Abstract
In Lactococcus lactis IL1403, 14 genes are under the control of the copper-inducible CopR repressor. This so-called CopR regulon encompasses the CopR regulator, two putative CPx-type copper ATPases, a copper chaperone, and 10 additional genes of unknown function. We addressed here the function of one of these genes, ytjD, which we renamed cinD (copper-induced nitroreductase). Copper, cadmium, and silver induced cinD in vivo, as shown by real-time quantitative PCR. A knockout mutant of cinD was more sensitive to oxidative stress exerted by 4-nitroquinoline-N-oxide and copper. Purified CinD is a flavoprotein and reduced 2,6-dichlorophenolindophenol and 4-nitroquinoline-N-oxide with kcat values of 27 and 11 s−1, respectively, using NADH as a reductant. CinD also exhibited significant catalase activity in vitro. The X-ray structure of CinD was resolved at 1.35 Å and resembles those of other nitroreductases. CinD is thus a nitroreductase which can protect L. lactis against oxidative stress that could be exerted by nitroaromatic compounds and copper.
Lactococcus lactis IL1403 is a Gram-positive lactic acid bacterium which is used for the manufacture of food and dairy products but also for an increasing number of biotechnological applications. Given the economical importance of this microorganism, it is often used as a model for molecular studies. Its genome has been sequenced (4), and its proteome has been extensively characterized (11). When applied to industrial processes, this bacterium has to face various stress conditions, such as low pH, high temperature, osmotic shock, and metal stress (44). For instance, in traditional cheese making in Switzerland, L. lactis is exposed to copper released from the copper vats.
Copper is an essential micronutrient for both prokaryotes and eukaryotes. The two oxidation states of copper, Cu+ and Cu2+, allow its participation in many important biological functions. More than 30 enzymes are known to use copper as a cofactor, such as superoxide dismutase (SOD), cytochrome c oxidase, or lysyl oxidase (20). The redox activity of copper can also lead to the generation of free radicals, which cause cellular damage (42, 43). Recently, alternative copper toxicity mechanisms have been demonstrated in bacteria in which copper interferes with the formation of catalytic iron-sulfur clusters (6, 22). Whatever the mechanism of copper toxicity, maintenance of copper homeostasis by controlling the uptake, accumulation, detoxification, and removal of copper is critical for living organisms.
Copper homeostasis in L. lactis has not yet been investigated in great detail but appears to resemble the well-characterized copper homeostatic system of Enterococcus hirae (34). L. lactis possesses a copRZA operon, which provides copper resistance. It encodes the CopA copper export ATPase, the CopR copper-inducible repressor, and the CopZ copper chaperone (23). CopR regulates not only the copRZA operon but also an additional 11 genes. This so-called CopR regulon also includes copB, encoding a second putative copper ATPase; lctO, encoding lactate oxidase; and the ydiDE, yahCD-yaiAB, and ytjDBA operons of unknown function. Of all the genes and operons constituting the CopR regulon, the ytjDBA operon was most strongly induced by copper (23). Based on sequence comparison, the first gene of this operon, ytjD, encodes an oxygen-insensitive nitroreductase, which we renamed cinD for copper-induced nitroreductase.
Nitroreductases are called oxygen insensitive when they can catalyze the two-electron reduction of nitro compounds in the presence of oxygen. Such enzymes are widespread in nature and are able to reduce a wide range of substrates, such as furazones, nitroaromatic compounds, flavins, and ferricyanide, using NADH or NADPH as the reductant. They are flavoproteins of 22 to 24 kDa and form homodimers with one flavin mononucleotide cofactor per monomer. Although oxygen-insensitive nitroreductases have been extensively studied, their in vivo function remains largely unknown. The closest relative of CinD, which has functionally been studied, is FRP of Vibrio harveyi, with 29% sequence identity to CinD. FRP is not a typical nitroreductase but appears to function as an NADH flavin oxidoreductase which provides reduced flavin to luciferase (19). The next closest relative of CinD, NfsA of Escherichia coli, with 23% sequence identity, exhibits the broad substrate specificity typical of most nitroreductases (48). The structure of this enzyme has been solved at a resolution of 1.7 Å (17). It closely resembles the structures of other enzymes which belong to the oxygen-insensitive nitroreductase family. NfsA has recently been shown to participate in the degradation of 2,4,6-trinitrotoluene (10). This suggests that an important function of nitroreductases could be the metabolism of xenobiotics.
We investigated here the structure and function of CinD of L. lactis. CinD was induced by copper, cadmium, and silver and protected L. lactis from oxidative stress exerted by 4-nitroquinoline-N-oxide (NQO). The purified enzyme is a flavoprotein and exhibited nitroreductase activity on NQO and a variety of other substrates, using NADH as the reductant. CinD also possesses catalase activity and is thus able to defend cells against oxidative stress exerted by hydrogen peroxide, xenobiotics, or copper. The three-dimensional structure of CinD was resolved at a 1.35-Å resolution and exhibits a typical nitroreductase structure.
MATERIALS AND METHODS
Chemicals.
Medium components were obtained from Axon Lab AG, Dättwil, Switzerland. Chemicals for protein extractions and enzymatic assays were supplied by Merck (Darmstadt, Germany), DNA nucleases were supplied by Roche Biochemicals (Switzerland), the plasmid pProExHTa and recombinant tobacco etch virus (rTEV) protease were supplied by Invitrogen, Carlsbad, CA, and the Ni-nitrilotriacetic acid-agarose (Ni-NTA-agarose) chromatographic matrix was supplied by Qiagen. Oligonucleotides were synthesized by Microsynth (Balgach, Switzerland). Proofreading DNA polymerases were obtained from Takara Bio Inc. (Otsu, Japan), Roche Biochemicals (Switzerland), and Stratagene (La Jolla, CA). NQO (Sigma-Aldrich, St. Louis, MO) was dissolved in dimethyl sulfoxide (DMSO) at 100 mM and stored at −20°C. Working solutions were diluted from this stock in DMSO on the day of use.
Bacterial strains and culture conditions.
L. lactis IL1403 was obtained from Emmanuelle Maguin (INRA, Jouy-en-Josas, France) and was grown semianaerobically (air-saturated media in sealed bottles), in M17 media (39) at 30°C or on plates containing M17 media with 1.5% agar (AppliChem, Darmstadt, Germany). Milk for growth of L. lactis was prepared by autoclaving a 10% solution of milk powder (Difco) at 121°C for 15 min. Growth in milk was followed either by plating and assessing the number of CFU or by clarifying samples by the addition of 4 volumes of 15 mM Na-EDTA, pH 12, and measuring the absorption at 600 nm. E. coli Top10 (Invitrogen) or DH5α (Stratagene, La Jolla, CA) cells used for cloning were transformed according to manufacturer's instructions. E. coli strains were cultivated aerobically at 37°C in LB media (32) with appropriate antibiotics. To determine growth inhibition zones, 200 μl of stationary-phase cultures was spread on M17 plates, and 1.5-cm-diameter cellulose filter disks with the required chemicals were applied to the plates. Plates were incubated at 30°C, and the growth inhibition zones were measured after 16 h. To determine the growth rates of L. lactis in liquid cultures, 1 ml of M17 media in capped, disposable spectrophotometric cuvettes was inoculated with a 1/50 volume of cells which had been frozen in the logarithmic growth phase in 17% glycerol at −70°C. After growth for 1 h at 30°C, growth inhibitors were added, and growth was monitored at 600 nm with a Lambda 16 spectrophotometer (PerkinElmer Life Sciences). For competition assays between L. lactis strains, 25-ml cultures in M17 media were grown for 26 generations by four successive 100-fold dilutions into fresh media every 24 h. After four transfers, the numbers of CFU of wild-type and ΔcinD cells were determined by plating serial dilutions on M17 plates with and without erythromycin and counting the numbers of CFU.
