Abstract
Clostridium difficile is a spore-forming bacterium that causes Clostridium difficile-associated disease (CDAD). Intestinal microflora keeps C. difficile in the spore state and prevents colonization. Following antimicrobial treatment, the microflora is disrupted, and C. difficile spores germinate in the intestines. The resulting vegetative cells are believed to fill empty niches left by the depleted microbial community and establish infection. Thus, germination of C. difficile spores is the first required step in CDAD. Interestingly, C. difficile genes encode most known spore-specific protein necessary for germination, except for germination (Ger) receptors. Even though C. difficile Ger receptors have not been identified, taurocholate (a bile salt) and glycine (an amino acid) have been shown to be required for spore germination. Furthermore, chenodeoxycholate, another bile salt, can inhibit taurocholate-induced C. difficile spore germination. In the present study, we examined C. difficile spore germination kinetics to determine whether taurocholate acts as a specific germinant that activates unknown germination receptors or acts nonspecifically by disrupting spores' membranes. Kinetic analysis of C. difficile spore germination suggested the presence of distinct receptors for taurocholate and glycine. Furthermore, taurocholate, glycine, and chenodeoxycholate seem to bind to C. difficile spores through a complex mechanism, where both receptor homo- and heterocomplexes are formed. The kinetic data also point to an ordered sequential progression of binding where taurocholate must be recognized first before detection of glycine can take place. Finally, comparing calculated kinetic parameters with intestinal concentrations of the two germinants suggests a mechanism for the preferential germination of C. difficile spores in antibiotic-treated individuals.
Clostridium difficile is a Gram-positive, rod-shaped, spore-forming, obligately anaerobic bacterium. Under stress conditions, vegetative C. difficile cells differentiate into infectious spores (26). Like other bacilli and clostridia, C. difficile spores are metabolically inactive and resistant to most environmental insults. Spores can revert to toxin-producing bacteria (a process called germination) in nutrient-rich environments, such as the mammalian host (25, 33).
C. difficile spores are carried asymptomatically by up to 5% of healthy human adults (15). In the absence of antibiotics, normal intestinal microflora interferes with colonization, and C. difficile remains in its quiescent spore state (8, 31). Following antimicrobial treatment, however, the normal bacterial flora is disrupted, and it is believed that C. difficile spores germinate in the intestines. The resulting vegetative cells fill empty niches in the depleted microbial community, where they multiply and produce toxins (12).
Germination of C. difficile spores in the gastrointestinal (GI) tract of immunocompromised patients is required for C. difficile-associated disease (CDAD), a condition responsible for approximately 25% of all cases of antibiotic-associated diarrhea (39). Prompt empirical antimicrobial treatment is mandatory for cancer patients with severe neutropenia caused by antineoplastic treatment (22) and in the postoperative period after organ transplantation (5). Unfortunately, these antimicrobial regimes also favor colonization by C. difficile (41). Furthermore, C. difficile is the most common identifiable cause of diarrhea in HIV patients (7). Thus, CDAD is a major complication that increases morbidity and mortality in cancer chemotherapy, transplant, and AIDS patients.
Even though germination of C. difficile spores is the first required step in CDAD establishment, little is known about this process. The mechanisms of spore germination have been studied mainly in bacilli. In these cases, the germination process is commonly triggered by the initial detection of low-molecular-weight germinants by proteinaceous germination (Ger) receptors (25, 33). Ger receptors generally consist of three membrane-bound proteins encoded by tricistronic operons (16, 19). Each Ger receptor recognizes a cognate germinant, such as amino acids, nucleosides, sugars, or salts (25, 33).
Proteins involved in germination are remarkably conserved in both bacilli and clostridia. Basic Local Alignment Search Tools (BLAST) searches of spore-specific proteins reveal analogs in all sequenced sporulating bacteria. Interestingly, C. difficile genes encode analogs for most spore-specific proteins, except for Ger receptors and coat proteins (32). Since C. difficile spores must germinate, C. difficile germination receptors may be too divergent from other sporulating bacteria. Alternatively, C. difficile spores may use a different protein set to detect its germinants (32). Indeed, a novel germination mechanism that does not require Ger receptors has been recently discovered (34). In this case, Bacillus subtilis spores use a serine/threonine protein kinase (PrkC) instead of Ger receptors to germinate in the presence of peptidoglycans (34). A search of genome sequences reveals the presence of a putative prkC homologue in C. difficile (28). Whether PrkC-like proteins contribute to the germination of C. difficile spores has not been determined.
Although the mechanism of C. difficile spore germination has not been elucidated, previous studies have shown that addition of bile salts increases the recovery of vegetative cells from C. difficile spores (21, 44). Furthermore, a recent article showed that C. difficile spores recognize glycine (an amino acid) and taurocholate (a bile salt) as germinants (36). Furthermore, another bile salt, chenodeoxycholate, was shown to inhibit taurocholate-induced C. difficile spore germination (37). Neither glycine nor taurocholate has been previously described as germinants for spores of bacilli or clostridia.
Taurocholate could induce germination by two alternate mechanisms. Since bile salts are involved in the emulsification of fats (9), taurocholate could trigger germination by nonspecifically permeabilizing spore membranes. This type of germination mechanism has been previously described for nisin-type antibiotics (23). On the other hand, taurocholate could be specifically recognized as a germinant by unknown receptors. Activation of receptors by small molecules is the most common germination mechanism utilized by Bacillus and Clostridium spores (16, 19, 27).
