Abstract
IS256 is the founding member of the IS256 family of insertion sequence (IS) elements. These elements encode a poorly characterized transposase, which features a conserved DDE catalytic motif and produces circular IS intermediates. Here, we characterized the IS256 transposase as a DNA-binding protein and obtained insight into the subdomain organization and functional properties of this prototype enzyme of IS256 family transposases. Recombinant forms of the transposase were shown to bind specifically to inverted repeats present in the IS256 noncoding regions. A DNA-binding domain was identified in the N-terminal part of the transposase, and a mutagenesis study targeting conserved amino acid residues in this region revealed a putative helix-turn-helix structure as a key element involved in DNA binding. Furthermore, we obtained evidence to suggest that the terminal nucleotides of IS256 are critically involved in IS circularization. Although small deletions at both ends reduced the formation of IS circles, changes at the left-hand IS256 terminus proved to be significantly more detrimental to circle production. Taken together, the data lead us to suggest that the IS256 transposase-mediated circularization reaction preferentially starts with a sequence-specific first-strand cleavage at the left-hand IS terminus.
IS256 is an insertion sequence widespread in the genomes of multiresistant enterococci and staphylococci (3). The element, which is 1,324 bp in size, consists of a single open reading frame encoding a transposase protein flanked by noncoding regions (NCRs) harboring imperfect inverted repeats (IRs) (see Fig. 1A). IS256 occurs in multiple free copies in its host genomes but is also known to form the ends of composite transposon Tn4001 conferring aminoglycoside resistance (29). In Staphylococcus epidermidis, IS256 has been identified as a typical marker of hospital-acquired multiresistant and biofilm-forming clones causing opportunistic infections in immunocompromised patients (11, 20-22, 26, 34). The element has been shown to trigger heterogeneous biofilm expression by reversible transposition into biofilm-associated genes and regulators (4, 5, 19, 49, 56). Also, IS256 has the capacity to influence antibiotic resistance, either by insertion into regulatory genes or by modulating antibiotic resistance gene expression through formation of strong hybrid promoters resulting from transposition into the neighborhood of antibiotic resistance genes (6, 18, 31, 32). Finally, multiple genomic IS256 copies may serve as crossover points for homologous recombination events and thereby play an important role in genome flexibility, adaptation, and evolution of staphylococcal and enterococcal genomes (29, 42, 55).
FIG. 1.
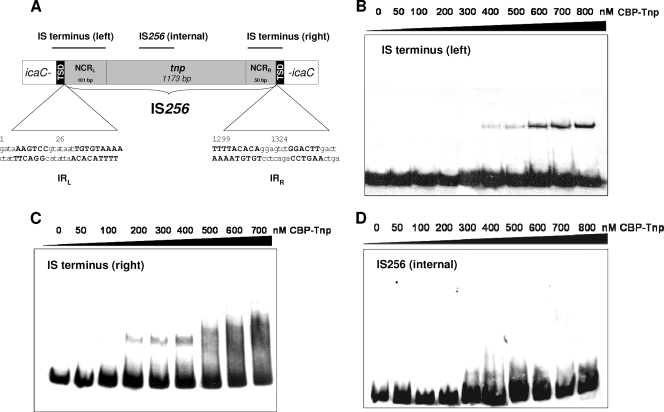
IS256 transposase binding to IS termini. (A) Genetic organization of IS256. The transposase gene (tnp) is flanked by NCRs that harbor imperfect IRs (IRL and IRR) at the ends of the element. The nucleotide sequence of the IRs is indicated by uppercase boldface letters, with nucleotide numbering referring to GenBank accession no. M18086. Insertion of IS256 into the S. epidermidis icaC gene on plasmid pIL2 (27) is shown, and black boxes mark the 8-bp target site duplications (TSDs) generated upon transposition of the element. Black bars at the top indicate localizations of DNA fragments used in the EMSAs presented in panels B to D. (B to D) EMSAs of purified IS256 transposase protein (CBP-Tnp) with various IS256-specific DNA fragments. A 15.5 nM concentration of an IS terminus (left)-carrying DNA fragment (B) or an IS terminus (right)-carrying DNA-fragment (C), as well as an interal IS256 fragment (D), were used with increasing amounts of protein. All experiments were performed in the presence of unspecific competitor [50 μg of poly(dI-dC) ml−1]. Molar ratios between DNA and protein comprised a range of 1:3 (50 nM CBP-Tnp) to 1:52 (800 nM CBP-Tnp).
Given its important biological role, it is surprising that very little is known about the molecular function of IS256 and its lifestyle. Empirical analyses of IS256 insertion sites in various bacterial genomes and loci did not reveal nucleotide sequence specificity for target site selection (3, 29, 56). Typically, IS256 generates 8- or 9-bp target site duplications (TSDs) upon transposition that are caused by staggered nicks of the target DNA and refill of the resulting gaps by the host repair system (43). In the course of phase variation events, IS256 TSDs can be completely removed, with the original host sequence being restored (56). Such precise IS256 excisions are caused by an illegitimate recombination event that requires fully intact TSDs but no functional IS256 transposase (14). IS256 transposition itself was found to involve the formation of double-stranded circular IS256 molecules in which the insertion sequence (IS) ends abut, bridged by a few base pairs of host DNA originating from the original insertion site (27, 39). IS256 circle formation is a strictly transposase-dependent process and IS circles are regarded as transposition intermediates which are likely to be relinearized during transposition. However, details of the transposition reaction, including circle formation, putative relinearization, target site selection, and insertion of the element are far from being understood at the molecular level. We experimentally addressed here, for the first time for a bacterial transposase of the IS256 family, the DNA-binding properties of this protein. We identified a DNA-binding domain in the N-terminal region of the protein. The domain contains a putative classical helix-turn-helix (HTH) motif that is demonstrated to be involved in sequence-specific interactions of the IS256 transposase with the IRs present in the NCRs of the element. Moreover, we suggest a role for the terminal nucleotides of the IS256 nucleotide sequence in first-strand cleavage and subsequent circularization of the element.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