Real-time quantitative PCR.
For RNA isolation, fresh mid-log cultures of L. lactis IL1403 in M17 medium were induced as indicated below in Results for 45 min at 30°C. Cells from 1 ml of culture were harvested by centrifugation, and the pellet was used immediately for RNA isolation with the RNeasy kit from Qiagen, according to the manufacturer's instructions. The optional treatment with DNase I (RNase free) for 30 min at room temperature was included. RNA concentrations were measured with a NanoDrop ND-1000 spectrophotometer. RNA quality was assessed on 1.2% agarose-formaldehyde gels stained with ethidium bromide. RNA was stored at −70°C. cDNA was reverse transcribed from 1 μg of total RNA per reaction using the iScript cDNA synthesis kit (Bio-Rad), according to the manufacturer's protocol. Real-time quantitative PCR was performed on the LightCycler (Roche), using SYBR Premix Ex Taq (Takara Bio Inc., Otsu, Japan), according to the manufacturer's instructions. Quantitative PCR conditions used were as follows: 1 cycle at 95°C for 10 s plus 40 cycles of denaturation at 95°C for 6 s, annealing at 60°C for 20 s, and extension at 72°C for 45 s. After the LightCycler PCRs, the products were analyzed for specificity and homogeneity by a melting curve analysis and additionally by electrophoresis on 1.2% agarose gels. For quantitative PCR, primers ytjD3 (5′-GCACAAGGTGTTCCTGAATC) and ytjD4 (5′-TCAGCAAGAGCAGTCCATAC) were used to measure cinD gene expression. 16S RNA was determined with primers dm7 (5′-GTGGCTCAACCATTGTATGC) and dm8 (5′-AGCCTCAGTGTCAGTTACAG). Real-time quantitative PCR results were expressed relative to 16S RNA.
Construction of the CinD expression vector pHYDSA1.
All plasmids were purified from E. coli DH5α using the large-scale NucleoBond AX plasmid isolation kit (Macherey-Nagel, Oensingen, Switzerland). The cinD gene of L. lactis IL1403 was cloned by PCR amplification of genomic L. lactis IL1403 DNA with the primers fm1 (5′-ATATGGCGCCATGTCATTCATTAAATCATTAG) and fm2 (5′-GCATGCAGCTTTTAAGAGCTCAACTTTAC). The resultant PCR product was cut with NarI and SacI and ligated into the histidine tag expression vector pProExHTa, resulting in pHYD2. This construct contained an A69S mutation in the cinD gene, which was mutated back to the wild-type sequence with the Chameleon mutagenesis kit (Invitrogen), as detailed by the manufacturer, resulting in pHYDSA2. The vector expressed a CinD fusion protein with an N-terminal six-histidine tag and a cleavage site for rTEV protease to remove the His tag. Cleaved CinD had the N-terminal methionine replaced by the sequence Gly-Ala-Arg.
CinD expression and purification.
E. coli DH5α (Invitrogen) transformed with expression plasmid pHYDSA2 was grown at 37°C in Luria-Bertani broth containing 100 μg/ml ampicillin to mid-log phase, cooled to 30°C, and induced with 1 mM isopropyl-β-d-thiogalactoside. Growth was continued for 4 h before the cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C. Cell pellets were suspended and washed twice with 140 mM NaCl and 20 mM NaPi, pH 7. All buffers used in subsequent steps were supplemented with 10 μM flavin mononucleotide (FMN) to prevent loss of the cofactor. The final pellet was resuspended in buffer A (50 mM Na-HEPES, 50 mM K2SO4 at pH 7.5, 1 mM β-mercaptoethanol), and a 1/100 volume of a protease inhibitor cocktail (100 mM N-α-p-tosyl-l-lysine-chloromethyl ketone, 100 mM N-p-tosyl-l-phenylalanine-chloromethyl ketone, 100 mM p-aminobenzamidine, 100 mM phenyl-methyl-sulfonyl-fluoride dissolved in dimethyl sulfoxide) was added. Cells were broken by three passages through a French press at 40 MPa. Debris was removed by centrifugation for 10 min at 3,000 × g in the cold. The supernatant was centrifuged at 90,000 × g for 30 min at 4°C, filtered, and loaded onto a 2- by 8-cm Ni-nitrilotriacetic acid (Ni-NTA)-agarose column, preequilibrated with buffer A. The column was washed with 5 column volumes of buffer A containing 20 mM imidazole, and CinD was eluted with 10 column volumes of a linear 0- to 200-mM imidazole gradient in buffer A. The fractions were analyzed by electrophoresis on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels and stained with Coomassie blue as described previously (18). The CinD-containing fractions were pooled, dialyzed against 50 mM Na-HEPES and 50 mM K2SO4, pH 7.4, concentrated 10-fold with a Centricon concentrator, and frozen at −70°C. For removal of the histidine tag, 600 μg of purified CinD was incubated with 100 U of rTEV protease for 4 h at 30°C. The proteolysis reaction mixture was passed through an Ni-NTA-agarose column in 50 mM Na-HEPES and 50 mM K2SO4, pH 7.0, and the flowthrough containing cleaved CinD was collected. For crystallization, CinD was further purified by gel filtration on a Superdex S75 column (GE Healthcare, Switzerland) in 300 mM NaCl, 25 mM Tris-Cl at pH 7.5, and 5 mM dithiothreitol. The purified protein was concentrated to 20 mg/ml and stored at −70°C. Samples for crystallization trials were supplemented with 1 mM FMN. Protein concentrations were determined with the Bio-Rad protein assay, using bovine serum albumin as a standard, except for that of pure CinD, which was estimated spectrophotometrically at 280 nm, using the extinction coefficient ɛ of 1.66 mg−1 cm−1.
Construction of the ΔytjD mutant strain.
An internal fragment of the cinD gene was PCR amplified from plasmid pHYD2 using primers fm33 (5′-GGCGGAAGCTTCACCAACAGCTTTCAATTC) and fm34 (5′-CGCGACTAGTTAGCAACAGCTTCGTCAATC). The resultant 399-bp product was digested with HindIII-SpeI and ligated into plasmid pTE1 (3), digested with the same enzymes. The resulting plasmid, pTEYD1, was propagated in E. coli DH5α in LB media with 40 μg/ml of kanamycin or 500 μg/ml of erythromycin. Plasmid integrity was checked by restriction analysis. For gene knockout, L. lactis IL1403 was transformed with pTEYD1 by electroporation as described previously (9). Transformants were selected at 30°C on agar plates with brain heart infusion media (Difco Laboratories, Detroit, MI), containing 5 μg/ml of erythromycin. Clones were screened for ΔcinD mutants by PCR amplification of genomic DNA, using primer fm18 (5′-GCTGCACCAGAAGGAGTGAAAGC), located upstream of the cinD gene, and primer fm23 (5′-AAGCTAAGGGCGAATTCCAGCAC), located on the pTEYD1 plasmid. The correctness of the junction between plasmid and genomic DNA was verified by restriction analysis of the resultant PCR fragments.
Western blot analysis of CinD expression.
