and that only a small amount of the 20S proteasome was generated without PAC1.
To test whether the degradation of native proteins by the 26S proteasome was impaired in the PAC1-deficient liver, we measured the degradation rate of ornithine decarboxylase, which is degraded by the 26S proteasome in an antizyme- and ATP-dependent manner, using the 26S proteasome fractions separated in Fig. 4B. This experiment exhibited no significant difference between the mutant and control livers (Fig. 4G), indicating that not only artificial small peptide-hydrolyzing activity but also the native protein-degrading activity of the 26S proteasome was maintained in the PAC1-deficient liver.
Taken together, these results suggest that not only the 26S proteasome but also the latent 20S proteasome is involved in the degradation of native proteins and possibly ubiquitinated proteins, considering the accumulation of ubiquitinated proteins in the PAC1-deficient liver.
Senescence-like phenotypes in the PAC1-deficient liver.
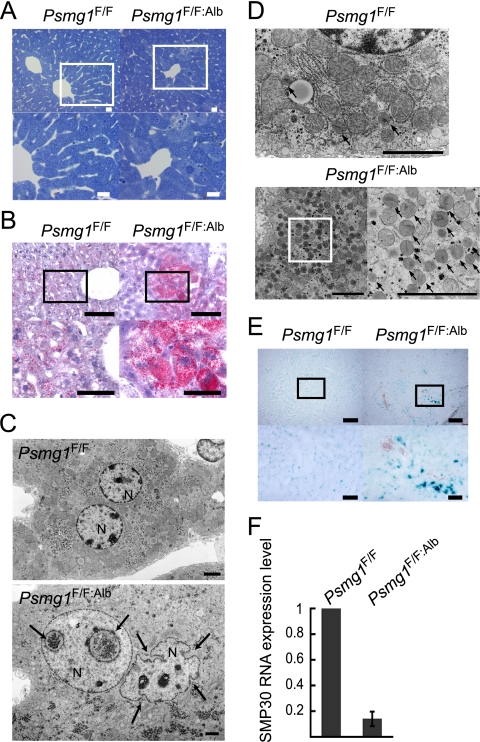
To investigate whether histological alterations were occurring in the PAC1-deficient liver, we performed microscopic analysis of liver sections from 6-month-old mice. Toluidine blue staining and light microscopy revealed that although specific disease-related pathological abnormalities, such as necrosis, apoptosis, fibrosis, and inflammation, were not observed, PAC1-deficient livers exhibited enlarged hepatocytes, accompanying irregular hepatic cords and narrow sinusoids (Fig. 5 A). Further analysis by oil red O staining detected intracellular accumulation of excess lipids in some of the mutant hepatocytes (Fig. 5B); these have been reported to accumulate due to metabolic disturbances associated with aging (3, 48).
FIG. 5.
Premature senescence-like phenotypes in Psmg1F/F:Alb liver. (A) Toluidine blue staining of Psmg1F/F and Psmg1F/F:Alb livers at 6 months of age. The lower panels are higher-magnification images of the regions outlined by white rectangles in the upper panels. Scale bars, 20 μm. (B) Cryosections of 1-year-old livers were stained with oil red O. The lower panels are higher-magnification images of the regions outlined by the rectangles in the upper panels. Scale bars, 100 μm (upper panels) and 50 μm (lower panels). (C) Electron-micrographic examination of hepatocyte nuclei in Psmg1F/F and Psmg1F/F:Alb livers at 6 months of age. The invagination of the nuclear envelope is indicated by arrows. N, nucleus. Scale bar, 2 μm. (D) Electron micrographs of hepatocyte peroxisomes in Psmg1F/F:Alb livers at 6 months of age. The lower right panel is a higher-magnification image of the region outlined by the rectangle in the lower left panel. Scale bar, 2 μm. (E) Detection of senescence-associated β-galactosidase activity on cryosections of Psmg1F/F and Psmg1F/F:Alb livers at 1 year of age. The lower panels are higher-magnification images of the regions outlined by the rectangles in the upper panels. Scale bar, 200 μm (upper panels) and 50 μm (lower panels). (F) Expression of senescence marker protein 30 (SMP-30) mRNA. The relative amount of SMP-30 mRNA in Psmg1F/F:Alb liver at 3 months of age was measured by real-time PCR analysis. The data represent means ± standard deviations (SD) from three independent experiments.
Ultrastructural examination demonstrated highly aberrant nuclear morphology in PAC1-deficient hepatocytes (Fig. 5C). Such morphological abnormality is well known in Hutchinson-Gilford progeria syndrome and often is observed in aging cells (3, 50, 69). We also noted the aberrant proliferation of peroxisomes, the deregulation of which is implicated in aging-related degenerative diseases (60), in PAC1-deficient hepatocytes (Fig. 5D). Collectively, these histological analyses suggest that PAC1 deficiency is associated with senescence.