Bacterial strains and plasmids used and generated in the present study are listed in Table 1. Primer oligonucleotide sequences and PCR conditions are summarized in Table S1 in the supplemental material.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Properties | Source or reference |
|---|---|---|
| Strains | ||
| E. coli BL21(DE3) | F−ompT hsdSB(rB− mB−) dcm gal λ(DE3) | Merck Biosciences |
| S. aureus RN4220 | 23 | |
| Plasmids | ||
| pIL2 | Wild-type IS256::icaC insertion cloned into Gram-positive/Gram-negative shuttle vector pRB473 | 27 |
| pIL2 IS256Δtnp | pIL2 carrying IS256Δtnp::icaC with an internal 20-bp deletion of the transposase gene | 27 |
| pIL2-NCR LΔ2 | pIL2 vectors carrying IS256 copies with 2- and 4-bp deletions at the left-hand (NCR L) and right-hand (NCR R) IS256 termini, respectively | This study |
| pIL2-NCR LΔ4 | ||
| pIL2-NCR RΔ2 | ||
| pIL2-NCR RΔ4 | ||
| pIL2-NCR LΔ2/NCR RΔ2 | ||
| pIL2-NCR LΔ2/NCR RΔ4 | ||
| pIL2-NCR LΔ4/NCR RΔ2 | ||
| pIL2-NCR LΔ4/NCR RΔ4 | ||
| pCAL-n-FLAG | Expression vector encoding the CBP and FLAG epitope; ori pBR322 blaZ | Stratagene |
| pCAL-n-FLAG-tnp | pCAL-n-FLAG expressing IS256 transposase | This study |
| pCAL-n-FLAG-tnp1-130 | pCAL-n-FLAG expressing the N terminus of the IS256 transposase (aa 1 to 130) | This study |
| pCAL-n-FLAG-tnp100-230 | pCAL-n-FLAG expressing an internal fragment of the IS256 transposase (aa 100 to 230) | This study |
| pCAL-n-FLAG-tnp200-390 | pCAL-n-FLAG expressing the C terminus of the IS256 transposase (aa 200 to 390) | This study |
| pCAL-n-FLAG-tnp L103P | pCAL-n-FLAG vectors expressing IS256 transposase derivatives with various amino acid substitutions | This study |
| pCAL-n-FLAG-tnp Y111A | ||
| pCAL-n-FLAG-tnp G114W | ||
| pCAL-n-FLAG-tnp T117A | ||
| pCAL-n-FLAG-tnp R118A | ||
| pCAL-n-FLAG-tnp L127P | ||
| pACYC184 | Cloning vector, ori p15A cat tet; compatible with ColE1- and pMB-1-related plasmids | Fermentas |
| pACYC184-Δtnp | pACYC184 vector carrying the transposase inactivated IS256 copy from pIL2 IS256Δtnp | This study |
Expression and purification of IS256 transposase.
Full-length IS256 transposase and N-terminal, C-terminal, and internal fragments, as well as derivatives with amino acid substitutions, were expressed and purified as calmodulin-binding peptide (CBP) fusion proteins, using the Affinity LIC cloning and purification kit (Stratagene, La Jolla, CA). Briefly, tnp-specific DNA fragments were amplified using the primers and conditions summarized in Table S1 in the supplemental material and introduced into p-CAL-n-FLAG by ligation-independent cloning (LIC). Single amino acid substitutions were achieved by recombination PCRs using the p-CAL-n-FLAG-tnp plasmid as a template, respectively (53). The integrities of the nucleotide sequences were checked by sequencing, and plasmids were transformed into Escherichia coli BL21(DE3), respectively. Induction of protein expression occurred at 30°C with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), respectively, except for the N-terminal transposase fragment, for which expression was induced at 18°C. Cell lysis and protein purification was performed according to the protocol provided by the manufacturer (Stratagene). Protein purifications were performed to near homogeneity as verified by SDS-PAGE (see Fig. 2 and also Fig. SA in the supplemental material).
FIG. 2.
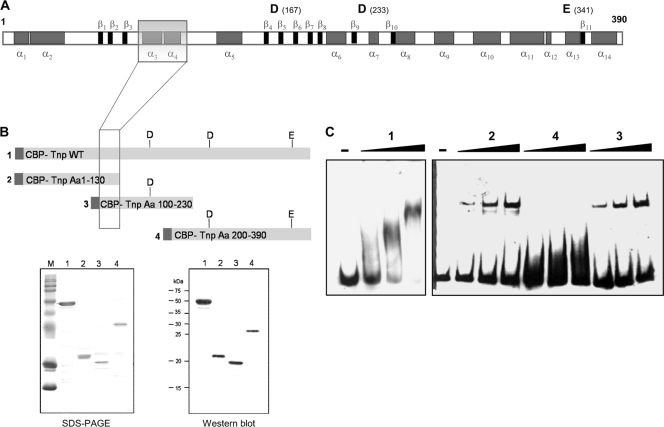
Identification of the DNA-binding domain of IS256 transposase. (A) Secondary structure prediction of the 390-aa transposase protein of IS256 based on PSIPRED analysis (http://bioinf.cs.ucl.ac.uk/psipred/psiform.html). Gray boxes indicate α-helices. β-Sheets are illustrated as black boxes. The three acidic residues of the catalytic DDE triad and their positions (in brackets) are shown above the diagram. The transparent box indicates the position of the putative DNA-binding domain identified in panels B and C. (B) Heterologous expression and purification of overlapping IS256 transposase fragments of various lengths. Dark gray boxes mark the CBP tag. Lanes: 1, full-length wild-type transposase; 2, N-terminal transposase fragment covering aa 1 to 130; 3, internal transposase fragment comprising aa 100 to 230; 4, C-terminal transposase fragment (aa 200 to 390). Proteins were overexpressed in E. coli, and purified fragments 1 to 4 were analyzed by SDS-PAGE (bottom left) and Western blotting with an anti-FLAG antibody (bottom right). (C) EMSAs using 15.5 nM DNA of the right-end IS256 terminus DNA as a substrate with increasing amounts (i.e., 200, 500, and 1,000 nM) of purified transposase fragments 1 to 4, respectively. The experiments were performed with 50 μg of poly(dI-dC) ml−1 as a nonspecific competitor, and the molar DNA/protein ratios were 1:13 (200 nM protein), 1:32 (500 nM protein), and 1:64 (1,000 nM protein), respectively.
EMSA.
