Abstract
Magnetotactic bacteria synthesize specific organelles, the magnetosomes, which are membrane-enveloped crystals of the magnetic mineral magnetite (Fe3O4). The biomineralization of magnetite involves the uptake and intracellular accumulation of large amounts of iron. However, it is not clear how iron uptake and biomineralization are regulated and balanced with the biochemical iron requirement and intracellular homeostasis. In this study, we identified and analyzed a homologue of the ferric uptake regulator Fur in Magnetospirillum gryphiswaldense, which was able to complement a fur mutant of Escherichia coli. A fur deletion mutant of M. gryphiswaldense biomineralized fewer and slightly smaller magnetite crystals than did the wild type. Although the total cellular iron accumulation of the mutant was decreased due to reduced magnetite biomineralization, it exhibited an increased level of free intracellular iron, which was bound mostly to a ferritin-like metabolite that was found significantly increased in Mössbauer spectra of the mutant. Compared to that of the wild type, growth of the fur mutant was impaired in the presence of paraquat and under aerobic conditions. Using a Fur titration assay and proteomic analysis, we identified constituents of the Fur regulon. Whereas the expression of most known magnetosome genes was unaffected in the fur mutant, we identified 14 proteins whose expression was altered between the mutant and the wild type, including five proteins whose genes constitute putative iron uptake systems. Our data demonstrate that Fur is a regulator involved in global iron homeostasis, which also affects magnetite biomineralization, probably by balancing the competing demands for biochemical iron supply and magnetite biomineralization.
Iron is an essential element for almost all bacteria, since iron-loaded metalloenzymes are integral parts of important biological pathways and processes like respiration, photosynthesis, N2 fixation, methanogenesis, and DNA synthesis (5). Beside being indispensable, iron can be toxic in excess due to its ability to catalyze the production of highly deleterious oxygen species via the Fenton reaction (77). Therefore, bacteria have to control their intracellular iron concentration in response to external iron availability. Iron homeostasis is typically controlled by iron-responsive transcriptional regulators, such as the ferric uptake regulator (Fur), which is the global regulator of iron metabolism in Escherichia coli (40). Fur serves as a sensor of intracellular iron concentration, and the regulation of gene expression by Fur proceeds via binding of a Fe2+-bound Fur dimer to an operator site in the promoter region of the regulated genes, thereby repressing transcription. In E. coli, this operator site consists of a 19-bp palindromic consensus sequence termed the “iron box” (18). Since Fur homologues can be found in a variety of Gram-negative and Gram-positive bacteria, a general mechanism for iron-responsive regulation has been suggested (19). However, work with other bacteria showed deviations from the classical model of Fur with respect to metal selectivity and biological functions. For example, several members of the Fur family of metalloregulators exhibit functional specialization (37), including responsiveness to zinc (Zur [46]), nickel (Nur [2]), manganese (Mur [47]), peroxide (PerR [12]), and heme (Irr [79]). Several bacteria possess global iron regulators that share no homology to regulators of the Fur family, including DtxR-like transcriptional regulators (IdeR) (67) and the RirA protein (69). Some alphaproteobacteria, which comprise the majority of cultivated magnetotactic bacteria (MTB), differ considerably from well-studied systems like E. coli or Pseudomonas aeruginosa with respect to the regulation of their iron metabolism (32, 52).
In addition to their biochemical iron requirement, MTB accumulate large amounts of iron for the synthesis of magnetosomes, which are specific intracellular organelles for magnetic navigation that are aligned in chains (31). Individual magnetosome crystals are composed of magnetite (Fe3O4) and enveloped by the magnetosome membrane (MM), which invaginates from the cytoplasmic membrane (33, 35) and consists of phospholipids and a set of specific proteins (23). The biomineralization of magnetosomes involves the uptake of large amounts of iron that may account for up to 4% of dry weight, intracellular sequestration of iron, and its crystallization (22). Although of central interest for the understanding of magnetite biomineralization, only few studies have addressed the connection of the MM with general iron metabolism and homeostasis of MTB. Early studies of the alphaproteobacterium Magnetospirillum gryphiswaldense demonstrated that magnetite biomineralization is tightly coupled to iron uptake (8), which proceeds by a fast, energy-dependent mechanism (57, 58). Recently, Rong et al. (51) showed that the disruption of the ferrous iron transporter FeoB1 leads to a reduction of magnetosome size and number in M. gryphiswaldense, which suggested a link between general iron metabolism and magnetosome biomineralization, although distinct pathways for magnetite formation and biochemical iron uptake were suggested by Faivre et al. (20).