Antibodies to CinD were raised by injecting rats with an initial subcutaneous injection of 60 μg of gel-purified, cleaved CinD, followed by two intravenous booster injections of 60 μg CinD each. Serum samples were coagulated overnight at 4°C, and the clear supernatant was used for Western blotting. Cultures in 10 ml of M17 media were grown to an optical density at 546 nm (OD546) of 0.6 to 0.8, followed by induction with 1 mM CuSO4 for 90 min when required. Cell lysates were prepared by centrifuging the cultures and treating the cell pellets with 200 μl of 10 mg/ml lysozyme, 1 mM EDTA, and 10 mM Tris-Cl at pH 8 on ice for 15 min. The lysates were treated with 40 μg of DNase I in 15 mM MgCl2 for 10 min at 0°C and centrifuged. The supernatants were denatured in SDS sample buffer, and 10 μg of each extract was resolved by electrophoresis on a 15% Tricine-SDS-polyacrylamide gel (33). Western blots were prepared as previously described (41), using a horseradish peroxidase-coupled goat anti-rat IgG secondary antibody (Santa Cruz). Bands were visualized by chemiluminescence using homemade reagents. Chemiluminescence signals were captured with a Fuji LAS-1000 imaging system (Fuji Photo Film, Tokyo, Japan).
Nitroreductase assay.
The reductase activity of purified CinD was measured spectrophotometrically at 25°C. The reaction mixture contained in 1 ml of 50 mM Tris-Cl at pH 7.5, 0.05 mM NADH, and 0.05 mM substrate. The reaction was started by the addition of enzyme, and the change in absorption of either NADH or substrate was monitored over time. The following molar absorption coefficients were used: 6,220 M−1 cm−1 for NADH at 340 nm, 20,600 M−1 cm−1 for 2,6-dichlorophenolindophenol (DCPIP) at 600 nm, and 1,000 M−1 cm−1 for K3[Fe(CN)6]3− at 420 nm. Self-absorption at 340 nm of substrates was corrected as follows: ɛNADH/|[ɛNADH + (ɛSox - ɛSred)]|, where ɛSox and ɛSred are the extinction coefficients of the oxidized and reduced substrates, respectively.
Oxygen measurements.
The catalase activity of CinD was measured by recording the production of oxygen from hydrogen peroxide with a Clark electrode. The reactions were performed at 25°C in a 1-ml chamber containing 50 mM Tris-Cl at pH 7.5 and various amounts of H2O2. Reactions were started by adding 3 μg of purified CinD.
Crystallization and structure determination.
Crystals were grown at 20°C by vapor diffusion with hanging drops, mixing 2 volumes of protein to 1 volume of crystallization buffer (100 mM Tris-Cl at pH 8, 200 mM MgCl2, 20% [wt/vol] polyethylene glycol 3350, 2% 2-methyl-2,4-pentanediol). Crystals were cryoprotected using the crystallization buffer supplemented with 20% 2-methyl-2,4-pentanediol and flash cooled in liquid nitrogen. Diffraction data were collected at 100 K at the Swiss Light Source (Paul Scherer Institute, Villigen, Switzerland) at beamline X06DA to a 1.35-Å resolution and processed using XDS (15). The structure was solved by molecular replacement using the program PHASER (25) and the nitroreductase structure from Streptococcus mutans as the search model (Protein Data Bank [PDB] accession number 2IFA) (Northeast Structural Genomics Consortium, unpublished data). Data collection and refinement statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameter | Value(s)a |
|---|---|
| Space group properties | |
| Space group | C2221 |
| a, b, c (Å) | 54.10, 122.22, 69.05 |
| α, β, γ (Å) | 90.0, 90.0, 90.0 |
| No. of molecules/ASUb | 1 |
| Data collection statistics | |
| Beamline | X06DA |
| Wavelength (Å) | 1.000 |
| Resolution range (Å) | 50.0-1.35 |
| No. of observations | 433,802 |
| No. of unique reflections | 48,865 |
| Completeness (%) | 99.2 (97.3) |
| Rmerge (%)c | 9.4 (58.4) |
| I/σ(I) | 12.36 (1.9) |
| Refinement statistics | |
| Resolution range (Å) | 32.13-1.35 |
| No. of reflections, working set | 47,384 |
| No. of reflections, test set | 1,466 |
| R/Rfree (%) | 13.15/15.55 |
| No. of atoms | |
| Protein | 3,253 |
| Ligand | 51 |
| Water | 300 |
| RMS deviations | |
| Bond length (Å) | 0.007 |
| Bond angle (°) | 0.863 |
| Ramachandran plot (%) | |
| Most favored | 98.33 |
| Allowed | 1.67 |
| Disallowed | 0.00 |
| B-factors (Å) | |
| Main chain | 19.1 |
| Side chain and water | 27.7 |
The values in parentheses for the resolution range, completeness, Rmerge, and I/σ(I) correspond to the outermost-resolution shell. Data collection and refinement statistics values were measured using XDS and Phenix, respectively.
ASU, asymmetric units.
Rmerge = ΣhklΣj |I(hkl; j) − {I(hkl)}|/[ΣhklΣj {I(hkl)}], where I(hkl; j) is the jth measurement of the intensity of the unique reflection (hkl) and {I(hkl)} is the mean overall symmetry-related measurement.
Protein structure accession number.
The CinD structure was deposited in the PDB under accession number 2WQF.
RESULTS
Regulation of CinD expression by metal ions.
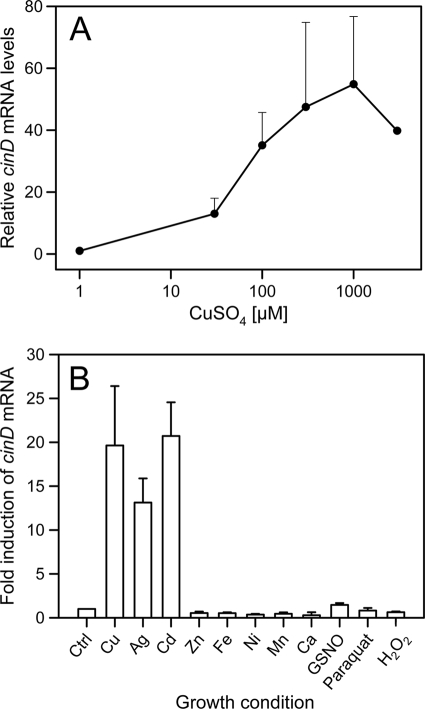
A global proteomics and bioinformatics analysis of the response of L. lactis IL1403 to copper exposure led to the identification of a copper-inducible nitroreductase, encoded by the ytjD gene (GenBank accession number NP_268070), which we here rename cinD (23). To learn more about the function of CinD in copper homeostasis and stress response, we analyzed the induction of the cinD gene in more detail. By real-time quantitative PCR, dose-dependent upregulation of cinD expression by copper was observed. Maximal induction was measured in cells induced with 1 mM Cu2+ (Fig. 1 A). Among other metals tested, 200 μM Cd2+ induced cinD to an extent similar to that of copper, while Ag+ at the same concentration induced cinD to about 60% of these levels (Fig. 1B). Zn2+, Fe2+, Ni2+, Mn2+, and Ca2+ failed to stimulate cinD expression. The cinD gene is under the control of CopR (23), a repressor homologous to CopY of E. hirae. Induction of genes under the control of CopR proceeds via formation of a binuclear Cu(I)-thiolate cluster which displaces the bound Zn2+ and releases the repressor from the DNA. Other soft metals with a preponderance to form thiolates, like Ag+ and Cd2+, were also found to induce the closely related CopY repressor of Enterococcus hirae (30). Induction of cinD by copper, silver, and cadmium is thus in line with the expectations for a gene under the control of the CopY-like repressor CopR. No significant or marginal induction of cinD by nitrosative (S-nitrosoglutathione [GSNO]) or oxidative stress (paraquat, H2O2) was observed, suggesting that oxidative or nitrosative stress does not directly induce cinD.