DNA substrates for electrophoretic mobility shift assays (EMSAs) were amplified by using the primers and conditions listed in Table S1 in the supplemental material and labeled by adding digoxigenin-11 (DIG)-ddUTP to the 3′ DNA end by terminal transferase using a DIG Gelshift kit (Roche, Mannheim, Germany). Labeling efficiency was determined according to the Gelshift kit protocol by comparing a dilution series of a DIG-labeled control oligonucleotide with that of the labeled DNA probe. DNA probes were regarded as efficiently labeled and used for downstream experiments when at least the 1 fmol/μl spot was clearly detectable. DNA-protein binding reactions were assessed according to the DIG Gelshift protocol: 2 μl of 5× binding buffer [100 mM HEPES (pH 7.6), 5 mM EDTA, 50 mM (NH4)2SO4, 5 mM dithiothreitol, 1% (wt/vol) Tween 20, 150 mM KCl], 0.5 μl of poly(dI-dC) (1 μg/μl), 0.5 μl of poly-l-lysine (0.1 μg/μl), 1 to 5 μl of protein (various concentrations), and double-distilled water (9 μl) were mixed together. Then, 1 μl of DIG-labeled DNA was added (15.5 fmol/μl). After incubation at room temperature for 20 min, the samples were mixed with 2 μl of loading buffer (0.2% [wt/vol] bromphenol blue, 40% glycerol, 60% [wt/vol] 1× Tris-borate-EDTA [TBE]) and separated by gel electrophoresis in 6% native 0.5× TBE polyacrylamide gels. The gels were electroblotted onto nylon membranes, and DNA was cross-linked by using UV light. Chemiluminescent detection was performed by using anti-digoxigenin antibody conjugated with alkaline phosphatase and CSPD chemiluminescent substrate as outlined in the DIG Gelshift kit protocol.
PCR detection of IS256 circular molecules and quantification by qPCR.
For the detection and quantification of IS256 circles, nonchromosomal DNA was isolated from early-exponential-phase cultures (optical density at 600 nm of 2) of Staphylococcus aureus RN4220 (23) bearing pIL2 (as a positive control), pIL2 IS256Δtnp (as a negative control), or pIL2 derivatives with deletions at the IS256 termini, respectively (Table 1). PCR assays to detect IS256 circles were performed as described previously using the outward primers 1 and 2 (27). For quantitative PCR (qPCR), the MyiQ single-color real-time PCR detection system and the IQ SYBR green Supermix (Bio-Rad, Munich, Germany) were used according to the instructions of the manufacturer. qPCR was performed with appropriate dilutions of nonchromosomal DNA of S. aureus RN4220(pIL2) and RN4220(pIL2) derivatives, respectively. IS256 circles were quantified by amplifying a 112-bp product specific for circular forms of IS256 using 500 nM outward primers IRL EMSA rev and IRR for CJ (see Table S1 in the supplemental material). A 132-bp icaC sequence amplified from the vector using the primers icaCintf and icaCLCrev served as an endogenous standard. The data were analyzed according to the model described by Pfaffl (37), and the quantities of circular IS256 DNA were calculated as relative amounts versus the IS256 wild type on plasmid pIL2.
RESULTS
Heterologous expression and purification of a functional IS256 transposase protein.
To investigate the DNA-binding properties of the IS256 transposase, the 390-amino-acid (aa) polypeptide was expressed and purified as a CBP fusion protein in E. coli. Overexpression of the CBP-transposase (CBP-Tnp) fusion protein upon IPTG induction, as well as purification of CPB-Tnp, is shown elsewhere (see Fig. SA, lanes 1 to 5, in the supplemental material). The N-terminal CPB tag used for affinity purification was left in place since it was not removable by enterokinase treatment. Also, replacement of the original enterokinase cleavage site by a factor Xa cleavage site did not result in proteolysis, which led us to conclude that the protease target region might be buried and inaccessible for proteolytic enzymes (data not shown). To verify whether or not the CBP tag impairs the function of the transposase, the protein was tested for its ability to accomplish circularization of an IS256 DNA substrate. Circular IS256 molecules can be readily detected by an inverse PCR using outward-directed IS256-specific primers (see Fig. 5B) (27). IS256 circle formation is transposase dependent but is supposed, as in other circle-forming elements, to involve host factors as well (27, 28, 48). Therefore, an in vivo dual vector system was assessed in E. coli. The CBP-Tnp protein was expressed from vector pCAL-n-FLAG-tnp in trans, while a second plasmid, pACYC184-IS256Δtnp, with a compatible origin of replication, harbored an IS256 copy with an inactivated transposase gene as DNA substrate (see Fig. SB in the supplemental material). No IS256 circular molecules were formed when the pACYC184-IS256Δtnp vector was present in E. coli alone or when, in the presence of pACYC184-IS256Δtnp, the CBP-Tnp expression remained uninduced (see Fig. SC in the supplemental material). In contrast, IS256 circles were clearly detectable in pACYC184-IS256Δtnp-bearing cells upon the induction of the CBP-Tnp transposase protein from the pCAL-n-FLAG-tnp vector (see Fig. SC in the supplemental material, 2 and 7 h), demonstrating that the CPB-Tnp protein is active.
FIG. 5.
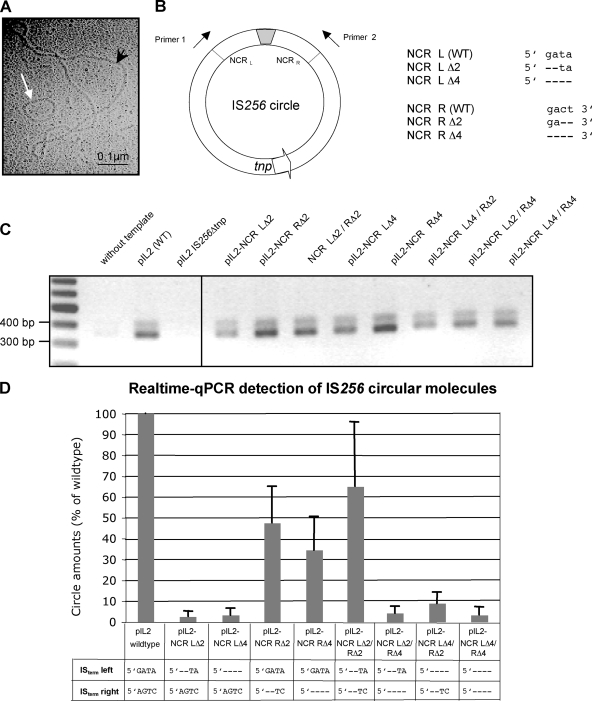
Effect of terminal IS deletions on IS256 circle formation. (A) Electron micrograph of an IS256 circular DNA molecule (white arrow). The picture was taken from an E. coli plasmid preparation of vector pIL2 carrying a wild-type IS256 insertion. The black arrowhead marks a 9.3-kb pIL2 vector molecule. (B) Illustration of an IS256 circle with abutted IS ends separated by a short stretch of foreign DNA (gray box). Arrows indicate the direction of primers used for PCR detections of IS256 circles in panels C and D. The orientation of the transposase gene tnp is marked by an arrow. The chart on the right details the nucleotide sequence of the last four nucleotides at the IS256 ends and their deletions introduced into the IS256 copy on vector pIL2, respectively. (C) Agarose gel electrophoresis of IS256 circle-specific PCR products amplified by using primers 1 and 2 and plasmid preparations of pIL2 (as a positive control) and pIL2 IS256Δtnp (as a negative control), as well as pIL2-derived mutants, with deletions in the IS256 termini as templates, respectively. (D) Comparison of the circle amounts detected in pIL2, carrying an IS256 wild-type copy, and various IS termini mutants, respectively, as determined by qPCR.