Transposon mutagenesis has been used in Bacillus species to identify germination receptors and their cognate ligands (6, 11). However, access to C. difficile mutants has been impeded by the lack of appropriate genetic tools (24). Conjugative transposons Tn916 and Tn5397 have been used to mutagenize C. difficile. Unfortunately, Tn916 tends to integrate multiple copies of the transposon in every bacterium genome (20). Furthermore, Tn5397 nonrandomly targets specific sites in the C. difficile genome (42).
More recently, a mariner-based transposon system has been reported for the in vivo mutagenesis of C. difficile (10). This new system could allow single transposon insertion at random genomic loci. However, phenotypic screening of a C. difficile random mutant library yielded a single sporulation/germination defective clone and a single pyrimidine auxotroph (10).
As an alternative to genetic approaches, we have used kinetic methods and molecular probes to study Bacillus and Clostridium spore germination (1, 3, 14, 30). These techniques allow proposing possible mechanisms for spore germination even when the identity of the germination receptors is not known (30).
In the present study, we examined C. difficile spore germination kinetics to distinguish whether taurocholate activates unknown germination receptors or causes spore membrane disruption. Kinetic analysis was consistent with a receptor-mediated mechanism for C. difficile spore germination. The kinetic data obtained can be best fitted to a complex ordered sequential mechanism where taurocholate must be recognized first, before binding of glycine can take place. Finally, comparing calculated kinetic parameters with intestinal concentrations of taurocholate and glycine suggested a mechanism for the difference in germination profile of C. difficile spores in healthy and antibiotic-treated individuals.
MATERIALS AND METHODS
Materials.
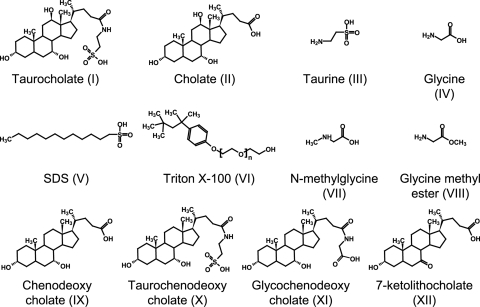
HistoDenz, taurocholate analogs, and glycine analogs (Fig. 1) were purchased from Sigma-Aldrich Corporation (St. Louis, MO). C. difficile strain 630 and strain VPI 10463 were obtained from the American Type Culture Collection (ATCC).
FIG. 1.
Compounds tested as agonists and antagonists of C. difficile spore germination. Taurocholate (compound I), cholate (compound II), taurine (compound III), glycine (compound IV), sodium dodecyl sulfate (SDS; compound V), Triton X-100 (compound VI), N-methylglycine (compound VII), glycine methyl ester (compound VIII), chenodeoxycholate (compound IX), taurochenodeoxycholate (compound X), glycochenodeoxycholate (compound XI), and 7-ketolithocholate (compound XII).
Bacterial strains and spore preparations.
C. difficile strains were individually plated onto brain heart infusion (BHI) agar supplemented with 1% yeast extract, 0.1% l-cysteine HCl, and 0.05% sodium taurocholate to yield single cell clones. Individual C. difficile colonies were grown in BHI broth and replated to obtain bacterial lawns. Plates were incubated for 5 days at 37°C in an anaerobic environment (5% CO2, 10% H2, and 85% N2). The resulting bacterial lawns were collected by flooding with ice-cold deionized water. Spores were pelleted by centrifugation and resuspended in fresh deionized water. After two washing steps, spores were separated from vegetative and partially sporulated cells by centrifugation through a 20% to 50% HistoDenz gradient (1). The spore pellet was washed five times with water, resuspended in sodium thioglycolate (0.5 g/liter) and stored at 4°C. Spores in all preparations were more than 95% pure as determined by microscopy observation of Schaeffer-Fulton-stained aliquots.
Kinetics of C. difficile spore germination.
Changes in light diffraction during spore germination (optical density at 580 nm [OD580]) were monitored using a Tecan Infinite M200 96-well plate reader set at 580 nm (Tecan Group, Männedorf, Switzerland). C. difficile spores were heat activated at 65°C for 30 min (13). After heat activation, spores were cooled to room temperature and resuspended in germination buffer (100 mM sodium phosphate buffer, pH 6.0, 0.5% NaHCO3) to an OD580 of 1. The spore suspension was monitored for autogermination for 30 min. Germination experiments were carried out with spores that did not autogerminate. Experiments were performed in triplicate on at least two different occasions with no fewer than two different spore preparations. All germination experiments were carried out in 96-well plates at a 200 μl/well final volume. Spores were allowed to germinate upon exposure to various concentrations of taurocholate (compound I [Fig. 1]) and glycine (compound IV). For taurocholate titrations, spores were exposed to various concentrations of taurocholate (8, 10, 14, 20, and 30 mM) at specific constant glycine concentrations (8, 9, 10, 13, and 16 mM). For glycine titrations, spores were exposed to various concentrations of glycine (8, 10, 12, 14, and 18 mM) at specific constant taurocholate concentrations (6, 7, 9, 12, and 15 mM). The concentration ranges for both cogerminants were selected to avoid data clusters in double reciprocal plots. Spore germination rates were evaluated based on the decrease in OD580. Relative OD580 values were derived by dividing each OD580 reading obtained at different times by the OD580 obtained at the beginning of germination. All measurements showed standard deviations of less than 10%. Germination rates (v) were determined for the various combinations of germinants used. Germination rates were calculated from the initial linear region of the germination curves (3). The resulting data were plotted as double reciprocal plots of 1/v versus 1/[variable germinant] or as squared double reciprocal plots of 1/v versus 1/[variable germinant]2. Maximum rate of germination (Vmax) parameters were determined from the intercept with the y axis of the best-fitted lines of the squared double reciprocal plots. Similarly, the intercept of the fitted lines with the x axis allowed the determination of apparent dissociation constants (K′). The data obtained were also plotted as Hill plots to obtain Hill numbers (n) and apparent dissociation constants (K′) at saturating concentrations of the cogerminant. All plots were fitted using the linear regression analysis from the SigmaPlot v.9 software.