To confirm that the PAC1-deficient hepatocytes underwent premature senescence, we examined the activity of senescence-associated β-galactosidase (SA-β-Gal). As shown in Fig. 5E, the PAC1-deficient liver of a 1-year-old mouse exhibited a denser and larger area of blue staining than that of the control liver. Furthermore, we observed a decrease in mRNA expression of the senescence marker protein-30 (SMP-30), which is a hepatic senescence marker that has been shown to decline with aging (22, 36) (Fig. 5F). These data further support the notion that the loss of PAC1 and the resultant disappearance of the free latent 20S proteasomes caused early senescence in the mouse liver.
Activation of oxidative and DNA damage responses and the ARF-p53 senescence pathway in the PAC1-deficient liver.
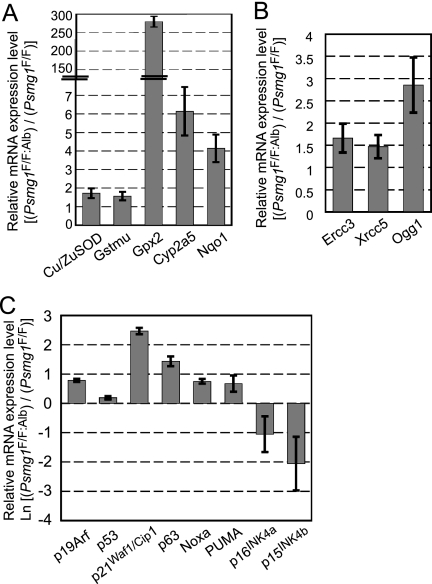
Accumulating evidence supports the hypothesis that free radicals and oxidative stress causes molecular damage that contributes to cell senescence (20, 25, 75). To examine whether the PAC1-deficient liver suffered from oxidative stress, we measured mRNA expression levels of genes involved in antioxidant defenses induced in response to oxidative stress. All of the genes examined were significantly upregulated in the PAC1-deficient liver, including Cu/Zn-superoxide dismutase (SOD), glutathione S-transferase (GST) mu, glutathione peroxidase 2 (Gpx2), cytochrome P450 2a5 (Cyp2a5), and NAD(P)H:quinone oxidoreductase (Nqo1), all of which are induced by the activation of the transcription factor Nrf2 or FoxO following oxidative stress (1, 4, 19, 46, 52, 79) (Fig. 6 A). This result suggests that the loss of PAC1 caused oxidative stress in the liver, which is consistent with previous reports (7, 13).
FIG. 6.
Activation of the oxidative stress response and a senescence pathway in Psmg1F/F:Alb liver. Relative mRNA levels in Psmg1F/F:Alb liver compared to those in Psmg1F/F liver by real-time RT-PCR. Data represent means ± standard deviations (SD) from experiments on three pairs of littermates. (A) Analysis of oxidative stress-related genes. (B) Analysis of DNA repair-related genes. (C) Analysis of senescence-associated genes. Note that the data are shown on a natural logarithmic scale.
Free radicals can damage various cellular molecules, including DNA (6, 20, 25). We therefore tested the expression levels of molecules involved in DNA repair that are induced upon DNA damage. Excision repair cross-complementing rodent repair deficiency, complementation group 3 (Ercc3), X-ray repair complementing defective repair in Chinese hamster cells 5 (Xrcc5), and 8-oxoguanine DNA-glycosylase 1 (Ogg1), which play a role in nucleotide excision repair, double-strand break repair, and base excision repair, were induced in PAC1-deficient livers (38) (Fig. 6B). This suggests that the DNA damage response pathway is activated following genotoxic stress in the PAC1-deficient liver.
Senescence is induced by multiple stimuli, including DNA damage and mitogenic signals, which activate the ARF-p53 and p16INK4a-pRb pathways to induce growth arrest by inhibiting cyclin-dependent kinases (10, 16, 31). Accordingly, we examined the expression of molecules involved in these pathways. The activation of the ARF-p53 pathway was demonstrated by increased mRNA levels of ARF, p53, and molecules regulated by p53, such as p21Waf1/Cip1, p63, Noxa, and Puma (18, 28, 42, 47, 72, 80) (Fig. 6C). Curiously, however, the expression of the key molecules of the other pathway, p16INK4a and p15INK4b, was markedly reduced (Fig. 6C). Since it is known that the ARF-p53 and p16INK4a-pRb pathways are largely coregulated (10, 16, 43), senescence caused by proteasome dysfunction might be characteristic in that it selectively regulates the former pathway, although the responsible mechanism remains unclear.
Nuclear accumulation of the 26S proteasome upon premature senescence in the PAC1-deficient liver.