Purified IS256 transposase binds to the noncoding DNA ends of the element in vitro.
Transposition of an IS requires release of the element from its donor DNA backbone, a reaction that is usually executed by the element's transposase. To initiate donor strand cleavage, the transposase protein of an IS element is required to recognize and to bind to its own IS ends. IS termini mostly consist of NCRs which often carry IRs as putative protein binding sites. Figure 1A shows the organization of IS256 inserted into a target gene (i.e., icaC from S. epidermidis). IS256 contains NCRs of different lengths on either side, a left-hand NCR of 101 bp and a right-hand NCR of 50 bp with imperfect IRs, respectively. The IRs consist of a stretch of six perfectly matching bases interrupted by seven heterogenous nucleotides, followed by another nine matching bases (Fig. 1 A). To analyze whether or not the IS256 transposase binds to the IS256 ends, various IS256-specific DNA fragments were PCR amplified from the IS256::icaC insertion site on vector pIL2 (27) and used as DNA substrates in EMSAs with purified CBP-Tnp protein. The DNA substrates covered the NCRs, along with the 8-bp target site duplication and a few nucleotides of the icaC gene, respectively (Fig. 1A). As indicated in Fig. 1B and C, the CBP-Tnp protein was found to bind to the left and to the right IS termini, respectively. Binding to the 147-bp IS terminus (left) occurred at an CBP-Tnp concentration of 400 nM (Fig. 1B) and at 200 nM with the 126-bp IS terminus (right) (Fig. 1C). In contrast, no transposase binding was detectable when an internal IS256 fragment was used as the DNA substrate (Fig. 1D). The data demonstrate that purified CBP-Tnp has DNA-binding activity in vitro and specifically interacts with the cognate IS256 termini.
The DNA-binding region is located in the N-terminal domain of the transposase.
Next, the identification of the DNA-binding domain(s) within the IS256 transposase protein was addressed. At first, an in silico analysis of the amino acid sequence was performed. The polypeptide consists of 390 aa residues with a molecular mass of 45.7 kDa and a pI of 9.08. Secondary structure predictions using the PSIPRED algorithm predicted the IS256 transposase as a predominantly α-helical protein (33). In addition to 15 α-helices, 11 short β-sheets were detected that are, apart from β1 to β3, located in the C-terminal portion of the protein that also harbors the catalytic DDE motif (Fig. 2 A). The acidic amino acid residues aspartate D167, aspartate D233, and glutamate E341 are assumed to form, in the tertiary structure, a pocket for divalent cation binding that is required for DNA cleavage (12). By bioinformatic means, no standard DNA-binding structures such as HTH motifs (9), zinc fingers (36), or leucine zippers (2) could be identified in the IS256 amino acid sequence. Thus, we aimed at an experimental determination of the DNA-binding region. Overlapping IS256 transposase fragments of various lengths were expressed and purified as CBP fusion proteins and then used in EMSAs using the IS terminus (right) as a DNA substrate, respectively (Fig. 2B and C). The protein fragments covered the full-length wild-type transposase (fragment 1), an N-terminal portion of the protein (fragment 2), and an internal part (fragment 3), as well as the C terminus of the transposase (fragment 4) (Fig. 2B). As demonstrated in Fig. 2C, fragments 1 to 3 were able to bind to IS terminus (right)-specific DNA, while the C-terminal fragment 4 exhibited no detectable binding capacity. The results suggest that the DNA-binding domain(s) of the IS256 transposase is likely to be located in the N-terminal portion of the protein.
Substitutions of amino acids in the putative DNA-binding region alter transposase-DNA interactions.
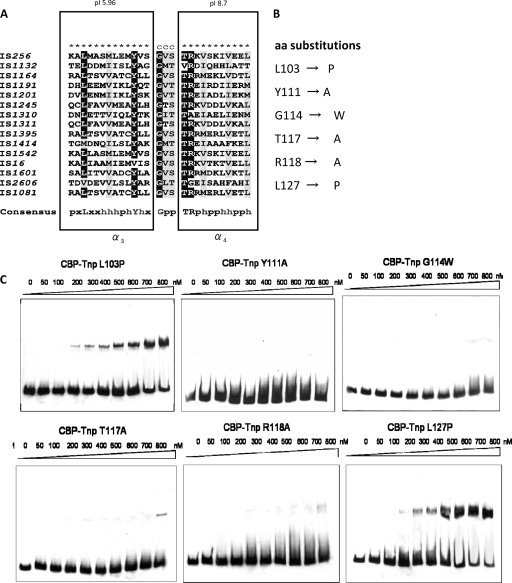
The N-terminal part of the transposase comprising aa 100 to 130, which are common to fragments 1 to 3, was analyzed in more detail. PSIPRED secondary structure analysis predicted the existence of two α-helices separated by a short coiled region of 3 aa residues (transparent box in Fig. 2A and 4A). The α3-helix which covers aa 101 to 113 is predominantly hydrophobic and has a negative net charge (pI 5.96), while helix α4 (aa 117 to 127) is hydrophilic and basic (pI 8.7) (24). Multiple sequence alignment of the IS256 transposase with a range of other bacterial IS256 transposases revealed conservation of this motif throughout IS256 family members and identified a number of highly conserved residues (Fig. 3 A) (25). To determine their impact on DNA binding, the conserved amino acid residues of the motif were exchanged in the CBP-Tnp protein by site-directed mutagenesis of the pCAL-n-FLAG-tnp vector nucleotide sequence and subsequent expression and purification of altered transposase proteins (Fig. 3B). The CBP-Tnp variants were then analyzed for their DNA-binding properties in EMSAs using again the IS terminus (right) as the DNA substrate (Fig. 3C). As indicated in Fig. 3C, substitution of L103 and L127 by helix-breaking proline residues at the N terminus of the α3-helix and the C terminus of the α4-helix did not affect CBP-Tnp binding. However, the DNA-binding capacity of the CBP-Tnp transposase protein was abolished or significantly reduced when any of the conserved amino acid residues in the two helices (i.e., Y111, T117, and R118) were substituted by alanine (Fig. 3C). Also, alteration of the predicted turn region by introducing a large tryptophan residue instead of the small glycine residue at position G114 resulted in a complete loss of CBP-Tnp interaction with the DNA substrate.