Inhibition analysis of C. difficile spore germination.
Spore aliquots were treated with saturating concentrations (20 mM) of glycine (compound IV) and individually supplemented with various concentrations (0.2, 0.4, 0.5, 0.6, and 0.8 mM) of chenodeoxycholate (compound IX). Spore suspensions were incubated for 15 min at room temperature while monitoring OD580. Taurocholate (compound I) was then added to 4, 6, 8, or 12 mM final concentration. Germination rates (v) were determined as described above. The resulting data were plotted as double reciprocal plots of 1/v versus 1/[taurocholate]2 at different chenodeoxycholate concentrations and as Dixon plots of 1/v versus [chenodeoxycholate]2 at different taurocholate concentrations. The various x-axis intercepts from the Dixon plots were further plotted against taurocholate concentrations to determine the apparent inhibition constant (Ki).
Testing of bile salts as activators and inhibitors of C. difficile spore germination.
To test for agonist of germination, C. difficile spores were incubated with glycine (12 mM final concentration) for 10 min. The resulting solutions were individually treated with taurocholate (compound I), chenodeoxycholate (compound IX), taurochenodeoxycholate (compound X), glycochenodeoxycholate (compound XI), and 7-keto-lithocholate (compound XI) to a 6 mM final concentration. Germination was monitored by the decrease in optical density, as described above. To test for inhibitors of germination, C. difficile spores were individually incubated for 10 min with glycine (12 mM final concentration) and either chenodeoxycholate (compound IX), taurochenodeoxycholate (compound X), glycochenodeoxycholate (compound XI), or 7-keto-lithocholate (compound XII). The resulting suspensions were treated with 6 mM taurocholate (compound I), and germination was monitored by the decrease in optical density, as described above.
Detergent effect for C. difficile spore germination.
C. difficile spores were treated with 12 mM glycine (compound IV) and individually supplemented with taurocholate (compound I), sodium dodecyl sulfate (compound V), or Triton X-100 (compound VI) to a 6 mM final concentration. Germination was monitored by the decrease in optical density, as described above.
C. difficile spore germination with taurocholate and glycine analogs.
To determine whether the components of taurocholate could activate germination, C. difficile spores were incubated with 12 mM glycine (compound IV) for 10 min. The resulting solutions were individually treated with cholate (compound II), taurine (compound III), or equimolar amounts of cholate and taurine to a 6 mM final concentration. Germination was monitored by the decrease in optical density, as described above.
To determine specific interactions with the glycine moiety, C. difficile spores were incubated with 6 mM taurocholate (compound I) for 10 min. To these solutions was added glycine (compound IV), N-methylglycine (compound VII), or glycine methyl ester (compound VIII) to a 12 mM final concentration, and germination was monitored by the decrease in optical density, as described above.
RESULTS
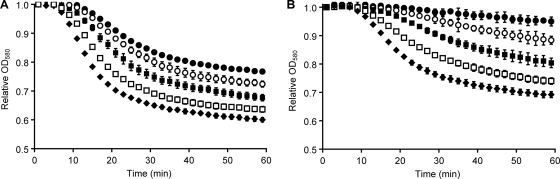
Taurocholate (compound I) is a bile salt formed by the condensation of cholate (compound II) and taurine (compound III). In the human lower intestine, taurocholate is deconjugated back to cholate and taurine (35). As reported previously (36), mixtures of taurocholate (compound I) and glycine (compound IV) were able to induce C. difficile spore germination (Fig. 2A and B). Similarly, chenodeoxycholate (compound IX) was able to inhibit taurocholate-mediated germination (37). Taurochenodeoxycholate (compound X), glycochenodeoxycholate (compound XI), and 7-keto-lithocholate (compound XII) were unable to induce or inhibit C. difficile spore germination (data not shown).
FIG. 2.
C. difficile spore germination in the presence of taurocholate and glycine mixtures. (A) C. difficile spores were incubated with 8 mM taurocholate and titrated with 8 (•), 9 (○), 10 (▪), 13 (□), and 16 (⧫) mM glycine concentrations. (B) C. difficile spores were incubated with 8 mM glycine and titrated with 6 (•), 7 (○), 9 (▪), 12 (□), and 15 (⧫) mM taurocholate concentrations.
Even though taurocholate/glycine mixtures were efficient germinants, replacing taurocholate (compound I) with cholate (compound II), taurine (compound III), or cholate/taurine combinations failed to efficiently induce germination of glycine-treated C. difficile spores in the time frame of the experiments (data not shown). Similarly, when taurocholate (compound I) was replaced with SDS (compound V) or Triton X-100 (compound VI), no germination was observed for glycine-treated C. difficile spores (data not shown).
Glycine (compound IV), the other cogerminant of C. difficile spores, is a common amino acid with a hydrogen side chain. Methylation of the amino (N-methylglycine [compound VII]) or carboxy group (glycine methyl ester [compound VIII]) resulted in glycine analogs unable to effectively trigger germination of taurocholate-treated C. difficile spores (data not shown).