Due to the toxicity of iron, there is a strong need for MTB to sustain a strict iron homeostasis. However, it is not clear how iron uptake and storage are regulated and balanced with the biochemical iron requirement and biomineralization. In M. gryphiswaldense, transcription of several magnetosome genes (mamGFDC and mms6) was increased in the presence of iron (56), indicating a regulatory effect of iron at the transcription level. Using a bioinformatic approach, Rodionov et al. predicted regulons of the putative iron-responsive regulators Fur and Irr in M. magneticum and M. magnetotacticum (50). In contrast to other alphaproteobacteria, such as the Rhizobiaceae, in these MTB a generic Fur protein was predicted to be the major global iron-responsive regulator, whereas Irr seems to have limited importance, regulating just single genes (50). However, an extension of the Fur regulon of other alphaproteobacteria was noted in M. magneticum and M. magnetotacticum, where in addition to multiple iron uptake genes, candidate Fur sites were observed upstream of genes related to magnetosome formation, such as mamGFDC and mms6. The hypothesis that Fur might be involved in the regulation of magnetosome biomineralization was further substantiated by the observed colocalization of fur homologues with magnetosome genes in M. magneticum and M. magnetotacticum as well as some uncultivated MTB (30). However, despite these indications for a putative role of Fur in controlling both iron homeostasis and magnetite synthesis, the mode of predicted iron regulation has remained unknown, since experimental analysis has been hampered by difficulties in genetic analysis of MTB.
In this study, we started to investigate components of general iron metabolism and their contribution to magnetite biomineralization in M. gryphiswaldense by the deletion of an identified fur-like gene. Subsequent analysis of intracellular iron metabolites and expression profiles in mutant and wild-type (WT) cells demonstrates that Fur is a global iron-responsive regulator in M. gryphiswaldense that also affects magnetosome biomineralization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids are described in Table 1. E. coli strains were routinely grown in lysogeny broth (LB) (10) supplemented with gentamicin (15 μg/ml), kanamycin (25 μg/ml), or ampicillin (50 μg/ml) at 37°C with vigorous shaking (200 rpm). For cultivation of strain BW29427, LB was supplemented with dl-α,ɛ-diaminopimelic acid to 1 mM. M. gryphiswaldense strains were grown in modified flask standard medium (FSM) with 50 μM ferric citrate (28) or in low-iron medium (LIM) (21) supplemented with 10 μM iron chelator 2,2′-dipyridyl, unless specified otherwise. Cultivation was carried out at 30°C with moderate agitation (120 rpm) under aerobic, microaerobic, or anaerobic conditions in 1-liter flasks containing 100 ml medium. For aerobic cultivation, cells were incubated in free gas exchange with air. To generate microaerobic conditions, flasks were sealed before autoclaving with butyl-rubber stoppers under a microaerobic gas mixture containing 2% O2 and 98% N2. For anaerobic conditions, O2 was omitted from the gas mixture. When necessary, media were supplemented with kanamycin (5 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Important feature(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F′ φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 | Invitrogen |
| BW29427 | hsdR17(rK− mK+)phoA supE44 thi-1 | K. Datsenko and B. L. Wanner, unpublished |
| H1717 | fhuF::lplacMu53 | 27 |