FIG. 4.
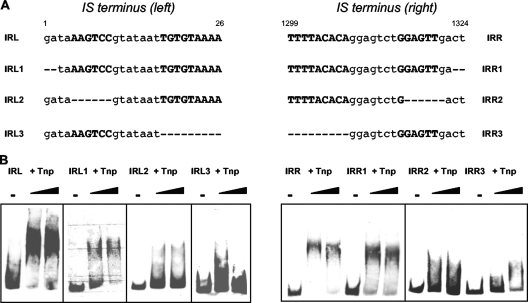
Influence of the integrity of the IRs on IS256 transposase binding. (A) Nucleotide sequence of IS256 ends harboring the IRs (uppercase boldface letters) and positions of nucleotide deletions (−) introduced into the IS terminus (left) and IS terminus (right) DNA substrates, respectively. (B) EMSAs in the presence of 50 μg of poly(dI-dC) ml−1 with 1.9 or 3.8 μM CBP-Tnp transposase and a 15.5 nM concentration of the mutated IS termini as DNA substrates, respectively. The molar DNA/protein ratios were 1:122 (1.9 μM CBP-Tnp) and 1:245 (3.8 μM CBP-Tnp), respectively.
FIG. 3.
Analysis of a putative HTH DNA-binding motif of the IS256 transposase. (A) Amino acid sequence alignment (25) of the putative DNA-binding region of IS256 (aa 100 to 130) and other bacterial members of the IS256 family. ★, α-helix; C, coiled region. Rectangles mark the predicted α-helices α3 and α4, respectively. Highly conserved and conserved residues are highlighted in black and gray, respectively. In the consensus sequence, “x,” “p,” and “h” indicate variable, polar, and hydrophobic amino acid residues, respectively. (B) Amino acid exchanges of highly conserved and conserved residues within the putative HTH motif of the CBP-Tnp protein. (C) EMSAs with 15.5 nM IS terminus (right)-specific DNA as substrate and increasing amounts of the altered CBP-Tnp proteins described in panels A and B. Binding assays were performed in the presence of 50 μg of poly(dI-dC) ml−1 as a nonspecific competitor, and molar DNA/protein ratios ranged from 1:3 (50 nM protein) to 1:52 (800 nM protein), respectively.
The IRs in the NCRs of IS256 are crucial for transposase binding.
After identification of the DNA-binding domain of the transposase, we were interested in the exact binding sites of the IS256 transposase at the IS termini. To establish whether the IRs within the NCRs serve as protein binding sites, the palindromic nucleotide sequences were mutated and tested for CBP-Tnp binding (Fig. 4 A). Figure 4 illustrates that CBP-Tnp transposase still binds to the IRs when the terminal two IS256 nucleotides, which do not belong to the IR, were deleted (Fig. 4B, IRL1 and IRR1). In contrast, transposase binding was impaired when any part of the IR sequence was affected. Thus, deletion of the outermost six matching base pairs of the imperfect IR resulted in a significant loss of transposase binding both in IS terminus (left) and in IS terminus (right), respectively (Fig. 4B, IRL2 and IRR2). The same effect was observed when the inner matching part of the IR (nucleotides [nt] 18 to 26; nt 1299 to 1307) was deleted (Fig. 4B, IRL3 and IRR3).
Terminal nucleotide deletions of IS256 affect the circularization efficiency of the element.
In a next set of experiments, we sought to determine the role of the terminal nucleotides of IS256 in circularization of the element. In some IS elements, transposition involves the generation of double-stranded circular IS molecules that are regarded as transposition intermediates, and IS256 was shown previously to use this alternative transposition pathway (27, 39). As illustrated in Fig. 5 A, such small circular IS256 molecules can be visualized by electron microscopy when propagating the element on a high-copy-number vector in E. coli. More conveniently, circle formation can be detected and quantified by PCR using outward-reading IS primers (Fig. 5B). The terminal four nucleotides at either end of IS256 do not belong to the transposase-binding IR nucleotide sequence. To analyze whether they have an impact on circularization, the terminal IS256 nucleotides were stepwise deleted in the wild-type IS256 copy on plasmid pIL2 (27). The resulting plasmids were propagated in S. aureus and analyzed for IS256 circle formation by semiquantitative and quantitative PCR, respectively. Figure 5C demonstrates that the deletions of two or four terminal nucleotides did not affect circle formation in general. Circles were still generated from all deletion mutants, even when all four nucleotides at both ends had been removed simultaneously (Fig. 5C, pIL2-NCR LΔ4/RΔ4). However, when quantifying the circle amounts by qPCR, significant differences were detected between the various deletion mutants (Fig. 5D). Compared to the circle amounts generated by the IS256 wild-type copy on pIL2, deletions of two or four nucleotides in the left-hand terminus resulted in a decrease in circle formation by ca. 97%, respectively (Fig. 5D, pIL2-NCR LΔ2 and pIL2-NCR LΔ4). Upon 2- and 4-bp deletions in the right-hand terminus, the circle amounts were also significantly reduced, but only by 53 and 66%, respectively (Fig. 5D, pIL-2 NCR RΔ2 and pIL2-NCR RΔ4). Simultaneous mutation of both IS ends revealed no significant reduction in circle formation when two base pairs were deleted at either end (Fig. 5D, pIL2-NCR LΔ2/RΔ2). However, the circle amounts dropped again dramatically by 91 to 97%, respectively, when 4 bp were removed from any of the two ends (Fig. 5D, pIL2-NCR LΔ4/RΔ2, pIL2-NCR LΔ2/RΔ4, and pIL2-NCR LΔ4/RΔ4). The combined data suggest that the terminal nucleotides of IS256 are not crucial for the circularization reaction itself, but their integrity has a significant impact on the efficiency of the process.