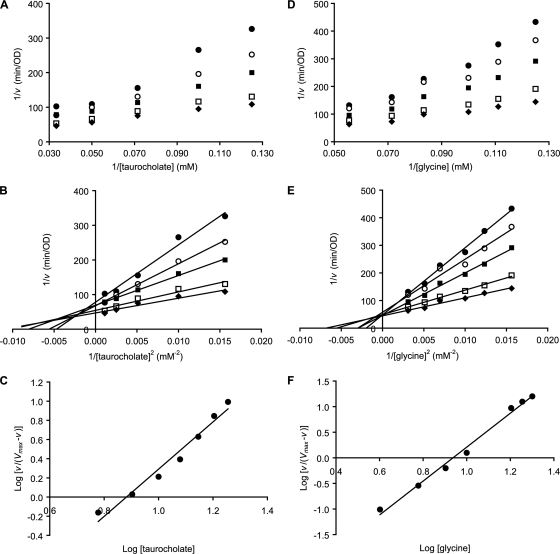
Kinetic analysis of C. difficile strain 630 spore germination with taurocholate (compound I) and glycine (compound IV) showed saturation kinetics. However, double reciprocal Lineweaver-Burk plots showed strong deviation from linearity for both taurocholate and glycine binding (Fig. 3A). Although the plots of 1/v versus 1/[taurocholate] were curved, the plots of 1/v versus 1/[taurocholate]2 were linear (Fig. 3B). Similarly, Hill plots for taurocholate yielded a Hill number (n) of 2.8 (Fig. 3C). Plotting 1/v versus 1/[taurocholate]3 resulted in parabolic graphs (data not shown).
FIG. 3.
Kinetics of Clostridium difficile spore germination in the presence of taurocholate and glycine. Germination rates were calculated from the linear segment of optical density changes over time. (A) Lineweaver-Burk plots of C. difficile spore germination at various concentrations of taurocholate (8, 10, 14, 20, and 30 mM) and 8 (•), 9 (○), 10 (▪), 13 (□), and 16 (⧫) mM glycine concentrations. (B) Squared Lineweaver-Burk plots of C. difficile spore germination at various concentrations of taurocholate (8, 10, 14, 20, and 30 mM) and 8 (•), 9 (○), 10 (▪), 13 (□), and 16 (⧫) mM glycine concentrations. (C) Hill plot for taurocholate binding at the saturating glycine concentration. (D) Lineweaver-Burk plots of C. difficile spore germination at various concentrations of glycine (8, 10, 12, 14, and 18 mM) and 6 (•), 7 (○), 9 (▪), 12 (□), and 15 (⧫) mM taurocholate concentrations. (E) Squared Lineweaver-Burk plots of C. difficile spore germination at various concentrations of glycine (8, 10, 12, 14, and 18 mM) and 6 (•), 7 (○), 9 (▪), 12 (□), and 15 (⧫) mM taurocholate concentrations. (F) Hill plot for glycine binding at the saturating taurocholate concentration.
The complexity of taurocholate binding to C. difficile spores prevented the calculation of the Michaelis-Menten constant (Km). Instead, intercepts of the x axis by the squared double reciprocal plots allowed calculating apparent dissociation constants (K′) that contained both Km and interaction factors for cooperative binding. For example, for two identical interacting binding sites, K′ = aKm2. Furthermore, the intercept of the Hill plot with the x axis allowed calculating K′ at saturating glycine concentrations (Fig. 3C). The apparent dissociation constant (K′) under this limiting condition was determined to be 7.7 mM2 for taurocholate binding.
The squared Lineweaver-Burk plots converged in the upper left quadrant (Fig. 3B). Thus, each plot intersected the y and x axes at different points. This shows that the maximum germination rate (Vmax) and the spores' affinity for taurocholate (K′) increased with increasing glycine concentrations. The K′ for taurocholate calculated at the highest and lowest glycine concentration tested was extrapolated to 82 and 180 mM2, respectively. This represents approximately a 2-fold increase in the affinity of spores for taurocholate over a 2-fold glycine concentration range.
Similar to taurocholate binding, the plots of 1/v versus 1/[glycine] were curved (Fig. 3D) and the plots of 1/v versus 1/[glycine]2 were linear (Fig. 3E). A Hill plot for glycine yielded a Hill number (n) of 3.3 (Fig. 3F). The apparent dissociation constant for glycine (K′) at saturating taurocholate concentrations was calculated to be 8.7 mM2.
Squared double reciprocal plot analysis of the effect of glycine on spore germination rate yielded a family of plots that converged on the y axis and intersected the x axis at different points (Fig. 3E). This shows that the maximum germination rate (Vmax) does not change with changing taurocholate concentrations. However, taurocholate does affect the affinity of spores for glycine. The K′ for glycine calculated at the highest and lowest taurocholate concentrations tested was extrapolated to 152 and 575 mM2, respectively. This represents an almost 4-fold increase in the affinity of spores for glycine over a 3-fold taurocholate concentration range.
Due to the complex response seen in the germination of C. difficile 630 spores, C. difficile VPI 10463 spores were also treated with combinations of taurocholate and glycine to assess possible interstrain variability. Similar to C. difficile 630 spores, germination of C. difficile VPI 10463 spores showed both cooperative behavior for individual germinants and synergy between taurocholate and glycine. Furthermore, the values for apparent dissociation constants (K′), maximum germination rates (Vmax), and Hill numbers (n) did not show significant variations between the two strains (Table 1).
TABLE 1.
Kinetic parameters for C. difficile spore germination
| Strain | Value for: |
|||||
|---|---|---|---|---|---|---|
| Taurocholate |
Glycine |
|||||
| Hill no. (n) | K′ (mM2)a | Vmax (OD/h)b | Hill no. (n) | K′ (mM2) | Vmax (OD/h) | |
| 630 | 2.8 (0.4) | 7.7 (1.5) | 1.7 (0.2) | 3.3 (0.9) | 8.7 (0.9) | 1.3 (0.2) |
| VPI 10463 | 2.3 (0.3) | 8.1 (0.6) | 1.8 (0.5) | 2.4 (0.5) | 7.5 (0.3) | 1.6 (0.3) |
K′ was determined at the saturating concentration of the second cogerminant.