| H1780 | fiu::lplacMu53 fur | 27 |
| M. gryphiswaldense | ||
| R3/S1 | Wild type, but Rifr Smr | 60 |
| MSR-1B | Spontaneous nonmagnetic mutant of MSR-1 | 55 |
| RU-1 | R3/S1 Δfur | This study |
| M. magneticum | ||
| AMB-1 | Wild type | 34 |
| Plasmids | ||
| pGEM-T Easy | Cloning vector; Ampr | Promega |
| pGEMPmamDC | pGEM-T Easy plus PmamDC | This study |
| pGEMPmamAB | pGEM-T Easy plus PmamAB | This study |
| pGEMPmms16 | pGEM-T Easy plus Pmms16 | This study |
| pGEMPrplK | pGEM-T Easy plus PrplK | This study |
| pGEMPfhuF | pGEM-T Easy plus PfhuF | This study |
| pGEMPmgr4079 | pGEM-T Easy plus Pmgr4079 | This study |
| pGEMfurup | pGEM-T Easy plus fur 2-kb upstream region | This study |
| pGEMfurdown | pGEM-T Easy plus fur 2-kb downstream region | This study |
| pJET1.2/blunt | Cloning vector; Ampr | Fermentas |
| pJETEcfur | pJET1.2/blunt plus fur from E. coli | This study |
| pJETamb1009 | pJET1.2/blunt plus amb1009 from M. magneticum | This study |
| pJETamb4460 | pJET1.2/blunt plus amb4460 from M. magneticum | This study |
| pJETmgr1314 | pJET1.2/blunt plus mgr1314 from M. gryphiswaldense | This study |
| pBBR1MCS-2 | Mobilizable broad-host-range vector; Kmr | 36 |
| pBBR1MCS-2fur | pBBR1MCS-2 containing fur | This study |
| pBBR1MCS-5 | Mobilizable broad-host-range vector; Gmr | 36 |
| pBBR1Ptet | Mobilizable broad-host-range vector; Kmr, Ptet | C. Lang, unpublished |
| pBBR1MCS-5Ptet | Mobilizable broad-host-range vector; Gmr, Ptet | This study |
| pBBR1MCS-5Ptet/Ecfur | pBBR1MCS-5Ptet with fur from pJETEcfur | This study |
| pBBR1MCS-5Ptet/amb1009 | pBBR1MCS-5Ptet with fur from pJETamb1009 | This study |
| pBBR1MCS-5Ptet/amb4460 | pBBR1MCS-5Ptet with fur from pJETamb4460 | This study |
| pBBR1MCS-5Ptet/mgr1314 | pBBR1MCS-5Ptet with fur from pJETmgr1314 | This study |
| pCM184 | Broad-host-range allelic exchange vector | 39 |
| pCM184furup | pCM184 with 2-kb fragment from pGEMfurup | This study |
| pCM184Δfur | pCM184furup with 2-kb fragment from pGEMfurdown | This study |
Molecular and genetic techniques.
Unless specified otherwise, molecular techniques were performed using standard protocols (54). DNA was sequenced using BigDye terminator v3.1 chemistry on an ABI 3700 capillary sequencer (Applied Biosystems, Darmstadt, Germany). Sequence data were analyzed using 4Peaks software (http://mekentosj.com/4peaks). All oligonucleotide primers (see Table S1 in the supplemental material) were purchased from Sigma-Aldrich (Steinheim, Germany).
Isolation of total RNA and qualitative reverse transcriptase PCR (RT-PCR).
For isolation of total cellular RNA, M. gryphiswaldense was grown in 100 ml LIM under microaerobic and anaerobic conditions as well as in FSM supplemented with 100 μM FeCl2 or MnCl2 under microaerobic conditions to mid-logarithmic growth phase. Cells were harvested and washed in 1 ml of phosphate-buffered saline (150 mM NaCl, 10 mM sodium phosphate, pH 7), and total RNA was isolated using an RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. Isolated RNA was incubated with 10 U of RNase-free DNase I (MBI Fermentas, St. Leon Roth, Germany) for 30 min at 37°C and quantified by spectrophotometric measurements using an ND-1000 spectrophotometer (NanoDrop Technologies, DE). cDNA was synthesized from RNA templates using random hexamer primers (Roche) and RevertAid H Minus M-MuLV reverse transcriptase (Fermentas) according to the manufacturer's instructions. Transcription of fur was monitored by PCR using the primers mgr1314fwRT and mgr1314revRT (see Table S1 in the supplemental material).
Fur titration assay (FURTA).