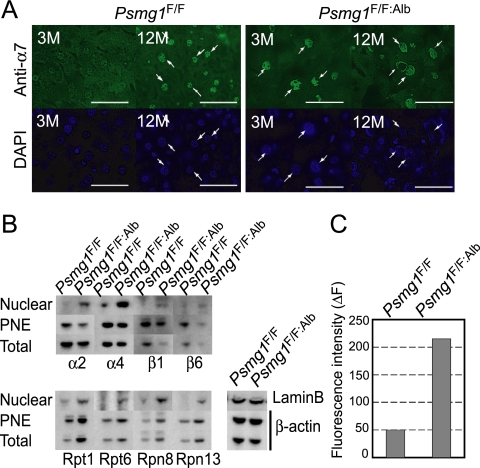
Aging and oxidative stress are known to enhance the production of methylglyoxal and glyoxal, which in turn modify proteins, lipids, and DNA to generate what are called advance glycation end products (20, 64). There is an intriguing report showing that nuclear proteasome activities were increased when cells were treated with glyoxal (11). Therefore, we investigated whether the intracellular distribution of the proteasome was affected in the PAC1-deficient liver by immunostaining liver sections with an antibody against α7, one of the subunits of the 20S proteasome. At 3 months of age, α7 was distributed nearly homogenously in both the cytosol and nucleus in control livers, whereas it accumulated in the nucleus in the PAC1-deficient hepatocytes (Fig. 7 A). At 12 months of age, this nuclear accumulation of the 20S proteasome was observed not only in the PAC1-deficient hepatocytes but also in the control hepatocytes, indicating that nuclear proteasome accumulation occurs as one of the normal aging processes and that the PAC1-deficient liver underwent accelerated senescence (Fig. 7A).
FIG. 7.
Age-dependent nuclear import of the 26S proteasomes and its acceleration in the absence of PAC1. (A) Immunofluorescent staining of Psmg1F/F and Psmg1F/F:Alb livers at 3 and 12 months of age using anti-α7 antibody. 4′,6-Diamidino-2-phenylindole (DAPI) was used for nuclear counterstaining. Scale bar, 50 μm. (B) Immunoblot analysis of nuclear fractions, postnuclear fraction (PNE), and total lysates from Psmg1F/F and Psmg1F/F:Alb livers at 6 months of age using antibodies against the indicated proteins. Immunoblots for lamin B and β-actin are shown as loading controls. (C) Suc-LLVY-MCA-hydrolyzing activity of nuclear proteasomes. The nuclear fractions shown in panel B were fractionated by glycerol density gradient centrifugation followed by measurements of peptidase activity in the fraction containing the 26S proteasome in the absence of SDS. The data represent the means of two independent experiments.
To confirm this observation and to investigate whether 19S RP also was accumulated in the nucleus of PAC1-deficient hepatocytes, we fractionated liver lysates from 3-month-old mice into nuclear fractions and postnuclear fractions, which then were subjected to immunoblot analysis using antibodies for proteasome subunits. Consistently with the immunofluorescence analysis, α2, β1, and β6, which are subunits of the 20S proteasome, all were enriched in the PAC1-deficient liver despite their decrease in total lysates (Fig. 7B). The amount of the nuclear 19S RP subunits, irrespective of whether they were the base subunits (Rpt1 amd Rpn13) or the lid subunits (Rpn8), also was much larger in the PAC1-deficient liver than in the control liver (Fig. 7B).
The increase in nuclear 19S RP in PAC1-deficient livers may simply reflect the increased expression of the 19S RP subunits in the PAC1-deficient liver, as revealed by the immunoblot of total lysates and postnuclear fractions. Whether these were accumulated in the nucleus as active 26S proteasomes is not clear. The analysis of peptidase activity revealed that the nuclear fraction of the PAC1-deficient liver exhibited 26S proteasome activity more than 4-fold higher than that in the control liver, indicating that the PAC1-deficient liver accumulated active 26S proteasomes in the nucleus (Fig. 7C).
DISCUSSION
Until now, nine proteasome-dedicated chaperones (PAC1/Pba1, PAC2/Pba2, PAC3/Pba3, PAC4/Pba4, POMP/Ump1, p28/Nas6, p27/Nas2, S5b/Hsm3, and PAAF1/Rpn14) involved in proteasome assembly have been identified both in mammals and in yeast (51, 56, 58). None of these, however, has been analyzed in the context of mammalian development as well as the maintenance of differentiated tissues. In this paper, we demonstrated for the first time the significance of the chaperone-assisted biogenesis of proteasomes in mouse development, central nervous system development, and liver homeostasis.