FIG. 1.
Real-time quantitative PCR analysis of cinD mRNA expression. (A) The induction of cinD mRNA was measured as a function of the copper concentration in media. Cells were grown semianaerobically and induced for 45 min with copper at the concentrations indicated on the abscissas. The error bars show standard deviations based on three independent experiments. (B) Induction of cinD mRNA was measured in response to different stress conditions. Cells were grown semianaerobically and induced for 45 min with the chemicals indicated on the abscissas. The following metal salts were used at 200 μM: CuSO4, AgNO3, CdSO4, ZnSO4, FeCl2, NiSO4, MnCl2, and CaSO4. GNSO, paraquat, and H2O2 were used at concentrations of 4 mM, 20 mM, and 1 mM, respectively. The error bars show standard deviations based on three independent experiments. All values are statistically different (P < 0.01) from each other (Kruskal-Wallis one-way analysis of variance on ranks).
Overexpression and purification of CinD.
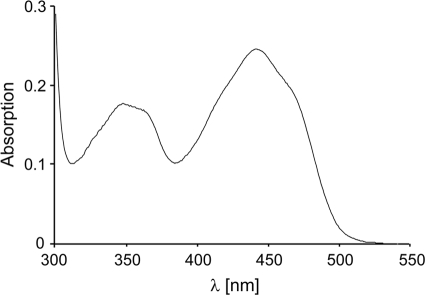
To determine the catalytic activity of CinD, an E. coli expression system was set up by constructing the plasmid pHYDSA2. It contained an isopropyl-β-d-thiogalactoside-inducible fusion of CinD with a cleavable N-terminal 6×His tag. This allowed for the expression of 6×His-CinD in copious amounts. It was purified by Ni-NTA-agarose affinity chromatography, followed by cleavage of the 6×His tag with rTEV protease. CinD could be purified to better than 95% purity in a single affinity purification step, with an average yield of 10 mg of recombinant protein per liter of culture (Fig. 2). Purified CinD was highly soluble and had the expected molecular mass of 22.6 kDa. CinD solutions were yellow and exhibited a typical FMN absorption spectrum (Fig. 3). Based on an extinction coefficient of 12,500 M−1 cm−1 for FMN, purified CinD contained 0.91 FMN molecules per monomer. This is reasonably close to the expected stoichiometry of one FMN per monomer.
FIG. 2.
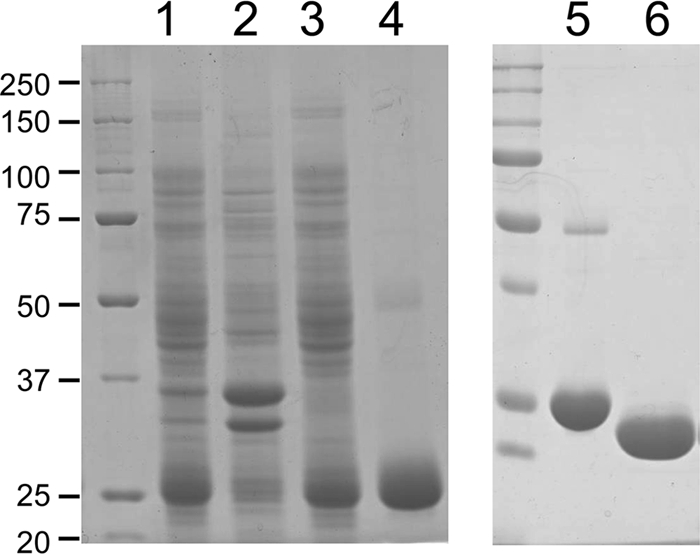
Affinity purification of CinD and His tag cleavage. Proteins were resolved on 12% sodium dodecyl sulfate gels and stained with Coomassie blue. Lane 1, 10 μg of crude bacterial lysate; lane 2, 10 μg of protein from the pellet obtained by ultracentrifugation of the lysate; lane 3, 10 μg of protein from the supernatant from ultracentrifugation; lane 4, 6 μg of protein eluted from a Ni-NTA-agarose column with 200 mM imidazole; lane 5, 6 μg of uncleaved 6×His-CinD; lane 6, 6 μg of rTEV-cleaved CinD after dialysis. The sizes of molecular mass markers are indicated in kDa on the left.
FIG. 3.
Absorption spectrum of CinD. The spectrum was recorded with 23 μM of purified CinD in 50 mM Tris-Cl, pH 8, at 25°C.
Characterization of the activity of CinD.
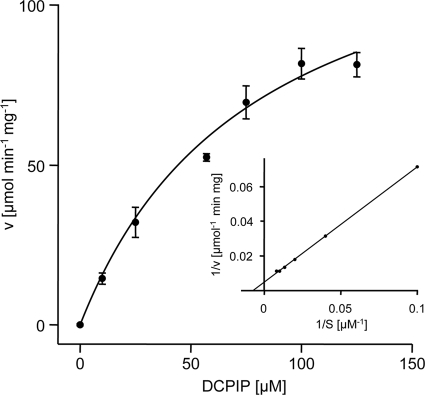
The sequence similarity between CinD and bacterial flavoproteins of the nitroreductase family suggested that CinD could have a similar function. A range of substrates typical for this class of enzymes was tested, using NADH as the reductant. The highest kcat value, 27 s−1, was measured with DCPIP. The Km value for this reaction was 79 μM, and the Vmax value was 383 μmol min−1 mg−1 (Fig. 4). A substantial kcat value of 11 s−1 was also measured with NQO, while activities below 10% of the maximal activity for a range of other substrates were recorded, including methylene blue (2.6 s−1), 4-chloro-7-nitrobenzofurazan (0.83 s−1), and K3[Fe(CN)6] (0.69 s−1). No substrate reduction in the presence of 50 μM FMN but without enzyme was observed. CinD failed to reduce many known substrates of nitroreductases, like nitrofurazone, 4-nitroanisol, 4-nitrophenol, chloramphenicol, GW9662, nitroprusside, methyl-3-nitro-1-nitrosoguanidin, 2,4,6-trinitrophenol, a range of differently halogenated nitrobenzenes, nitroaniline, 3-nitro-l-tyrosine, sulfafurazol, nitrosocysteine, nitrosoglutathione, nitrophenylphosphate, flavin adenine dinucleotide (FAD), and FMN. Both NADH and NADPH could serve as electron donors, but NADH resulted in four times higher specific activities. Thus, CinD is an oxygen-insensitive nitroreductase and exhibits rather narrow substrate specificity. CinD was also found to possess significant catalase activity (Fig. 5). This reaction was NAD(P)H independent, was not catalyzed by FMN alone, and exhibited a Km of 31 mM and a Vmax of 68 μmol min−1 mg−1. While the Km for this reaction is relatively low, the activity may still be biologically relevant; L. lactis and some related organisms do not possess catalase and accumulate hydrogen peroxide in millimolar concentrations (4, 45).
FIG. 4.
Kinetic analysis of DCPIP reduction by CinD. DCPIP reduction by 3 μg of CinD was monitored by following the oxidation at 340 nm of NADH. Reduction rates were fitted with Michaelis-Menten kinetics using SigmaPlot. (Inset) Lineweaver-Burk plot of the same data.