Vmax was determined at the saturating concentration of both cogerminants.
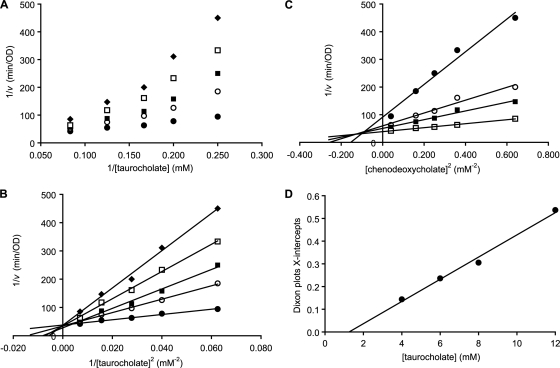
Chenodeoxycholate (compound IX) has been shown to inhibit taurocholate/glycine-mediated germination of C. difficile spores (37). As with taurocholate and glycine, Lineweaver-Burk plots of 1/v versus 1/[taurocholate] at different chenodeoxycholate concentrations were nonlinear (Fig. 4A). In contrast, double reciprocal inhibition plots of 1/v versus 1/[taurocholate]2 at different chenodeoxycholate concentrations were linear and converged on the y axis (Fig. 4B). This shows that chenodeoxycholate does not affect the maximum germination rate (Vmax), but affects taurocholate binding affinity (K′).
FIG. 4.
Kinetic analysis of chenodeoxycholate inhibition of C. difficile spore germination. Germination rates were calculated from the linear segment of optical density changes over time. (A) Lineweaver-Burk plots of C. difficile spore germination at various concentrations of taurocholate (4, 6, 8, and 12 mM) and 0.2 (•), 0.4 (○), 0.5 (▪), 0.6 (□), and 0.8 mM (⧫) chenodeoxycholate concentrations. (B) Squared Lineweaver-Burk plots of C. difficile spore germination at various concentrations of taurocholate (4, 6, 8, and 12 mM) and 0.2 (•), 0.4 (○), 0.5 (▪), 0.6 (□), and 0.8 mM (⧫) chenodeoxycholate concentrations. (C) Dixon plot of C. difficile spore germination at various concentrations of chenodeoxycholate (0.2, 0.4, 0.5, 0.6, and 0.8 mM) and 4 (•), 6 (○), 8 (▪), and 12 (□) mM taurocholate. (D). Replot of Dixon plot x intercepts versus taurocholate concentrations was fitted to a straight line and yielded a Ki of 1.3 mM2 for chenodeoxycholate binding.
Dixon plots of 1/v versus [chenodeoxycholate] were curved, but plots of 1/v versus [chenodeoxycholate]2 were linear and converged in the upper left quadrant (Fig. 4C). Thus, Dixon plots intercepted the x axis at different values, showing that the taurocholate concentration affected the affinity of spores for chenodeoxycholate. Replotting of the variable x-axis intercepts versus taurocholate concentrations allowed calculating an inhibition constant (Ki) of 1.3 mM2 for chenodeoxycholate (Fig. 4D). Similarly, chenodeoxycholate was tested for the germination inhibition of C. difficile strain VPI 10463 spores. The Ki for this strain was determined to be 0.9 mM2.
DISCUSSION
The C. difficile genome does not encode any known Ger receptors (32). However, C. difficile spores must be able to germinate to complete their life cycle. Recent reports have implicated glycine and taurocholate as cogerminants for C. difficile spore germination (36, 43). Since other bacilli and clostridia have shown interstrain variability in germination response (4, 19, 27, 29), we tested two different C. difficile strains (630 and VPI 10463) to determine qualitative and quantitative differences in their germination profiles. These strains were selected for analysis because they are used as reference strains. C. difficile strain 630 is a virulent and multidrug-resistant strain, and its genome has been sequenced (32). C. difficile strain VPI 10463 is generally used as a reference toxigenic strain (38). Both C. difficile strains showed similar germination kinetics with combinations of taurocholate (compound I) and glycine (compound IV) (Table 1). The correspondence in the germination profiles of both C. difficile strains 630 and VPI 10463 suggests that the germination mechanism is conserved across this species.
Taurocholate is a bile salt involved in the emulsification of fats (9). Thus, taurocholate could trigger germination by either permeabilizing the spore membranes or activating a cognate receptor. Kinetic analysis provides evidence that C. difficile spores possess unknown taurocholate and glycine germination receptors. First, glycine is a common amino acid with no known membrane-disrupting activity. Hence, the function of glycine as a cogerminant suggests the presence of a glycine-specific germination receptor. The specificity of glycine recognition is further highlighted by the inability of the glycine analogs N-methylglycine (compound VII) and glycine methyl ester (compound VIII) to substitute as coactivators of spore germination. Similarly, the inability of cholate (compound II) and taurine (compound III), alone or in combination, to efficiently induce germination points to specific interactions for taurocholate recognition.
The presence of germination receptors is consistent with the parabolic saturation kinetics observed for both taurocholate and glycine during C. difficile spore germination. This is characteristic of systems where specific binding steps are required for receptor activation and has been observed for Ger receptor-mediated germination in other sporulating bacteria (1, 3). Kinetic analysis of C. difficile spore germination also pointed to complex interactions in the recognition of both germinants. Lineweaver-Burk and Hill plots suggested that there are at least two interacting taurocholate-binding sites. Similarly, there seems to be at least two glycine-interacting binding sites. These results are consistent with putative taurocholate and glycine receptor homocomplexes that are activated cooperatively. In these cases, binding of a first taurocholate (or glycine) molecule increases the affinity for a second taurocholate (or glycine) molecule at an identical but separate binding site. We have seen similar cooperative effects for the binding of inosine by Bacillus cereus spores and the binding of l-arginine and l-phenylalanine by Clostridium sordellii spores (1, 30).