Mice that systemically lacked PAC1 died around E6.0. However, this phenotype is still milder than that of mice deficient in proteasome subunits Rpt3 and Rpt5, which died before development into blastocysts (∼E3.5) (67). This is consistent with the results observed in mammalian cells, where the loss of PAC1 was not as severe as the loss of the proteasome subunits, and supports the notion that proteasome biogenesis is not entirely dependent on the assembly chaperones. Nevertheless, efficient proteasome biogenesis assisted by PAC1 is needed to develop beyond the blastocyst stage and thus is essential for mouse development.
In terms of the central nervous system-specific deletion of PAC1, the development of the glial cells was profoundly affected compared to Purkinje cell development. Although we cannot rule out the possibility that this difference is dependent on cell type, it is more likely due to differences in stages of active proliferation. When the Purkinje cells developed, the proteasome activity was, for the most part, retained. However, when glial cells explosively proliferated, the proteasome activity of the mutant brain was reduced to 70 to 80% of that of the control brain, as far as we could assess using a short peptide substrate. This suggests that only a 20 to 30% reduction in proteasome activity is critical for rapidly proliferating cells during development.
In contrast to Psmg1F/F:Nes mice, Psmg1F/F:Alb mice, in which the hepatocyte-specific deletion of PAC1 occurs mostly after birth, grew up without gross abnormality. This enabled us to assess the roles of PAC1 in terminally differentiated and quiescent cells, which revealed the biological significance of latent 20S proteasomes. In normal mammalian tissues, the molar amount of the 20S proteasome is approximately 2- to 3-fold greater than that of 19S RP, and therefore both the 26S proteasome and the free latent 20S proteasome not associated with 19S RP exist in cells, and very few free 19S RPs are observed. Profiles of the proteasome activity of PAC1-deficient livers in glycerol gradient analysis revealed that they lost the free 20S proteasome but displayed a normal amount of the 26S proteasome and hence regular activity. However, ubiquitinated proteins were accumulated in these cells, suggesting that the free 20S proteasome participates in the turnover of ubiquitinated proteins.
As there has been no previous report showing that the 20S proteasome directly recognizes and degrades ubiquitinated proteins, some proteins that usually are degraded directly by the 20S proteasome in a ubiquitin-independent manner, such as natively unfolded proteins and oxidized proteins (8, 39), may accumulate and subsequently be ubiquitinated. This increase of additional ubiquitinated proteins might exceed the capacity of the 26S proteasomes and thus lead to the accumulation observed in PAC1-deficient livers. It also can be speculated that the dissociation of the 20S proteasome from 19S RP occurs during the degradation of a ubiquitinated protein, so that 19S RP is able to attach to another latent 20S proteasome for the degradation of another ubiquitinated substrate, while the substrate-loaded 20S proteasome is capped by PA28 for the acceleration of peptide release. Although some reports have shown that the 26S proteasome is stable during protein degradation (32, 44), we observed that free 19S RP increased while the 20S-PA28 complex drastically increased upon the inhibition of the 20S proteasome by MG132, which is thought to mimic degradation intermediates of substrates during proteolysis (our unpublished data) (71).
The PAC1-deficient liver exhibited several phenotypes typical for senescence, such as increased SA-β-gal staining, oxidative stress response, and the activation of the ARF-p53 pathway. There have been many reports proposing a link between decreased proteasome activity and aging, indicating that the connection between proteasome activity and senescence is not just a secondary effect of cellular aging. Findings have not only shown that proteasome inhibition in cells by specific inhibitors and the RNA interference-mediated knockdown of proteasome subunits induces cellular senescence (9, 14) but also that the elevation of proteasome activity by the overexpression of proteasome subunits or transcription factor Nrf2, which increases the expression of the proteasome subunits, actively relieved senescence and even extended life spans in Drosophila melanogaster (12, 13, 15, 41, 77). Therefore, proteasome activity might be a primary regulatory mechanism involved in mammalian senescence. However, many missing links between proteasome inactivation, oxidative stress, irregular nuclear structure, and the nuclear accumulation of the proteasome still remain.
The observation that the PAC1-deficient liver maintained 26S proteasome formation while the PAC1-deficient brain did not probably was due to a difference in the proliferation status of the two tissues. The adult liver is a quiescent organ in which the rapid production of the proteasome is not needed because of its long half-life (76). Therefore, even the PAC1-independent, inefficient biogenesis of the 20S proteasome would suffice for maintaining the amount of the 26S proteasome that matches the protein turnover in quiescent cells. In contrast, developing brains as well as embryos are rapidly proliferating and demand the efficient production of the proteasome. Thus, PAC1-dependent, efficient assembly of the 20S proteasome is a prerequisite for maintaining 26S proteasome activity at a level sufficient for active protein turnover in proliferating cells.