Putative promoter regions PmamDC, PmamAB, Pmms16, Pmgr4079, and PrplK were PCR amplified with Taq polymerase (Fermentas) from genomic DNA of M. gryphiswaldense R3/S1. The Fur-regulated promoter PfhuF was amplified from whole cells of E. coli DH5α. The resulting PCR fragments were 200 to 400 bp long and included the intergenic region upstream from the start codon to the next open reading frame. The PCR products were cloned into pGEM-T Easy, sequenced, and transformed into E. coli H1717. For examination of Fur regulation, plasmid-carrying E. coli H1717 strains were streaked on MacConkey lactose agar supplemented with ampicillin and 100 μM 2,2′-dipyridyl or 30 μM FeCl3 and cultivated overnight at 37°C.
Heterologous transcomplemention of an E. coli fur mutant.
For expression of Fur-like proteins, a 1.9-kb NcoI/SacI fragment from pBBR1Ptet bearing an anhydrotetracycline-inducible promoter was inserted into pBBR1MCS-5, which had been cut with the same restriction enzymes to generate pBBR1MCS-5Ptet. fur-like genes from M. gryphiswaldense and M. magneticum as well as fur from E. coli were PCR amplified with Taq polymerase (Fermentas) using primers adding an NdeI restriction site on the 5′ end and a SacI restriction site on the 3′ end of the corresponding gene (see Table S1 in the supplemental material). PCR products were ligated into a pJET1.2/blunt cloning vector using a CloneJET PCR cloning kit (Fermentas) according to the manufacturer's recommendations. Subsequently, the genes were cloned into pBBR1MCS-5Ptet using the restriction sites NdeI and SacI. The resulting plasmids were transformed into strain H1780. For transcomplementation analysis, plasmid-carrying strains were grown in LB supplemented with gentamicin under iron-replete (100 μM FeCl2) and iron-depleted (200 μM 2,2′-dipyridyl) conditions to early log phase, induced by the addition of anhydrotetracycline at a final concentration of 100 ng/ml, and incubated for another 2 to 3 h at 37°C. Determination of ß-galactosidase activity was carried out as described previously (42).
Generation of a fur deletion strain.
A two-step, cre-lox-based method was used to generate an unmarked deletion of fur (39). For the generation of an unmarked M. gryphiswaldense fur mutant, 2-kb fragments of the up- and downstream regions of Mgfur (M. gryphiswaldense Mgr1314; see Results) were amplified by PCR using Phusion polymerase (NEB) (for primers, see Table S1 in the supplemental material), cloned into pGEM-T Easy, and sequenced. Vector pGEMfurup was digested with MunI and NotI. The resulting 2-kb fragment was inserted into MunI/NotI-digested pCM184 to yield pCM184furup. Subsequently, pGEMfurdown and pCM184furup were digested with AgeI. The resulting 2-kb fragment from pGEMfurdown was then ligated into pCM184furup to yield pCM184Δfur. After verification of the correct orientation of the deletion construct by PCR, pCM184Δfur was transferred to M. gryphiswaldense R3/S1 by conjugation as described previously (61). Putative kanamycin-resistant fur mutants were isolated on LIM agar after incubation for 10 days at 30°C and 1% O2 and analyzed by PCR. Correct genomic recombination could be verified in four of six candidate mutants by Southern blot analysis (see Fig. S1 in the supplemental material). One of them was subjected to conjugation with pCM157, a plasmid coding for Cre recombinase. After two passages in FSM, we found two clones that were no longer kanamycin resistant, due to the excision of the loxP site-flanked kanamycin resistance marker by Cre recombinase. One clone was cured from the cre expression plasmid pCM157 by repeated passaging in fresh FSM and was designated RU-1.
Analytical methods.