Aside from cooperative interactions for individual germinants, we also detected synergy between glycine and taurocholate binding. In these cases, the affinity of either germinant increases when the complementary germinant is bound at its specific site. The change in affinity for one germinant in the presence of the other is best explained by a receptor-mediated process where putative taurocholate and glycine receptors interact to form heterocomplexes and affect their respective binding abilities. We have detected similar synergy in the binding of inosine and l-alanine by Bacillus anthracis spores (3).
Lastly, a membrane disruptive mechanism would be nonspecific. Hence, detergents and other bile salts should be equally able to activate spore germination. However, SDS (compound V), Triton X-100 (compound VI), and chenodeoxycholate (compound IX) were unable to induce C. difficile spore germination. Instead, both Lineweaver-Burk inhibition and Dixon plots showed that chenodeoxycholate, a bile salt with potential detergent properties, is bound cooperatively and is a competitive inhibitor against taurocholate activation of C. difficile spore germination (37). These results suggest that chenodeoxycholate and taurocholate are recognized specifically by the same binding site. The specificity of taurocholate as a C. difficile germinant is further highlighted by the inability of taurochenodeoxycholate (compound X), glycochenodeoxycholate (compound XI), and 7-keto-lithocholate (compound XII) to activate or inhibit C. difficile spore germination.
In the absence of Ger receptors, C. difficile spores must use other proteins to recognize glycine and taurocholate. Recently, a serine/threonine kinase (PrkC) was shown to activate germination in B. subtilis in response to peptidoglycans (34). Although the C. difficile genome encodes a PrkC homologue, the molecular structures of peptidoglycans and bile salts are dissimilar. It would be interesting to determine if PrkC is active in C. difficile spores and if it is able to recognize bile salts instead of peptidoglycans as germination signals.
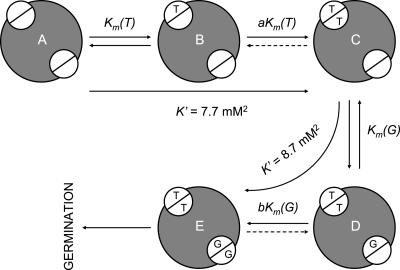
The maximum germination rate (Vmax) increases when taurocholate is titrated at increasing glycine concentrations (Fig. 3B). On the other hand, Vmax does not change when glycine is titrated at increasing taurocholate concentrations (Fig. 3E). These results are consistent with a sequential ordered mechanism for germinant binding (Fig. 5). In this model, C. difficile spore germination would require binding of both ligands. However, resting spores (Fig. 5A) could bind only taurocholate to form a taurocholate-spore complex (Fig. 5B). The cooperative behavior of taurocholate binding would rapidly saturate other taurocholate-binding sites (Fig. 5C). The taurocholate-activated spores would be the only spore form able to bind glycine (Fig. 5D). Cooperative binding of glycine would allow rapid saturation of glycine-binding sites (Fig. 5E). Once both germinants are bound, the spores would be competent to complete the germination process.
FIG. 5.
Proposed kinetic mechanism for C. difficile spore germination. Spores are represented by gray circles. Cooperative germination receptors are represented by white split circles. Solid arrows represent fast processes. Dashed arrows represent slow processes. Km(T) is the apparent Michaelis-Menten constant for taurocholate binding, and a is the interaction factor between taurocholate binding sites. Km(G) is the apparent Michaelis-Menten constant for glycine binding, and b is the interaction factor between glycine binding sites. Dormant spores (A) must first detect a taurocholate molecule to start the germination process (B). The cooperative behavior of taurocholate binding rapidly saturates other taurocholate binding sites (C). The resulting taurocholate-spore complex is then competent to bind glycine (D). Cooperative binding of glycine allows rapid saturation of glycine binding sites to form a taurocholate-spore-glycine complex (E) that is activated for germination.
The apparent dissociation constants (K′) that were obtained from the Michaelis-Menten analysis of germination kinetics include both the Michaelis-Menten constant (Km), which represents the concentration of a compound required to obtain half the maximal germination rate, and interaction factors that define cooperative interactions. In order for a spore to germinate, however, germination receptors must be activated by their corresponding germinants. Thus, K′ also represented the concentration of a germinant required for half of the spore population to be fully complexed. As a result, environmental concentrations below K′ will result in a larger fraction of the spore population remaining in an uncomplexed, dormant form. Correspondingly, environmental concentrations above K′ will result in mostly complexed, germinating spores (30). Consequently, kinetic analysis of C. difficile spore germination allows postulating a mechanism for spore activation by comparing the germinants' apparent dissociation constants (K′) with their available concentrations in the intestinal luminal space.
The concentration of taurocholate in intestinal fluids has been reported to be in the millimolar range in the upper intestine (40). In contrast, in the lower intestine, where C. difficile is postulated to germinate (15), taurocholate is deconjugated into taurine and cholate by the intestinal flora (35). Hence, in the lower intestine, the concentration of active taurocholate is negligible (18). Although more variable, the concentration of glycine in intestinal mucosa has also been reported to be in the millimolar range (2).
The concentration of chenodeoxycholate in the GI tract follows an inverse relation to taurocholate concentration. In the normal upper intralumenal space, chenodeoxycholate is undetectable (40). However, two inactive bile salts, taurochenodeoxycholate (compound X) and glycochenodeoxycholate (compound XI) are deconjugated in the lower intestine, increasing chenodeoxycholate concentration. Indeed, in the human cecum chenodeoxycholate is the major component of bile salts (approximately 120 μM), while taurocholate is present at approximately 1 μM (18). Thus, in the lower intestine, chenodeoxycholate can act as a natural inhibitor of C. difficile spore germination.