FIG. 5.
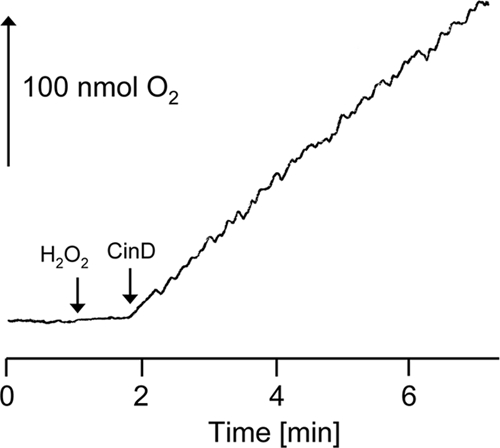
Catalase activity of CinD. Purified CinD (3 μg) was incubated with 10 mM H2O2, and oxygen evolution was measured with a Clark electrode. The arrows indicate the additions of H2O2 and CinD.
Effect of cinD gene inactivation.
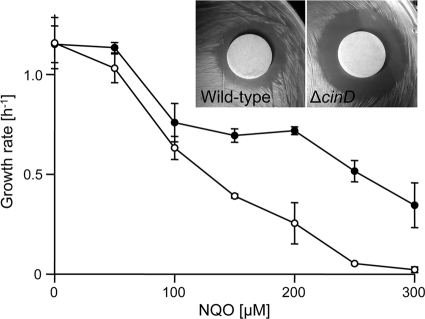
To obtain information about the function of CinD in vivo, a ΔcinD strain was constructed by homologous, Campbell-like recombination with plasmid pTEYD1, carrying a fragment of the cinD gene. Insertion of pTEYD1 into the genome at the correct site was confirmed by PCR, and the absence of the CinD protein in the knockout mutant was demonstrated by Western blotting with an antibody against CinD (Fig. 6). The ΔcinD strain exhibited increased sensitivity toward NQO (Fig. 7). There was complete growth inhibition of the ΔcinD mutant by 300 μM NQO, while wild-type cells still grew at a growth rate of 35% of that observed in the absence of NQO. The increased sensitivity of the ΔcinD strain to NQO was even more apparent on solid growth media by the large zone of growth inhibition around an NQO-soaked filter disk (Fig. 7, inset). NQO causes oxidative stress and is a good substrate of CinD in vitro; NQO reduction apparently diminishes the oxidative stress exerted by this agent. DCPIP, which was the best substrate of CinD in vitro, was essentially nontoxic to both the wild type and the ΔcinD mutant. NQO, like paraquat and hydrogen peroxide, did not induce expression of CinD (not shown).
FIG. 6.
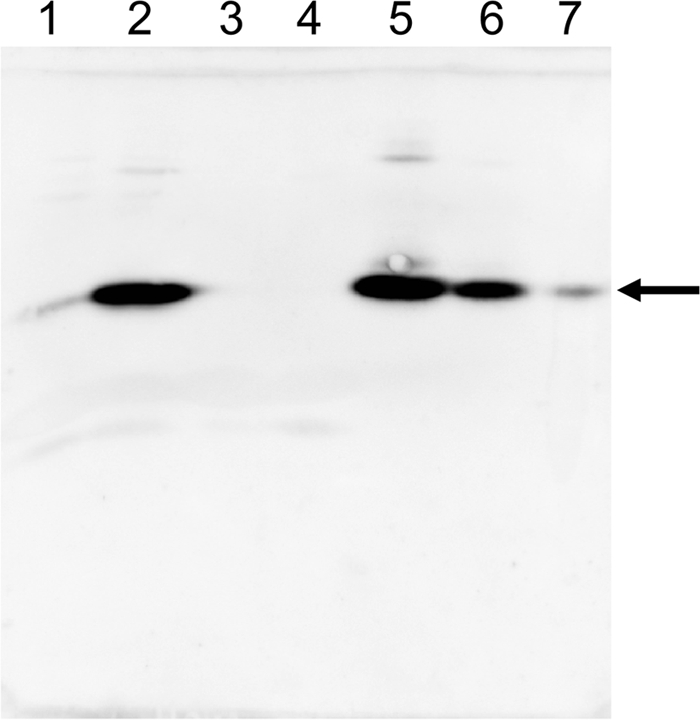
CinD expression in the wild type and the ΔcinD strain and induction by copper. CinD expression was assessed by Western blotting, developed with a CinD antibody. Lane 1, uninduced wild type; lane 2, wild type induced with 1 mM CuSO4; lane 3, uninduced ΔcinD; lane 4, ΔcinD induced with 1 mM CuSO4; lanes 5 to 7, 60, 6, and 0.6 ng of purified CinD, respectively. Induction in all cases was for 90 min. The arrow points to the position of CinD.
FIG. 7.
Effect of cinD inactivation on the sensitivity toward 4-nitroquinoline-N-oxide. L. lactis wild-type (•) and ΔcinD (○) cells were grown in 1-ml cuvettes containing M17 medium supplemented with 0 to 300 μM NQO. Standard deviations calculated from three independent experiments are depicted with error bars. (Inset) Phenotype of ΔcinD under oxidative stress. Filter disks soaked with 25 mM NQO were applied to bacterial lawns of L. lactis wild-type and ΔcinD cells. The plates were incubated overnight at 30°C.
Although NQO primarily causes oxidative stress, we also investigated a possible role of CinD in defense against nitrosative stress. Protein nitrosylation was indistinguishable in the wild type and the ΔcinD mutant, as assessed with an anti-nitrosocysteine antibody and the biotin switch detection method for nitrosylated proteins (8). There was also no phenotypic difference between the wild type and the cinD mutant strain when grown in the presence of agents that cause nitrosative stress, such as nitrosoglutathione, nitrosocysteine, nitrosocysteinyl-glycine, diethylamine, NONOate, and nitrite (not shown). So, a role of CinD in the defense against nitrosative stress appears unlikely.
Since CinD expression is regulated by copper, we examined the copper sensitivity of the ΔcinD strain. In standard growth experiments, growth inhibition of the wild type and the ΔcinD mutant by copper appeared to be the same. A greater sensitivity of the ΔcinD mutant toward copper could, however, be observed in growth competition experiments. Cultures in M17 media were inoculated with a 100-fold excess of the ΔcinD mutant over the wild type (1.5 × 109 and 1.5 × 107 CFU, respectively) and grown in the presence or absence of 0.5 mM copper for 26 generations by serial transfers. In the final cultures, the ratio of the wild type to ΔcinD was (5.82 ± 0.06) × 106 CFU when grown with copper and (1.8 ± 1.4) × 103 CFU when grown without copper. This clearly shows a growth advantage of the wild type over the ΔcinD mutant, which is strongly accentuated by copper. We also performed a series of growth experiments in milk to mimic a natural environment of L. lactis. In growth media prepared from milk powder, wild-type cells invariably exhibited a growth advantage and better survival over the ΔcinD mutant in the presence of copper. Unfortunately, these experiments were difficult to quantify and are thus not shown. The main problem was the different cell morphologies observed in milk. While the wild type preferentially grew as cell pairs, the ΔcinD mutant grew in chains of 8 to 12 cells. Copper shifted these morphologies to more single cells for the wild type and spherical clumps of many cells for the ΔcinD mutant. This made it difficult to enumerate living cells reliably, either by optical density or by plating and counting of the number of CFU. Yet, it appears clear that expression of CinD provides wild-type cells with a growth advantage under stress conditions exerted by copper and oxidative stress-inducing chemicals.