This observation may be indicative of a cancer therapy targeting proteasome assembly. The proteasome has become an attractive target for cancer therapy in recent years, since a proteasome inhibitor called bortezomib was found to be effective against refractory multiple myeloma (2). However, the inhibition of proteasome activity often leads to severe adverse effects in healthy organs. Considering that cancer cells are continuously producing proteasomes for their survival, the inhibition of proteasome assembly would be a much safer and yet effective anticancer therapy, because the quiescent cells that constitute most of our body should be much less sensitive to such an inhibitor. Our results clarified that chaperone-dependent proteasome biogenesis is essential for rapidly proliferating cells, while quiescent cells like hepatocytes can tolerate the loss of the pathway for at least up to 18 months. This should provide important information for developing inhibitors of proteasome assembly chaperones, including PAC1.
Supplementary Material
Acknowledgments
We thank Larissa Kogleck and Eri Katayama for valuable advice and comments on the manuscript.
This work was supported by grants to S.M. and K.T. from the Ministry of Education, Science and Culture of Japan and a grant to S.M. from the Mochida Memorial Foundation.
Footnotes
Published ahead of print on 24 May 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abu-Bakar, A., V. Lamsa, S. Arpiainen, M. R. Moore, M. A. Lang, and J. Hakkola. 2007. Regulation of CYP2A5 gene by the transcription factor nuclear factor (erythroid-derived 2)-like 2. Drug Metab. Dispos 35:787-794. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J. 2004. The proteasome: a suitable antineoplastic target. Nat. Rev. Cancer. 4:349-360. [DOI] [PubMed] [Google Scholar]
- 3.Andrew, W. 1962. An electron microscope study of age changes in the liver of the mouse. Am. J. Anat. 110:1-18. [DOI] [PubMed] [Google Scholar]
- 4.Banning, A., S. Deubel, D. Kluth, Z. Zhou, and R. Brigelius-Flohe. 2005. The GI-GPx gene is a target for Nrf2. Mol. Cell. Biol. 25:4914-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumeister, W., J. Walz, F. Zuhl, and E. Seemuller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92:367-380. [DOI] [PubMed] [Google Scholar]
- 6.Beckman, K. B., and B. N. Ames. 1998. The free radical theory of aging matures. Physiol. Rev. 78:547-581. [DOI] [PubMed] [Google Scholar]
- 7.Bieler, S., S. Meiners, V. Stangl, T. Pohl, and K. Stangl. 2009. Comprehensive proteomic and transcriptomic analysis reveals early induction of a protective anti-oxidative stress response by low-dose proteasome inhibition. Proteomics 9:3257-3267. [DOI] [PubMed] [Google Scholar]
- 8.Breusing, N., and T. Grune. 2008. Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol. Chem. 389:203-209. [DOI] [PubMed] [Google Scholar]
- 9.Byrne, A., R. P. McLaren, P. Mason, L. Chai, M. R. Dufault, Y. Huang, B. Liang, J. D. Gans, M. Zhang, K. Carter, T. B. Gladysheva, B. A. Teicher, H. P. Biemann, M. Booker, M. A. Goldberg, K. W. Klinger, J. Lillie, S. L. Madden, and Y. Jiang. Knockdown of human deubiquitinase PSMD14 induces cell cycle arrest and senescence. Exp. Cell Res. 316:258-271. [DOI] [PubMed]
- 10.Campisi, J., and F. d'Adda di Fagagna. 2007. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8:729-740. [DOI] [PubMed] [Google Scholar]
- 11.Cervantes-Laurean, D., M. J. Roberts, E. L. Jacobson, and M. K. Jacobson. 2005. Nuclear proteasome activation and degradation of carboxymethylated histones in human keratinocytes following glyoxal treatment. Free Radic Biol. Med. 38:786-795. [DOI] [PubMed] [Google Scholar]
- 12.Chondrogianni, N., and E. S. Gonos. 2007. Overexpression of hUMP1/POMP proteasome accessory protein enhances proteasome-mediated antioxidant defence. Exp. Gerontol. 42:899-903. [DOI] [PubMed] [Google Scholar]
- 13.Chondrogianni, N., F. L. Stratford, I. P. Trougakos, B. Friguet, A. J. Rivett, and E. S. Gonos. 2003. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J. Biol. Chem. 278:28026-28037. [DOI] [PubMed] [Google Scholar]
- 14.Chondrogianni, N., I. P. Trougakos, D. Kletsas, Q. M. Chen, and E. S. Gonos. 2008. Partial proteasome inhibition in human fibroblasts triggers accelerated M1 senescence or M2 crisis depending on p53 and Rb status. Aging Cell 7:717-732. [DOI] [PubMed] [Google Scholar]