Using estimated metabolite concentration gradients in the digestive tract (18, 40) together with our kinetic analysis, we hypothesize that in healthy individuals, C. difficile spores will remain dormant in the lower intestine regardless of the glycine concentration. In contrast, strong antibiotic treatment will deplete the gut flora. As a result, taurocholate will not be deconjugated to cholate and taurine, and the concentration of taurocholate in the lower intestine will increase. Concomitantly, the concentration of inhibitory chenodeoxycholate will decrease. Under these conditions, we predict that C. difficile spores will first bind undegraded taurocholate in the lower intestinal lumen. Taurocholate-activated spores can then bind glycine. The taurocholate/glycine-bound spores will be activated and will be able to complete the germination process. Once germinated, the outgrowing C. difficile cells can further produce 7-α-hydroxysteroid dehydrogenase, which can oxidize chenodeoxycholate into inactive 7-keto-lithocholate (compound XII), further suppressing chenodeoxycholate action (32).
The model proposed here is mostly consistent with the model proposed by Sorg and Sonenshein (36, 37). We agree that the normal microflora probably keeps C. difficile spores from germinating by controlling bile salt homeostasis (36). We further concur on the potential role of chenodeoxycholate as a deterrent of C. difficile colonization (37). However, in our study, cholate was an inefficient germinant. As such, we do not expect that cholate will be able to efficiently activate spores as they pass through the GI tract. Thus, we expect taurocholate to be the main germination signal.
Although our model is based on correlating in vivo germinant concentrations with in vitro kinetic data, a recent published article further supports our mechanistic predictions (17). This work showed that intestinal and cecal bile salt mixtures from antibiotic-treated mice were able to stimulate colony formation from C. difficile spores to greater levels than from untreated animals. This is consistent with differential metabolism of bile salts influencing C. difficile spore germination (17). Nevertheless, more research is necessary to establish the complete feasibility of our model.
Footnotes
Published ahead of print on 18 June 2010.
REFERENCES
- 1.Abel-Santos, E., and T. Dodatko. 2007. Differential nucleoside recognition during Bacillus cereus 569 (ATCC 10876) spore germination. New J. Chem. 31:748-755. [Google Scholar]
- 2.Ahlman, B., C.-E. Leijonmarck, C. Lind, E. Vinnars, and J. Wernerman. 1993. Free amino acids in biopsy specimens from the human colonic mucosa. J. Surgic. Res. 55:647-653. [DOI] [PubMed] [Google Scholar]
- 3.Akoachere, M., R. C. Squires, A. M. Nour, L. Angelov, J. Brojatsch, and E. V. Abel-Santos. 2007. Identification of an in vivo inhibitor of Bacillus anthracis Sterne spore germination. J. Biol. Chem. 282:12112-12118. [DOI] [PubMed] [Google Scholar]
- 4.Alberto, F., V. Broussolle, D. R. Mason, F. Carlin, and M. W. Peck. 2003. Variability in spore germination response by strains of proteolytic Clostridium botulinum types A, B and F. Lett. Appl. Microbiol. 36:41-45. [DOI] [PubMed] [Google Scholar]
- 5.Arslan, H., E. K. Inci, O. K. Azap, H. Karakayali, A. Torgay, and M. Haberal. 2007. Etiologic agents of diarrhea in solid organ recipients. Transpl. Infect. Dis. 9:270-275. [DOI] [PubMed] [Google Scholar]
- 6.Barlass, P. J., C. W. Houston, M. O. Clements, and A. Moir. 2002. Germination of Bacillus cereus spores in response to l-alanine and to inosine: the roles of gerL and gerQ operons. Microbiology 148:2089-2095. [DOI] [PubMed] [Google Scholar]
- 7.Bartlett, J. G. 2007. Changing trends in bacterial infections: Staphylococcus aureus, bacterial pneumonia, Clostridium difficile. Top. HIV Med. 15:94-98. [PubMed] [Google Scholar]
- 8.Borriello, P. S. 1990. The influence of the normal flora on Clostridium difficile colonization of the gut. Ann. Med. 22:61-67. [DOI] [PubMed] [Google Scholar]
- 9.Carey, M. C., and D. M. Small. 1970. The characteristics of mixed micellar solutions with particular reference to bile. Am. J. Med. 49:590-608. [DOI] [PubMed] [Google Scholar]
- 10.Cartman, S. T., and N. P. Minton. 2010. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 76:1103-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clements, M. O., and A. Moir. 1998. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J. Bacteriol. 180:6729-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloud, J., and C. P. Kelly. 2007. Update on Clostridium difficile associated disease. Curr. Opin. Gastroenterol. 23:4-9. [DOI] [PubMed] [Google Scholar]
- 13.Desrosier, N. W., and F. Heiligman. 1956. Heat activation of bacterial spores. Food Res. 21:54-62. [Google Scholar]
- 14.Dodatko, T., M. Akoachere, N. Jimenez, Z. Alvarez, and E. Abel-Santos. 2010. Dissecting interactions between nucleosides and germination receptors in Bacillus cereus 569 spores. Microbiology 156:1244-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fekety, R., and A. B. Shah. 1993. Diagnosis and treatment of Clostridium difficile colitis. JAMA 269:71-75. [PubMed] [Google Scholar]