DISCUSSION
IS256 is the prototype element of the IS256 family of insertion sequences that are present in a wide range of bacterial genera, including Staphylococcus, Enterococcus, Mycobacterium, Burkholderia, Rhizobium, Rhodococcus, Lactobacillus, and Yersinia (30). Members of this family have similar sizes (between 1.3 and 1.5 kb), generate 8- or 9-bp TSDs and carry related IRs (43). Generally, IS elements belonging to the IS256 family contain a single open reading frame that encodes a transposase with a conserved DDE catalytic domain signature motif. Interestingly, related transposases containing this DDE motif have also been identified in eukaryotic systems, such as the Mutator superfamily in plants (10, 15). The currently available information on transposase proteins of this important group of mobile genetic elements is mainly based on bioinformatic studies (15). This particularly applies to the bacterial members of IS256 family transposases, none of which has yet been characterized biochemically. To gain more insight into the properties of these proteins, we therefore sought to express and characterize recombinant forms of the IS256 transposase, the bacterial prototype enzyme of this family, focusing on its subdomain organization and functional properties.
Sequence-specific DNA binding and functional organization of the transposase protein.
Using gel shift assays and purified IS256 transposase protein, we were able to show that the purified enzyme binds to noncoding DNA regions at the IS256 termini but not to internal IS256 DNA fragments (Fig. 1). These data are in line with other transposases for which a specific binding to the cognate IS ends has been demonstrated (7, 35, 45, 54). Many bacterial transposases share a similar subdomain organization with the DNA-binding domain being located at the N terminus, whereas the catalytic domain occupies the C-terminal part of the protein (12). The IS256 transposase proved to be no exception to this rule. Three conserved acidic amino acid residues—D167, D233, and E341—in the C-terminal domain are predicted to form the catalytic site (12). Each of these residues has previously been demonstrated to be necessary for circularization of the IS element (27), suggesting that they are part of the classical DDE motif conserved across many prokaryotic and eukaryotic transposases and even retroviral integrases (12, 15). The results from our gel shift assays using N or C terminally truncated forms of the transposase conclusively showed that the DNA-binding region of the transposase resides in the N-proximal region of the protein (Fig. 2). Many transposases proteins act preferentially in cis by various mechanisms (8, 51). In some transposases such as IS10, the physical separation of DNA-binding and catalytic domains supports enzymatic activity in cis (17). Thus, after translation of the N-terminal DNA-binding domain, the nascent transposase protein may bind its target sequence on the IS and thus guide the catalytic domain, once translated, to the cleavage site on the IS DNA from which the transposase is expressed (17). Also, the functional interplay between N-terminal DNA-binding and C-terminal catalytic domains has been demonstrated in some ISs to reduce the DNA-binding activity of the latter (17, 52). Our observation that the two N-terminal fragments bound their DNA substrates with higher affinity than the full-length protein suggests a similar scenario for the IS256 transposase (Fig. 2C), but more detailed quantitative analyses are required to substantiate this hypothesis.
DNA binding by the IS256 transposase involves a putative HTH motif.
DNA binding by transposases and integrases may be mediated by various DNA-binding structures, such as leucine zippers, zinc fingers and, most commonly, HTH motifs (12, 16, 36). Our initial bioinformatic analysis did not provide convincing evidence for the presence of any of these motifs in the full-length IS256 transposase. Secondary structure predictions however suggested the presence of two α-helices connected by a short coiled region in the N-terminal portion of the protein (aa 100 to 130). As discussed above, our gel shift data had suggested a critical role of this region in DNA-binding activity (Fig. 2). More detailed analyses subsequently revealed structural features that resembled those of classical DNA-binding HTH motifs (13), a finding which was further corroborated by the identification of a number of very well conserved amino acid residues in the two helices (i.e., α3 and α4) and the putative turn region (Fig. 3A). Thus, protein analysis tools predicted helix α3 to be weakly acidic and hydrophobic, while helix α4 to be basic and hydrophilic. The putative turn region connecting the two helices contains the essential glycine residue (Fig. 3A). If we place this glycine residue (G114) at position 9 and limit the total length of this structural element to the typical size of 20 aa, the putative HTH motif would span from residues serine S106 to glutamate E125. The putative HTH structure appears to be conserved in a wide range of IS256 family members and site-directed mutagenesis of conserved amino acid residues further supported the idea of an HTH motif playing a major role in DNA binding by the transposase (Fig. 3A and B). Although being well conserved, leucine residues L103 and L127 seem to be located outside the actual DNA-binding helices as their replacement with proline did not affect DNA-binding activity, which is consistent with the predicted size of the motif (see above and the L103P and L127P panels in Fig. 3C). In contrast, substitution of the highly conserved tyrosine Y111 by alanine in the α3 helix significantly reduced DNA binding, supporting a key role of this residue in DNA binding (Fig. 3C, Y111A panel). Similar effects of mutations in the N-terminal helix have been described previously for HTH motifs of other IS transposases, such as IS1 and IS911 (36, 40, 47). The amino acid residues in the turn region of HTHs (small-hydrophobic-small) are thought to form a hinge that holds the two helices in the correct mutual orientation required for function (1). In line with this idea, substitution of glycine G114 in the predicted turn region of the IS256 transposase with tryptophan completely abolished DNA-binding (Fig. 3C, G114W panel). Finally, substitutions with alanine of two other conserved amino acid residues (T117 and R118) in the C-terminal α4 helix significantly reduced (but did not abolish) DNA-binding activity as weak interactions remained detectable at high protein concentrations (Fig. 3C, T117A and R118A panels). We consider it unlikely that the single amino acid replacements with alanine caused any major structural changes. Most likely, these replacements caused a decrease in DNA-binding affinity by reducing the number of hydrogen bonds that stabilize the interactions between the enzyme's “recognition” domain and its target DNA. Taken together, the data led us to suggest the presence of a classical HTH motif in the N-terminal part of the IS256 transposase that is critically involved in the specific binding of the IS ends.
IS256 transposase interaction with IRs.
Most IS elements harbor in their terminal NCRs palindromic nucleotide sequences which are used as major binding sites for the transposase protein (12). IS256 carries imperfect IRs at its ends (Fig. 1A). Deletions of the IRs at either IS end impaired transposase binding, suggesting that the IS256 transposase interacts specifically with these IR sequences (Fig. 4). Transposase binding of IS termini is usually followed by first-strand donor cleavage which, in circle-forming elements (e.g., IS911, the IS3 family, and IS30), is known to occur only at one end of the element (12). The released 3′ OH end is then transferred in a site-specific manner to the opposite IS terminus, where it attacks the same strand. The resulting nicked DNA molecule is resolved, releasing a circular copy of the IS (12, 38). Although the circle formation mechanism used by IS256 has not yet been elucidated in detail, it seems reasonable to think that it proceeds in a similar manner. In a previous study, we observed that IS256 is able to attack either end, while another study suggested that it is always the left-hand IS256 terminus that attacks the right-end of the element (27, 39). However, in both studies, only a small number of IS256 circle molecules were analyzed that largely precluded statistic analyses. The data reported here indicate that the IS256 transposase binds to both ends of the element (Fig. 1). It remains to be studied whether this similar IS256 transposase binding correlates with similar first-strand cleavage and strand transfer activities.