Crystal structure of CinD.
The crystal structure of the CinD-FMN complex was refined at a 1.35-Å resolution, with good statistics (Table 1). The structure is very well defined; only residues 75 to 86, forming a flexible loop, have very weak electron densities. A DALI search (13) revealed a hypothetical protein, SMU.260, with S. mutans (50% sequence identity, root mean square [RMS] deviation of 1.2 Å for 200 residues; PDB accession number 2IFA) and Bacillus cereus ATCC 14579 (41% sequence identity, RMS deviation of 1.3 Å; PDB accession number 1YWQ) having the most similar entries in the PDB. Other database entries classified as similar have sequence identities of 23% and less.
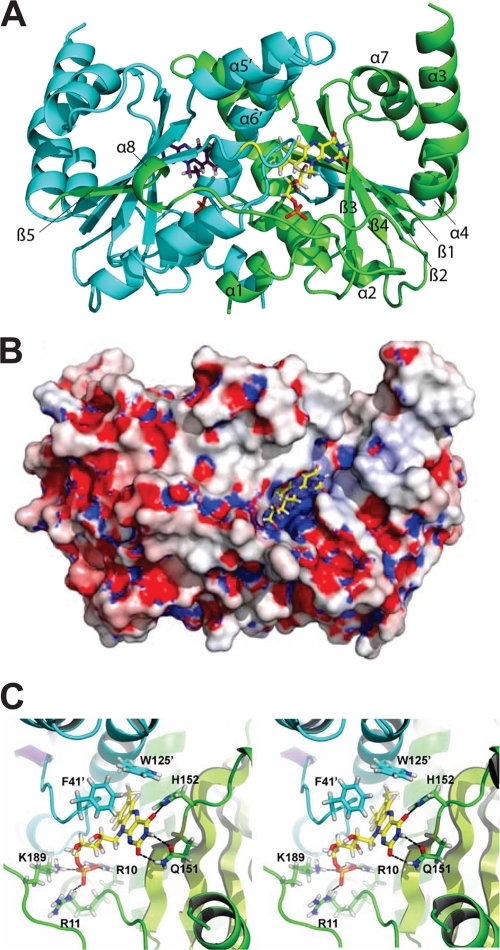
Like other nitroreductases, CinD forms a homodimeric structure in which the monomer possesses an α+β fold and the FMN is positioned at the dimer interface (Fig. 8 A), i.e., the active site is built by residues of both subunits. A total of 2,521 Å2 of solvent-accessible surface per monomer becomes buried upon dimer formation, with a total monomer surface of 10,891 Å2. Both termini of the CinD monomers extend into the direction of the dimer partner. The central β-sheet is formed by four antiparallel strands together with the parallel β5 strand from the C terminus of the other subunit. The five-stranded β-sheet is shielded by two and three α-helices, respectively.
FIG. 8.
Structure of CinD. (A) Cartoon representation with one subunit shown in light blue and purple and the other in green and lime. FMN molecules are depicted as stick models, and key residues discussed in the text are indicated. (B) Molecular surface representation with mapped electrostatic potential. The potential is drawn from −15 kT (red) to +15 kT (blue). Adaptive Poisson-Boltzmann Solver was used for the electrostatics calculation, assuming an ion concentration of 0.15 M (2). (C) Stereo view of the FMN binding site. Hydrogen bonds are indicated as black dashed lines, nitrogen atoms are colored in dark blue, oxygen atoms are in red, hydrogen atoms are in gray, and carbon atoms are in yellow.
The dimer interface is built up by hydrophobic interactions formed by the short α1-helices (Phe3-Glu8) and the α6-helices (Ala119-Ala141) from both subunits. Residues from the N-terminal-extending α1-helix (Phe3, Ser6, Leu7, and Glu8) are buried in a hydrophobic cleft of the neighboring subunit (formed by residues Phe3, Ser6, Phe36, Asn136, Thr139, Ala140, and Glu143), and the α6-helix from each subunit is placed in a hydrophobic cleft of the other subunit. As mentioned above, the C-terminal part (Glu188-Ala202) composed of the short α8-helix (Asp194-Arg197) and the β5 strand (Phe198-Ala201) extends over the surface to the neighboring monomer, where the β5 strand contributes to the central pleated β-sheet of the other monomer. Seven hydrogen bonds and two salt bridges (between Arg35 and Asp194 of the other subunit) contribute to the stability as well.
The cofactor FMN is bound in a deep cleft that is positively charged (Fig. 8B). The local electrostatic potential originates mainly from Arg10, Arg11, and Lys189, which coordinate the phosphate group (Fig. 8C). The si face of the isoalloxazine ring is backed by strand β1. From this side, the carboxylamide of Gln151 forms hydrogen bonds to O2 and HN3 of the pyrimidine moiety of FMN. A further hydrogen bond is donated by the side chain of His152. All residues mentioned belong to the same subunit. The re face of the isoalloxazine ring is covered by Phe41′ and Trp125′ of the other subunit. They are further apart and form part of the substrate binding site. The heterocycle of FMN is nonplanar, with a dihedral angle of about 18.5 degrees between the planes of the dimethylbenzoide and pyrimidine ring. This bend has been implied in favoring the reduction of FMN, owing to the fact that reduced FMN possesses a dihedral angle of about 25 degrees (12). Bent cofactors have been observed in many nitroreductases, but exceptions have also been described, namely, for E. coli YdjA (7) and Streptococcus mutans SMU.260 (PDB accession number 2IFA). The latter was the initial search model and shares 40% amino acid identity with CinD. Closer inspection of SMU.260 revealed a clear indication of a bent cofactor in this nitroreductase as well, but the bending was less pronounced than that in CinD. While the overall crystal structure of CinD resembles a typical nitroreductase, there is one profound difference between CinD and the majority of the reported nitroreductases. Two prominent α-helices that usually shield the active site on the re face of FMN are substantially shortened in CinD and Streptococcus mutans SMU.260 and are even completely missing in E. coli YdjA. These helices are believed to have an influence on substrate specificity and recognition (7).
Taken together, the present work shows the structure and the function of CinD nitroreductase of L. lactis. We showed that this enzyme can protect cells against oxidative stress by NQO but also function as a catalase. In a natural setting, CinD probably protects L. lactis against a range of xenobiotics and oxidative stress conditions.
DISCUSSION
CinD had been identified previously by proteomics as the most strongly copper-induced protein of the CopR regulon of L. lactis IL1403 (23). In the present study, we characterized CinD (YtjD) as a copper-induced oxygen-insensitive nitroreductase with a role in defense against oxidative stress. Purified CinD was most active in the reduction of DCPIP and NQO, a strong inducer of oxidative stress. The enzyme also exhibited significant catalase activity.