- 15.Chondrogianni, N., C. Tzavelas, A. J. Pemberton, I. P. Nezis, A. J. Rivett, and E. S. Gonos. 2005. Overexpression of proteasome beta5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J. Biol. Chem. 280:11840-11850. [DOI] [PubMed] [Google Scholar]
- 16.Collado, M., M. A. Blasco, and M. Serrano. 2007. Cellular senescence in cancer and aging. Cell 130:223-233. [DOI] [PubMed] [Google Scholar]
- 17.Crone, S. A., A. Negro, A. Trumpp, M. Giovannini, and K. F. Lee. 2003. Colonic epithelial expression of ErbB2 is required for postnatal maintenance of the enteric nervous system. Neuron 37:29-40. [DOI] [PubMed] [Google Scholar]
- 18.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 19.Essers, M. A., L. M. de Vries-Smits, N. Barker, P. E. Polderman, B. M. Burgering, and H. C. Korswagen. 2005. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science 308:1181-1184. [DOI] [PubMed] [Google Scholar]
- 20.Finkel, T., and N. J. Holbrook. 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408:239-247. [DOI] [PubMed] [Google Scholar]
- 21.Finley, D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita, T., T. Shirasawa, K. Uchida, and N. Maruyama. 1996. Gene regulation of senescence marker protein-30 (SMP30): coordinated up-regulation with tissue maturation and gradual down-regulation with aging. Mech. Ageing Dev. 87:219-229. [DOI] [PubMed] [Google Scholar]
- 23.Funakoshi, M., R. J. Tomko, Jr., H. Kobayashi, and M. Hochstrasser. 2009. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell 137:887-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillette, T. G., B. Kumar, D. Thompson, C. A. Slaughter, and G. N. DeMartino. 2008. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J. Biol. Chem. 283:31813-31822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giorgio, M., M. Trinei, E. Migliaccio, and P. G. Pelicci. 2007. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 8:722-728. [DOI] [PubMed] [Google Scholar]
- 26.Goldberg, A. L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426:895-899. [DOI] [PubMed] [Google Scholar]
- 27.Graus-Porta, D., S. Blaess, M. Senften, A. Littlewood-Evans, C. Damsky, Z. Huang, P. Orban, R. Klein, J. C. Schittny, and U. Muller. 2001. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron 31:367-379. [DOI] [PubMed] [Google Scholar]
- 28.Guo, X., W. M. Keyes, C. Papazoglu, J. Zuber, W. Li, S. W. Lowe, H. Vogel, and A. A. Mills. 2009. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat. Cell Biol. 11:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamazaki, J., S. Iemura, T. Natsume, H. Yashiroda, K. Tanaka, and S. Murata. 2006. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25:4524-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamazaki, J., K. Sasaki, H. Kawahara, S. Hisanaga, K. Tanaka, and S. Murata. 2007. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol. Cell. Biol. 27:6629-6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris, S. L., and A. J. Levine. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 24:2899-2908. [DOI] [PubMed] [Google Scholar]
- 32.Hendil, K. B., R. Hartmann-Petersen, and K. Tanaka. 2002. 26 S proteasomes function as stable entities. J. Mol. Biol. 315:627-636. [DOI] [PubMed] [Google Scholar]
- 33.Hirano, Y., H. Hayashi, S. Iemura, K. B. Hendil, S. Niwa, T. Kishimoto, M. Kasahara, T. Natsume, K. Tanaka, and S. Murata. 2006. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol. Cell 24:977-984. [DOI] [PubMed] [Google Scholar]
- 34.Hirano, Y., K. B. Hendil, H. Yashiroda, S. Iemura, R. Nagane, Y. Hioki, T. Natsume, K. Tanaka, and S. Murata. 2005. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 437:1381-1385. [DOI] [PubMed] [Google Scholar]
- 35.Hirano, Y., T. Kaneko, K. Okamoto, M. Bai, H. Yashiroda, K. Furuyama, K. Kato, K. Tanaka, and S. Murata. 2008. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 27:2204-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishigami, A., Y. Kondo, R. Nanba, T. Ohsawa, S. Handa, S. Kubo, M. Akita, and N. Maruyama. 2004. SMP30 deficiency in mice causes an accumulation of neutral lipids and phospholipids in the liver and shortens the life span. Biochem. Biophys. Res. Commun. 315:575-580. [DOI] [PubMed] [Google Scholar]
- 37.Jariel-Encontre, I., G. Bossis, and M. Piechaczyk. 2008. Ubiquitin-independent degradation of proteins by the proteasome. Biochim. Biophys. Acta 1786:153-177. [DOI] [PubMed] [Google Scholar]