- 16.Fisher, N., and P. Hanna. 2005. Characterization of Bacillus anthracis germinant receptors in vitro. J. Bacteriol. 187:8055-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giel, J. L., J. A. Sorg, A. L. Sonenshein, and J. Zhu. 2010. Metabolism of bile salts in mice influences spore germination Clostridium difficile. PLoS One 5:e8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton, J. P., G. Xie, J.-P. Raufman, S. Hogan, T. L. Griffin, C. A. Packard, D. A. Chatfield, L. R. Hagey, J. H. Steinbach, and A. F. Hofmann. 2007. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 293:G256-G263. [DOI] [PubMed] [Google Scholar]
- 19.Hornstra, L. M., Y. P. de Vries, M. H. Wells-Bennik, W. M. de Vos, and T. Abee. 2006. Characterization of germination receptors of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 72:44-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain, H. A., A. P. Roberts, and P. Mullany. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. J. Med. Microbiol. 54:137-141. [DOI] [PubMed] [Google Scholar]
- 21.Kamiya, S., K. Yamakawa, H. Ogura, and S. Nakamura. 1987. Effect of various sodium taurocholate preparations on the recovery of Clostridium difficile spores. Microbiol. Immunol. 31:1117-1120. [DOI] [PubMed] [Google Scholar]
- 22.Maschmeyer, G., and A. Haas. 2008. The epidemiology and treatment of infections in cancer patients. Intern. J. Antimicrob. Agents 31:193-197. [DOI] [PubMed] [Google Scholar]
- 23.Mazzotta, A. S., and T. J. Montville. 1999. Characterization of fatty acid composition, spore germination, and thermal resistance in a nisin-resistant mutant of Clostridium botulinum 169B and in the wild-type strain. Appl. Environ. Microbiol. 65:659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minton, N., G. Carter, M. Herbert, T. O'Keeffe, D. Purdy, M. Elmore, A. Ostrowski, O. Pennington, and I. Davis. 2004. The development of Clostridium difficile genetic systems. Anaerobe 10:75-84. [DOI] [PubMed] [Google Scholar]
- 25.Moir, A., B. M. Corfe, and J. Behravan. 2002. Spore germination. Cell Mol. Life Sci. 59:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paredes, C. J., K. V. Alsaker, and E. T. Papoutsakis. 2005. A comparative genomic view of Clostridial sporulation and physiology. Nature Rev. Microbiol. 3:969-978. [DOI] [PubMed] [Google Scholar]
- 27.Paredes-Sabja, D., J. A. Torres, P. Setlow, and M. R. Sarker. 2008. Clostridium perfringens spore germination: characterization of germinants and their receptors. J. Bacteriol. 190:1190-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peck, M. W., and K. P. Robert. 2009. Biology and genomic analysis of Clostridium botulinum. Adv. Microb. Physiol. 55:183-265. [DOI] [PubMed] [Google Scholar]
- 29.Plowman, J., and M. W. Peck. 2002. Use of a novel method to characterize the response of spores of nonproteolytic Clostridium botulinum types B, E and F to a wide range of germinants and conditions. J. Appl. Microbiol. 92:681-694. [DOI] [PubMed] [Google Scholar]
- 30.Ramirez, N., and E. Abel-Santos. 2010. Requirements for germination of Clostridium sordellii spores in vitro. J. Bacteriol. 192:418-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolfe, R. D., S. Helebian, and S. M. Finegold. 1981. Bacterial interference between Clostridium difficile and normal fecal flora. J. Infect. Dis. 143:470-475. [DOI] [PubMed] [Google Scholar]
- 32.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 33.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 34.Shah, I. M., M.-H. Laaberki, D. L. Popham, and J. Dworkin. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimada, K., K. S. Bricknell, and S. M. Finegold. 1969. Deconjugation of bile acids by intestinal bacteria: review of literature and additional studies. J. Infect. Dis. 119:73-81. [DOI] [PubMed] [Google Scholar]
- 36.Sorg, J. A., and A. L. Sonenshein. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorg, J. A., and A. L. Sonenshein. 2009. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 191:1115-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stabler, R. A., L. F. Dawson, L. T. H. Phua, and B. W. Wren. 2008. Comparative analysis of BI/NAP1/027 hypervirulent strains reveals novel toxin B-encoding gene (tcdB) sequences. J. Med. Microbiol. 57:771-775. [DOI] [PubMed] [Google Scholar]
- 39.Surowiec, D., A. G. Kuyumjian, M. A. Wynd, and C. E. Cicogna. 2006. Past, present, and future therapies for Clostridium difficile-associated disease. Ann. Pharmacother. 40:2155-2163. [DOI] [PubMed] [Google Scholar]
- 40.Vertzoni, M., H. Archontaki, and C. Reppas. 2008. Determination of intralumenal individual bile acids by HPLC with charged aerosol detection. J. Lipid Res. 49:2690-2695. [DOI] [PubMed] [Google Scholar]
- 41.von Baum, H., A. Sigge, M. Bommer, W. V. Kern, R. Marre, H. Dohner, P. Kern, and S. Reuter. 2006. Moxifloxacin prophylaxis in neutropenic patients. J. Antimicrob. Chemother. 58:891-894. [DOI] [PubMed] [Google Scholar]
- 42.Wang, H., M. C. M. Smith, and P. Mullany. 2006. The conjugative transposon Tn5397 has a strong preference for integration into its Clostridium difficile target site. J. Bacteriol. 188:4871-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wheeldon, L. J., T. Worthington, A. C. Hilton, T. S. J. Elliott, and P. A. Lambert. 2008. Physical and chemical factors influencing the germination of Clostridium difficile spores. J. Appl. Microbiol. 105:2223-2230. [DOI] [PubMed] [Google Scholar]
- 44.Wilson, K. H. 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol. 18:1017-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]