Role of the terminal IS256 nucleotides in circle formation.
Unlike in most other IS, the “tips” of the IS element are not identical in IS256 (Fig. 5B). The four nucleotides located at the extreme termini vary in IS256 (left, 5′-GATA-3′; right, 5′-AGTC-3′) and do not take part in transposase binding, but they seem to have a role in strand processing and circle formation. If 2 or 4 bp at either end were deleted, circle formation remained detectable, indicating that the extreme IS256 termini are not essential for the circularization reaction (Fig. 5C). The data also suggest some flexibility with respect to the target site selection for IS256 transposase-mediated first-strand cleavage. On the other hand, however, the integrity of the terminal four nucleotides clearly contributed to the efficiency of the process, and genetic manipulation of either of the two IS termini had differential effects on circle formation. Thus, deletions at the left-hand terminus resulted in a dramatic drop in circle production (up to 97%), even when only two nucleotides had been removed (Fig. 5D). Although manipulations at the right-hand terminus also reduced the number of circles, the effects were less dramatic than those seen at the left-hand terminus. A notable exception was pIL-NCR LΔ2/RΔ2. Although the outermost two nucleotides had been deleted from both ends, near-wild-type levels of circles were still being produced (Fig. 5D). On the basis of these data, we hypothesize that IS256 transposase-mediated strand processing occurs preferentially, albeit not exclusively, at the left-hand terminus, which then triggers the reactions leading to the circularization of the element. Asymmetric transposase activity has also been reported for other circle-forming elements, such as IS30 and IS911, and may occur at different stages of the transposition pathway (41, 44). Differential transposase activities at specific flanking or target sequences may involve a range of factors and mechanisms. Thus, specific host factors, such as IHF, or the DNA-methylation status were found to play a role in some elements (i.e., IS10 and IS50) (46, 50). Also, differential binding affinities of the enzyme to the transposon ends or subterminal sequences adjacent to the IRs can have an impact on the process (e.g., in IS911 and IS30) (41, 44). Clearly, more studies are required to answer the question of how the “biased” transposase reaction in IS256 is mediated at the molecular level. The combined data lead us to suggest that the few nucleotides at the very ends of the element play an important role in the circularization process, and there is even some initial evidence to suggest that the specific sequence, including a cytidine residue present at the very left end might contribute to the observed bias of transposase cleavage toward the left end. Again, more experimental work is needed to substantiate this hypothesis and elucidate this mechanism in more detail.
The present study provides the first experimental characterization of a transposase from the IS256 family and establishes the IS256 transposase as a DNA-binding protein. The protocols developed in the present study for the expression, purification, and characterization of the IS256 transposase should provide a solid basis for future studies of the molecular mechanisms involved in IS256 transposition and its major consequences for the biology and evolution of a wide range of natural hosts, including important human and animal pathogens.
Supplementary Material
Acknowledgments
We are grateful to Nadine Emrich and Katja Dietrich for excellent technical assistance.
This study was supported by the Deutsche Forschungsgemeinschaft (Transregional Collaborative Research Centre 34), the Federal Ministry of Education and Research (Network Pathogenomics-plus, grant PTJ-BIO//03U213B), and the Department for Employment and Learning (Northern Ireland) through its Strengthening the All-Island Research Base.
Footnotes
Published ahead of print on 11 June 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aravind, L., V. Anantharaman, S. Balaji, M. M. Babu, and L. M. Iyer. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231-262. [DOI] [PubMed] [Google Scholar]
- 2.Bornberg-Bauer, E., E. Rivals, and M. Vingron. 1998. Computational approaches to identify leucine zippers. Nucleic Acids Res. 26:2740-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361-367. [DOI] [PubMed] [Google Scholar]
- 4.Cafiso, V., T. Bertuccio, M. Santagati, V. Demelio, D. Spina, G. Nicoletti, and S. Stefani. 2007. agr-Genotyping and transcriptional analysis of biofilm-producing Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 51:220-227. [DOI] [PubMed] [Google Scholar]
- 5.Conlon, K., H. Humphreys, and J. O'Gara. 2004. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 186:6208-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couto, I., S. W. Wu, A. Tomasz, and H. de Lencastre. 2003. Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to the species. J. Bacteriol. 185:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derbyshire, K. M., and N. D. Grindley. 1992. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 11:3449-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derbyshire, K. M., and N. D. Grindley. 1996. Cis preference of the IS903 transposase is mediated by a combination of transposase instability and inefficient translation. Mol. Microbiol. 21:1261-1272. [DOI] [PubMed] [Google Scholar]
- 9.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen, J. A., M. I. Benito, and V. Walbot. 1994. Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22:2634-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu, J., H. Li, M. Li, C. Vuong, M. Otto, Y. Wen, and Q. Gao. 2005. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J. Hosp. Infect. 61:342-348. [DOI] [PubMed] [Google Scholar]
- 12.Haren, L., B. Ton-Hang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 13.Harrison, S. C., and A. K. Aggarwal. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 59:933-969. [DOI] [PubMed] [Google Scholar]
- 14.Hennig, S., and W. Ziebuhr. 2008. A transposase-independent mechanism gives rise to precise excision of IS256 from insertion sites in Staphylococcus epidermidis. J. Bacteriol. 190:1488-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua-Van, A., and P. Capy. 2008. Analysis of the DDE motif in the mutator superfamily. J. Mol. Evol. 67:670-681. [DOI] [PubMed] [Google Scholar]
- 16.Ivics, Z., Z. Izsvak, A. Minter, and P. B. Hackett. 1996. Identification of functional domains and evolution of Tc1-like transposable elements. Proc. Natl. Acad. Sci. U. S. A. 93:5008-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain, C., and N. Kleckner. 1993. Preferential cis action of IS10 transposase depends upon its mode of synthesis. Mol. Microbiol. 9:249-260. [DOI] [PubMed] [Google Scholar]