- 38.Jiang, Z., J. Hu, X. Li, Y. Jiang, W. Zhou, and D. Lu. 2006. Expression analyses of 27 DNA repair genes in astrocytoma by TaqMan low-density array. Neurosci. Lett. 409:112-117. [DOI] [PubMed] [Google Scholar]
- 39.Jung, T., and T. Grune. 2008. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 60:743-752. [DOI] [PubMed] [Google Scholar]
- 40.Kaneko, T., J. Hamazaki, S. Iemura, K. Sasaki, K. Furuyama, T. Natsume, K. Tanaka, and S. Murata. 2009. Assembly pathway of the mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 137:914-925. [DOI] [PubMed] [Google Scholar]
- 41.Kapeta, S., N. Chondrogianni, and E. S. Gonos. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J. Biol. Chem. 285:8171-8184. [DOI] [PMC free article] [PubMed]
- 42.Keyes, W. M., Y. Wu, H. Vogel, X. Guo, S. W. Lowe, and A. A. Mills. 2005. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 19:1986-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim, W. Y., and N. E. Sharpless. 2006. The regulation of INK4/ARF in cancer and aging. Cell 127:265-275. [DOI] [PubMed] [Google Scholar]
- 44.Kleijnen, M. F., J. Roelofs, S. Park, N. A. Hathaway, M. Glickman, R. W. King, and D. Finley. 2007. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 14:1180-1188. [DOI] [PubMed] [Google Scholar]
- 45.Komatsu, M., S. Waguri, T. Ueno, J. Iwata, S. Murata, I. Tanida, J. Ezaki, N. Mizushima, Y. Ohsumi, Y. Uchiyama, E. Kominami, K. Tanaka, and T. Chiba. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kops, G. J., T. B. Dansen, P. E. Polderman, I. Saarloos, K. W. Wirtz, P. J. Coffer, T. T. Huang, J. L. Bos, R. H. Medema, and B. M. Burgering. 2002. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419:316-321. [DOI] [PubMed] [Google Scholar]
- 47.Kruse, J. P., and W. Gu. 2009. Modes of p53 regulation. Cell 137:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuk, J. L., T. J. Saunders, L. E. Davidson, and R. Ross. 2009. Age-related changes in total and regional fat distribution. Ageing Res. Rev. 8:339-348. [DOI] [PubMed] [Google Scholar]
- 49.Le Tallec, B., M. B. Barrault, R. Courbeyrette, R. Guerois, M. C. Marsolier-Kergoat, and A. Peyroche. 2007. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell 27:660-674. [DOI] [PubMed] [Google Scholar]
- 50.Liu, B., J. Wang, K. M. Chan, W. M. Tjia, W. Deng, X. Guan, J. D. Huang, K. M. Li, P. Y. Chau, D. J. Chen, D. Pei, A. M. Pendas, J. Cadinanos, C. Lopez-Otin, H. F. Tse, C. Hutchison, J. Chen, Y. Cao, K. S. Cheah, K. Tryggvason, and Z. Zhou. 2005. Genomic instability in laminopathy-based premature aging. Nat. Med. 11:780-785. [DOI] [PubMed] [Google Scholar]
- 51.Matias, A. C., P. C. Ramos, and R. J. Dohmen. Chaperone-assisted assembly of the proteasome core particle. Biochem. Soc. Trans. 38:29-33. [DOI] [PubMed]
- 52.McWalter, G. K., L. G. Higgins, L. I. McLellan, C. J. Henderson, L. Song, P. J. Thornalley, K. Itoh, M. Yamamoto, and J. D. Hayes. 2004. Transcription factor Nrf2 is essential for induction of NAD(P)H:quinone oxidoreductase 1, glutathione S-transferases, and glutamate cysteine ligase by broccoli seeds and isothiocyanates. J. Nutr. 134:3499S-3506S. [DOI] [PubMed] [Google Scholar]
- 53.Meiners, S., D. Heyken, A. Weller, A. Ludwig, K. Stangl, P. M. Kloetzel, and E. Kruger. 2003. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J. Biol. Chem. 278:21517-21525. [DOI] [PubMed] [Google Scholar]
- 54.Murata, S., H. Kawahara, S. Tohma, K. Yamamoto, M. Kasahara, Y. Nabeshima, K. Tanaka, and T. Chiba. 1999. Growth retardation in mice lacking the proteasome activator PA28gamma. J. Biol. Chem. 274:38211-38215. [DOI] [PubMed] [Google Scholar]
- 55.Murata, S., H. Udono, N. Tanahashi, N. Hamada, K. Watanabe, K. Adachi, T. Yamano, K. Yui, N. Kobayashi, M. Kasahara, K. Tanaka, and T. Chiba. 2001. Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta. EMBO J. 20:5898-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murata, S., H. Yashiroda, and K. Tanaka. 2009. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10:104-115. [DOI] [PubMed] [Google Scholar]
- 57.Park, S., J. Roelofs, W. Kim, J. Robert, M. Schmidt, S. P. Gygi, and D. Finley. 2009. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 459:866-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park, S., G. Tian, J. Roelofs, and D. Finley. Assembly manual for the proteasome regulatory particle: the first draft. Biochem. Soc. Trans. 38:6-13. [DOI] [PMC free article] [PubMed]