By quantitative PCR as well as by Western blotting, it was shown that copper induced cinD in the order of 20- to 50-fold, in line with the 35-fold induction determined by proteomics (23). CinD was also induced by silver and cadmium, as would be expected for a gene under the control of CopR, which is homologous to CopY of E. hirae (30). Oxidative stress by NQO, paraquat, or hydrogen peroxide did not induce cinD expression, which contrasts with the oxidative stress-dependent activation of some other nitroreductases. nfsA of E. coli and nfrA of Staphylococcus aureus are nitroreductases that are under the control of SoxR (21) and PerR (37), respectively, which are regulators that respond to oxidative stress. The NfrA nitroreductase of Bacillus subtilis, on the other hand, is induced by paraquat and hydrogen peroxide (28). Oxidative stress and nitrosative stress are apparently interconnected, since overlapping sets of genes are induced by transcription factors sensing oxidative stress, like OxyR and SoxR, and by those sensing nitrosative stress, like NorR and Fur (29). In contrast, cinD induction in L. lactis was observed only with heavy metal ions. Chemicals which induce nitrosative stress, such as nitrosoglutathione, nitrosocysteine, or l-arginine, did not induce genes under the control of CopR. This provides support to the notion that cinD expression is governed solely by the copper-responsive CopR repressor. Further support for this comes from the promoter structure of CinD, which features two copies of the so-called “cop box” of consensus TACANNTGTA, which is a universal DNA binding site for CopR-like repressors (31). The cop box is present in one or two copies in all seven promoters of copper-regulated genes and operons we have studied so far (23, 36), and copper, silver, and cadmium are the only inducers of these genes/promoters which could be identified.
Oxygen-insensitive nitroreductases have been studied in microorganisms as different as Enterococcus cloacae (5), E. coli, and Vibrio harveyi (48), Vibrio fischeri (49), Bacillus subtilis (27), Staphylococcus aureus (37), and Synechocystis spp. (38). Nitroreductases catalyze the reduction of a large range of nitroaromatic compounds; therefore, they are generally assumed to be involved in detoxification processes. Indeed, the first enzyme described was isolated from bacteria growing in a weapons storage dump and could reduce the explosive trinitrotoluene (5). Because of their broad substrate specificity, nitroreductases have also received attention for biotechnological applications (26, 47). However, the biological function of this class of enzymes remains unclear. It has been proposed for Bacillus subtilis that nitroreductases are responsible for the maintenance of the cellular redox state, thereby protecting from oxidative stress. Indeed, we showed here that CinD can protect L. lactis from oxidative stress exerted by NQO. This chemical is quite toxic to L. lactis and is a good substrate for CinD, second only to DCPIP, which does not appear to be toxic to L. lactis. Detoxification of NQO by CinD is supported by the observations that it is reduced by CinD in vitro and that the ΔcinD strain is more sensitive to NQO than the wild type. In its natural environment, L. lactis may not encounter NQO but may encounter similar compounds with oxidative stress potential. If copper elicits or acerbates the oxidative stress potential of such chemicals, it would make sense that copper induces the CinD defense system.
Alternatively, or in addition, the catalase activity of CinD could offer an important protection against oxidative stress by decomposing hydrogen peroxide to oxygen and water. Fast-growing L. lactis produces considerable quantities of H2O2. This can occur by at least two mechanisms, namely, by NADH oxidase (45) and by the LctO lactate dehydrogenase. LctO converts lactate to pyruvate, under production of hydrogen peroxide. It has been argued that these two activities serve in the scavenging of molecular oxygen to reduce oxidative stress (3, 24). Since L. lactis is devoid of a “normal” catalase and of SOD, alternative mechanism dealing with hydrogen peroxide and oxidative stress must be present. NADH peroxidase can convert hydrogen peroxide to water, but this activity is apparently very low (45). Interestingly, LctO belongs to the CopR regulon, is upregulated by copper, but does not contribute to increased copper tolerance (3). The induction of LctO by copper appears counterproductive since this would lead to the production of H2O2, which in turn, could undergo a Fenton reaction with copper, leading to the generation of toxic OH. radicals. However, pyruvate (generated by LctO) can nonenzymatically react with hydrogen peroxide according to the following reaction: pyruvate + H2O2 → acetate + CO2 + H2O. Such a mechanism is supported by the accumulation of up to 4 mM pyruvate by L. lactis strain NZ2007, which is deficient in lactate dehydrogenase and does not produce detectable hydrogen peroxide during logarithmic growth (45). The induction of LctO under copper stress may thus serve in the elimination of hydrogen peroxide by nonenzymatic reaction with pyruvate. Pyruvate was also shown to exert a protective effect against copper-induced cysteine autoxidation in astrocytes (46) and to protect motor neurons expressing mutant forms of SOD1 (which cause amyotrophic lateral sclerosis) against copper toxicity (16).
CinD (YtjD) is the first gene of an operon originally designated ytjDBA. ytjB is predicted to encode a manganese transporter, and ytjA is predicted to encode a ribonucleotide-2(3)-phosphate reductase. Copper-induced manganese uptake by YtjB could provide an additional level of protection against oxidative stress. Although L. lactis IL1403 is devoid of a Mn-containing SOD, Mn2+ has also been claimed to dismutate hydrogen peroxide nonenzymatically (35). The reaction has been studied in detail in vitro (40), but recent evidence suggests that it does not play a role in vivo. Instead, manganese apparently can metallate mononuclear enzymes in lieu of iron, thereby precluding a Fenton reaction between iron and hydrogen peroxide (1). Induction of manganese uptake by hydrogen peroxide has been reported for many bacteria, and uptake can be accomplished by P-type ATPases, ABC-type permeases, or Nramp-type manganese uptake systems (for a review, see reference 14). YtjB of L. lactis resembles an Nramp-type transporter; whether it plays a role in manganese uptake and protection against oxidative stress is currently under investigation in our laboratory.
Clearly, the defense against copper toxicity and copper-associated oxidative stress takes place at several levels. Depending on the bacterial species, it involves copper export by ATPases, detoxification of superoxide by SOD, and scavenging of hydrogen peroxide enzymatically by catalase or maybe nonenzymatically by manganese, pyruvate, and other α-ketoacids. In addition, cytoplasmic manganese may metallate some enzymes in lieu of iron to curb the Fenton reaction. Intracellular glutathione can complex copper and protect against oxidative damage, and cytoplasmic zinc can also protect enzymes from copper toxicity. Finally, a number of regulatory systems respond to oxidative stress and prevent DNA and cell damage by other mechanisms. CinD with its nitroreductase and catalase activities adds yet another protective element to this network. Several more uncharacterized, copper-induced genes of the CopR regulon are likely to contribute to this complex network, and considerably more work is required to fully understand the defense of L. lactis against copper and oxidative stress.
Acknowledgments
This work was supported by grants 3100A0-122551 and A3100-120174 from the Swiss National Foundation, by a grant from the International Copper Association, and by a grant from the Swiss State Secretary for Education and Research.
Footnotes
Published ahead of print on 18 June 2010.