- 18.Jansen, A., M. Turck, C. Szekat, M. Nagel, I. Clever, and G. Bierbaum. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int. J. Med. Microbiol. 297:205-215. [DOI] [PubMed] [Google Scholar]
- 19.Kiem, S., W. S. Oh, K. R. Peck, N. Y. Lee, J. Y. Lee, J. H. Song, E. S. Hwang, E. C. Kim, C. Y. Cha, and K. W. Choe. 2004. Phase variation of biofilm formation in Staphylococcus aureus by IS256 insertion and its impact on the capacity adhering to polyurethane surface. J. Korean Med. Sci. 19:779-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koskela, A., A. Nilsdotter-Augustinsson, L. Persson, and B. Soderquist. 2009. Prevalence of the ica operon and insertion sequence IS256 among Staphylococcus epidermidis prosthetic joint infection isolates. Eur. J. Clin. Microbiol. Infect. Dis. 28:655-660. [DOI] [PubMed] [Google Scholar]
- 21.Kozitskaya, S., S. H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozitskaya, S., M. E. Olson, P. D. Fey, W. Witte, K. Ohlsen, and W. Ziebuhr. 2005. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 43:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 24.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 25.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 26.Li, M., X. Wang, Q. Gao, and Y. Lu. 2009. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J. Med. Microbiol. 58:456-461. [DOI] [PubMed] [Google Scholar]
- 27.Loessner, I., K. Dietrich, D. Dittrich, J. Hacker, and W. Ziebuhr. 2002. Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J. Bacteriol. 184:4709-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loot, C., C. Turlan, and M. Chandler. 2004. Host processing of branched DNA intermediates is involved in targeted transposition of IS911. Mol. Microbiol. 51:385-393. [DOI] [PubMed] [Google Scholar]
- 29.Lyon, B. R., M. T. Gillespie, and R. A. Skurray. 1987. Detection and characterization of IS256, an insertion sequence in Staphylococcus aureus. J. Gen. Microbiol. 133:3031-3038. [DOI] [PubMed] [Google Scholar]
- 30.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maki, H., N. McCallum, M. Bischoff, A. Wada, and B. Berger-Bachi. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1953-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maki, H., and K. Murakami. 1997. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:6944-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 34.Montanaro, L., D. Campoccia, V. Pirini, S. Ravaioli, M. Otto, and C. R. Arciola. 2007. Antibiotic multiresistance strictly associated with IS256 and ica genes in Staphylococcus epidermidis strains from implant orthopedic infections. J. Biomed. Mater. Res. A 83:813-818. [DOI] [PubMed] [Google Scholar]
- 35.Normand, C., G. Duval-Valentin, L. Haren, and M. Chandler. 2001. The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. J. Mol. Biol. 308:853-871. [DOI] [PubMed] [Google Scholar]
- 36.Ohta, S., E. Yoshimura, and E. Ohtsubo. 2004. Involvement of two domains with helix-turn-helix and zinc finger motifs in the binding of IS1 transposase to terminal inverted repeats. Mol. Microbiol. 53:193-202. [DOI] [PubMed] [Google Scholar]
- 37.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polard, P., and M. Chandler. 1995. An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev. 9:2846-2858. [DOI] [PubMed] [Google Scholar]
- 39.Prudhomme, M., C. Turlan, J. P. Claverys, and M. Chandler. 2002. Diversity of Tn4001 transposition products: the flanking IS256 elements can form tandem dimers and IS circles. J. Bacteriol. 184:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rousseau, P., E. Gueguen, G. Duval-Valentin, and M. Chandler. 2004. The helix-turn-helix motif of bacterial insertion sequence IS911 transposase is required for DNA binding. Nucleic Acids Res. 32:1335-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rousseau, P., C. Loot, C. Turlan, S. Nolivos, and M. Chandler. 2008. Bias between the left and right inverted repeats during IS911 targeted insertion. J. Bacteriol. 190:6111-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 43.Siguier, P., J. Perochon, L. Lestrade, J. Mahillon, and M. Chandler. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32-D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szabo, M., J. Kiss, Z. Nagy, M. Chandler, and F. Olasz. 2008. Sub-terminal sequences modulating IS30 transposition in vivo and in vitro. J. Mol. Biol. 375:337-352. [DOI] [PubMed] [Google Scholar]
- 45.Szabo, M., J. Kiss, and F. Olasz. 2010. Functional organization of the inverted repeats of IS30. J. Bacteriol. 192:3414-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomcsanyi, T., and D. E. Berg. 1989. Transposition effect of adenine (Dam) methylation on activity of O end mutants of IS50. J. Mol. Biol. 209:191-193. [DOI] [PubMed] [Google Scholar]
- 47.Ton-Hoang, B., C. Turlan, and M. Chandler. 2004. Functional domains of the IS1 transposase: analysis in vivo and in vitro. Mol. Microbiol. 53:1529-1543. [DOI] [PubMed] [Google Scholar]
- 48.Turlan, C., C. Loot, and M. Chandler. 2004. IS911 partial transposition products and their processing by the Escherichia coli RecG helicase. Mol. Microbiol. 53:1021-1033. [DOI] [PubMed] [Google Scholar]
- 49.Vuong, C., S. Kocianova, Y. Yao, A. B. Carmody, and M. Otto. 2004. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190:1498-1505. [DOI] [PubMed] [Google Scholar]
- 50.Ward, C. M., S. J. Wardle, R. K. Singh, and D. B. Haniford. 2007. The global regulator H-NS binds to two distinct classes of sites within the Tn10 transpososome to promote transposition. Mol. Microbiol. 64:1000-1013. [DOI] [PubMed] [Google Scholar]
- 51.Weinreich, M. D., A. Gasch, and W. S. Reznikoff. 1994. Evidence that the cis preference of the Tn5 transposase is caused by nonproductive multimerization. Genes Dev. 8:2363-2374. [DOI] [PubMed] [Google Scholar]
- 52.Wiegand, T. W., and W. S. Reznikoff. 1994. Interaction of Tn5 transposase with the transposon termini. J. Mol. Biol. 235:486-495. [DOI] [PubMed] [Google Scholar]
- 53.Yao, Z., D. H. Jones, and C. Grose. 1992. Site-directed mutagenesis of herpesvirus glycoprotein phosphorylation sites by recombination polymerase chain reaction. PCR Methods Appl. 1:205-207. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, M., and W. S. Reznikoff. 1997. Tn5 transposase mutants that alter DNA binding specificity. J. Mol. Biol. 271:362-373. [DOI] [PubMed] [Google Scholar]
- 55.Ziebuhr, W., K. Dietrich, M. Trautmann, and M. Wilhelm. 2000. Chromosomal rearrangements affecting biofilm production and antibiotic resistance in a Staphylococcus epidermidis strain causing shunt-associated ventriculitis. Int. J. Med. Microbiol. 290:115-120. [DOI] [PubMed] [Google Scholar]
- 56.Ziebuhr, W., V. Krimmer, S. Rachid, I. Loessner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.