- 59.Patrick, G. N., P. Zhou, Y. T. Kwon, P. M. Howley, and L. H. Tsai. 1998. p35, the neuronal-specific activator of cyclin-dependent kinase 5 (Cdk5) is degraded by the ubiquitin-proteasome pathway. J. Biol. Chem. 273:24057-24064. [DOI] [PubMed] [Google Scholar]
- 60.Périchon, R., J. M. Bourre, J. F. Kelly, and G. S. Roth. 1998. The role of peroxisomes in aging. Cell Mol. Life Sci. 54:641-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Postic, C., M. Shiota, K. D. Niswender, T. L. Jetton, Y. Chen, J. M. Moates, K. D. Shelton, J. Lindner, A. D. Cherrington, and M. A. Magnuson. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274:305-315. [DOI] [PubMed] [Google Scholar]
- 62.Rabl, J., D. M. Smith, Y. Yu, S. C. Chang, A. L. Goldberg, and Y. Cheng. 2008. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30:360-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radhakrishnan, S. K., C. S. Lee, P. Young, A. Beskow, J. Y. Chan, and R. J. Deshaies. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38:17-28. [DOI] [PMC free article] [PubMed]
- 64.Ramasamy, R., S. F. Yan, and A. M. Schmidt. 2006. Methylglyoxal comes of AGE. Cell 124:258-260. [DOI] [PubMed] [Google Scholar]
- 65.Roelofs, J., S. Park, W. Haas, G. Tian, F. E. McAllister, Y. Huo, B. H. Lee, F. Zhang, Y. Shi, S. P. Gygi, and D. Finley. 2009. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459:861-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saeki, Y., E. A. Toh, T. Kudo, H. Kawamura, and K. Tanaka. 2009. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell 137:900-913. [DOI] [PubMed] [Google Scholar]
- 67.Sakao, Y., T. Kawai, O. Takeuchi, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, K. Takeda, and S. Akira. 2000. Mouse proteasomal ATPases Psmc3 and Psmc4: genomic organization and gene targeting. Genomics 67:1-7. [DOI] [PubMed] [Google Scholar]
- 68.Salceda, S., and J. Caro. 1997. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 272:22642-22647. [DOI] [PubMed] [Google Scholar]
- 69.Scaffidi, P., and T. Misteli. 2006. Lamin A-dependent nuclear defects in human aging. Science 312:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt, M., J. Hanna, S. Elsasser, and D. Finley. 2005. Proteasome-associated proteins: regulation of a proteolytic machine. Biol. Chem. 386:725-737. [DOI] [PubMed] [Google Scholar]
- 71.Shibatani, T., E. J. Carlson, F. Larabee, A. L. McCormack, K. Fruh, and W. R. Skach. 2006. Global organization and function of mammalian cytosolic proteasome pools: implications for PA28 and 19S regulatory complexes. Mol. Biol. Cell 17:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shibue, T., S. Suzuki, H. Okamoto, H. Yoshida, Y. Ohba, A. Takaoka, and T. Taniguchi. 2006. Differential contribution of Puma and Noxa in dual regulation of p53-mediated apoptotic pathways. EMBO J. 25:4952-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sillitoe, R. V., and A. L. Joyner. 2007. Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu. Rev. Cell Dev. Biol. 23:549-577. [DOI] [PubMed] [Google Scholar]
- 74.Smith, D. M., S. C. Chang, S. Park, D. Finley, Y. Cheng, and A. L. Goldberg. 2007. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell 27:731-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sohal, R. S., and R. Weindruch. 1996. Oxidative stress, caloric restriction, and aging. Science 273:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tanaka, K., and A. Ichihara. 1989. Half-life of proteasomes (multiprotease complexes) in rat liver. Biochem. Biophys. Res. Commun. 159:1309-1315. [DOI] [PubMed] [Google Scholar]
- 77.Tonoki, A., E. Kuranaga, T. Tomioka, J. Hamazaki, S. Murata, K. Tanaka, and M. Miura. 2009. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell. Biol. 29:1095-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tronche, F., C. Kellendonk, O. Kretz, P. Gass, K. Anlag, P. C. Orban, R. Bock, R. Klein, and G. Schutz. 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23:99-103. [DOI] [PubMed] [Google Scholar]
- 79.Venugopal, R., and A. K. Jaiswal. 1996. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. U. S. A. 93:14960-14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Villunger, A., E. M. Michalak, L. Coultas, F. Mullauer, G. Bock, M. J. Ausserlechner, J. M. Adams, and A. Strasser. 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302:1036-1038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.