REFERENCES
- 1.Anjem, A., S. Varghese, and J. A. Imlay. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72:844-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barré, O., F. Mourlane, and M. Solioz. 2007. Copper induction of lactate oxidase of Lactococcus lactis: a novel metal stress response. J. Bacteriol. 189:5947-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, C., and M. DeLuca. 1991. Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae. J. Biol. Chem. 266:4119-4125. [PubMed] [Google Scholar]
- 6.Chillappagari, S., A. Seubert, H. Trip, O. P. Kuipers, M. A. Marahiel, and M. Miethke. 2010. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 192:2512-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, J. W., J. Lee, K. Nishi, Y. S. Kim, C. H. Jung, and J. S. Kim. 2008. Crystal structure of a minimal nitroreductase, ydjA, from Escherichia coli K12 with and without FMN cofactor. J. Mol. Biol. 377:258-267. [DOI] [PubMed] [Google Scholar]
- 8.Forrester, M. T., M. W. Foster, M. Benhar, and J. S. Stamler. 2009. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic. Biol. Med. 46:119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber, S. D., and M. Solioz. 2007. Efficient transformation of Lactococcus lactis IL1403 and generation of knock-out mutants by homologous recombination. J. Basic Microbiol. 47:281-286. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Perez, M. M., P. van Dillewijn, R. M. Wittich, and J. L. Ramos. 2007. Escherichia coli has multiple enzymes that attack TNT and release nitrogen for growth. Environ. Microbiol. 9:1535-1540. [DOI] [PubMed] [Google Scholar]
- 11.Guillot, A., C. Gitton, P. Anglade, and M. Y. Mistou. 2003. Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics 3:337-354. [DOI] [PubMed] [Google Scholar]
- 12.Haynes, C. A., R. L. Koder, A. F. Miller, and D. W. Rodgers. 2002. Structures of nitroreductase in three states: effects of inhibitor binding and reduction. J. Biol. Chem. 277:11513-11520. [DOI] [PubMed] [Google Scholar]
- 13.Holm, L., and C. Sander. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsburgh, M. J., S. J. Wharton, A. G. Cox, E. Ingham, S. Peacock, and S. J. Foster. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44:1269-1286. [DOI] [PubMed] [Google Scholar]
- 15.Kabsch, W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, H. J., J. M. Kim, J. H. Park, J. J. Sung, M. Kim, and K. W. Lee. 2005. Pyruvate protects motor neurons expressing mutant superoxide dismutase 1 against copper toxicity. Neuroreport 16:585-589. [DOI] [PubMed] [Google Scholar]
- 17.Kobori, T., H. Sasaki, W. C. Lee, S. Zenno, K. Saigo, M. E. Murphy, and M. Tanokura. 2001. Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution. J. Biol. Chem. 276:2816-2823. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. J. Biol. Chem. 80:575-599. [DOI] [PubMed] [Google Scholar]
- 19.Lei, B., and S. C. Tu. 1998. Mechanism of reduced flavin transfer from Vibrio harveyi NADPH-FMN oxidoreductase to luciferase. Biochemistry 37:14623-14629. [DOI] [PubMed] [Google Scholar]
- 20.Linder, M. C., and M. Hazegh Azam. 1996. Copper biochemistry and molecular biology. Am. J. Clin. Nutr. 63:797S-811S. [DOI] [PubMed] [Google Scholar]
- 21.Liochev, S. I., A. Hausladen, and I. Fridovich. 1999. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:3537-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macomber, L., and J. A. Imlay. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106:8344-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnani, D., O. Barré, S. D. Gerber, and M. Solioz. 2008. Characterization of the CopR regulon of Lactococcus lactis IL1403. J. Bacteriol. 190:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marty-Teysset, C., F. de la Torre, and J. Garel. 2000. Increased production of hydrogen peroxide by Lactobacillus delbrueckii subsp. bulgaricus upon aeration: involvement of an NADH oxidase in oxidative stress. Appl. Environ. Microbiol. 66:262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNeish, I. A., N. K. Green, M. G. Gilligan, M. J. Ford, V. Mautner, L. S. Young, D. J. Kerr, and P. F. Searle. 1998. Virus directed enzyme prodrug therapy for ovarian and pancreatic cancer using retrovirally delivered E. coli nitroreductase and CB1954. Gene Ther. 5:1061-1069. [DOI] [PubMed] [Google Scholar]
- 27.Morokutti, A., A. Lyskowski, S. Sollner, E. Pointner, T. B. Fitzpatrick, C. Kratky, K. Gruber, and P. Macheroux. 2005. Structure and function of YcnD from Bacillus subtilis, a flavin-containing oxidoreductase. Biochemistry 44:13724-13733. [DOI] [PubMed] [Google Scholar]
- 28.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 29.Mukhopadhyay, P., M. Zheng, L. A. Bedzyk, R. A. LaRossa, and G. Storz. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. U. S. A. 101:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odermatt, A., and M. Solioz. 1995. Two trans-acting metalloregulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. J. Biol. Chem. 270:4349-4354. [DOI] [PubMed] [Google Scholar]
- 31.Portmann, R., D. Magnani, J. V. Stoyanov, A. Schmechel, G. Multhaup, and M. Solioz. 2004. Interaction kinetics of the copper-responsive CopY repressor with the cop promoter of Enterococcus hirae. J. Biol. Inorg. Chem. 9:396-402. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 34.Solioz, M., and J. V. Stoyanov. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27:183-195. [DOI] [PubMed] [Google Scholar]
- 35.Stadtman, E. R., B. S. Berlett, and P. B. Chock. 1990. Manganese-dependent disproportionation of hydrogen peroxide in bicarbonate buffer. Proc. Natl. Acad. Sci. U. S. A. 87:384-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strausak, D., and M. Solioz. 1997. CopY is a copper-inducible repressor of the Enterococcus hirae copper ATPases. J. Biol. Chem. 272:8932-8936. [DOI] [PubMed] [Google Scholar]
- 37.Streker, K., C. Freiberg, H. Labischinski, J. Hacker, and K. Ohlsen. 2005. Staphylococcus aureus NfrA (SA0367) is a flavin mononucleotide-dependent NADPH oxidase involved in oxidative stress response. J. Bacteriol. 187:2249-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda, K., M. Iizuka, T. Watanabe, J. Nakagawa, S. Kawasaki, and Y. Niimura. 2007. Synechocystis DrgA protein functioning as nitroreductase and ferric reductase is capable of catalyzing the Fenton reaction. FEBS J. 274:1318-1327. [DOI] [PubMed] [Google Scholar]
- 39.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tikhonov, K. G., O. M. Zastrizhnaya, Y. N. Kozlov, and V. V. Klimov. 2006. Composition and catalase-like activity of Mn(II)-bicarbonate complexes. Biochemistry (Mosc.) 71:1270-1277. [DOI] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urbanski, N. K., and A. Beresewicz. 2000. Generation of *OH initiated by interaction of Fe2+ and Cu+ with dioxygen; comparison with the Fenton chemistry. Acta Biochim. Pol. 47:951-962. [PubMed] [Google Scholar]
- 43.Uriu-Adams, J. Y., and C. L. Keen. 2005. Copper, oxidative stress, and human health. Mol. Aspects Med. 26:268-298. [DOI] [PubMed] [Google Scholar]
- 44.van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 45.van Niel, E. W., K. Hofvendahl, and B. Hahn-Hagerdal. 2002. Formation and conversion of oxygen metabolites by Lactococcus lactis subsp. lactis ATCC 19435 under different growth conditions. Appl. Environ. Microbiol. 68:4350-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, X. F., and M. S. Cynader. 2001. Pyruvate released by astrocytes protects neurons from copper-catalyzed cysteine neurotoxicity. J. Neurosci. 21:3322-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, Y., J. F. Wu, H. Liu, S. J. Wang, S. J. Liu, and N. Y. Zhou. 2006. Characterization of genes involved in the initial reactions of 4-chloronitrobenzene degradation in Pseudomonas putida ZWL73. Appl. Microbiol. Biotechnol. 73:166-171. [DOI] [PubMed] [Google Scholar]
- 48.Zenno, S., H. Koike, A. N. Kumar, R. Jayaraman, M. Tanokura, and K. Saigo. 1996. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J. Bacteriol. 178:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zenno, S., and K. Saigo. 1994. Identification of the genes encoding NAD(P)H-flavin oxidoreductases that are similar in sequence to Escherichia coli Fre in four species of luminous bacteria: Photorhabdus luminescens, Vibrio fischeri, Vibrio harveyi, and Vibrio orientalis. J. Bacteriol. 176:3544-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]