Iron concentrations were determined by a modified version (74) of the ferrozine assay (65) or by using a flame atomic absorption spectrometer (FAAS) (model AA240; Varian). For determination of iron content, cell pellets were washed in 20 mM Tris-HCl, 5 mM EDTA, pH 7.4, and digested as described previously (28). Transmission electron microscopy (TEM) analyses were performed as described previously (30). Siderophore production was monitored by a modified chrome azurol S (CAS) agar plate assay (41), and culture supernatants were measured using a CAS decoloration assay as previously described (62). Protein concentrations were measured with a bicinchoninic protein quantification kit (Sigma, Munich, Germany) according to the manufacturer's instructions. The average magnetic orientation of cell suspensions (Cmag) was assayed as previously described (59). Briefly, cells were aligned at different angles relative to a light beam by means of an external magnetic field. The ratio of the resulting maximum and minimum scattering intensities (Cmag) is correlated with the average number of magnetic particles and can be used for a qualitative assessment of magnetite formation.
Transmission Mössbauer spectroscopy (TMS).
For the determination of intracellular iron metabolites, microaerobic precultures (100 ml) of M. gryphiswaldense WT and RU-1 were grown in iron-replete FSM. The fur mutant was alternatively precultured in LIM supplemented with 10 μM 2,2′-dipyridyl. After three passages in the corresponding medium, all cultures were transferred to fresh FSM. RU-1 was grown in microaerophilic 1-liter batch cultures supplemented with a mixture of 20 μM 57Fe(citrate)2 and 20 μM 56Fe(citrate)2. Cells of M. gryphiswaldense WT were incubated with 20 μM 57Fe(citrate)2 in an oxystat fermentor under defined microaerophilic conditions using a modified protocol of large-scale cultivation of M. gryphiswaldense (28).
For TMS, cells were harvested by centrifugation at 4,700 rpm and 4°C. Pellets were washed, weighed, transferred into Delrin Mössbauer sample holders, frozen in liquid nitrogen, and kept at this temperature until measurement. TMS was performed in constant acceleration mode. The spectrometer was calibrated against α-iron at room temperature. Samples were measured in a continuous-flow cryostat (Oxford Instruments) above the Verwey transition of magnetite at 130 K. The 57Co source exhibiting an activity of 0.19 GBq was sealed in an Rh matrix at room temperature and was mounted on a constant velocity drive. The detector consisted of a proportional counter filled with argon-methane (90:10). Spectral data were buffered in a multichannel analyzer and transferred to a personal computer for further analysis by employing the Vinda program on an Excel 2003 platform. Spectra were analyzed by least-square fits of Lorentzian line shapes to the experimental data (25).
Cell fractionation and preparation of protein extracts.
For proteomic analysis, M. gryphiswaldense R3/S1 and RU-1 were grown in 100 ml iron-rich FSM (50 μM ferric citrate) or iron-depleted LIM plus 10 μM 2,2′-dipyridyl (<1 μM iron) under anaerobic conditions to log phase in nine parallels. All parallels of each condition were pooled, and cells were pelleted at 9,200 × g, washed (20 mM Tris-HCl, pH 7.4, 5 mM EDTA), and resuspended into ice-cold 20 mM Tris-HCl, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, pH 7.4. Cell suspensions were lysed by three passages through a French press, and cellular debris was removed by low-speed centrifugation. Cleared cell lysates were subjected for 30 min to centrifugation at 265,000 × g to separate cellular membranes, magnetosomes, and empty magnetosome vesicles from the soluble protein fraction (24). Pelleted membrane proteins were resuspended in 20 mM Tris-HCl, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, pH 7.4, 1% sodium dodecyl sulfate (SDS). Nonmagnetic fractions were prepared by subjecting cell extracts to magnetic columns and sucrose cushion centrifugation using a protocol for the isolation of magnetosomes as described previously (24) but omitting EDTA from buffers. All protein fractions were stored at −80°C until analysis.
2D gel electrophoresis.
For isoelectric focusing (IEF), protein extracts from the soluble fraction (500 μg protein) were loaded onto commercially available immobilized pH gradient (IPG) strips (pH 3-10 NL; Amersham Biosciences) according to the method of Büttner et al. (14). In the second dimension, polyacrylamide gels of 12.5% acrylamide and 2.6% bisacrylamide were used. The resulting two-dimensional (2D) gels were stained with colloidal Coomassie brilliant blue (CBB) as described previously (75).
Protein digestion, mass spectrometry, and data analysis.
Spots were cut from the 2D gels and transferred into microtiter plates. Proteins were tryptically digested using an Ettan spot handling workstation (GE Healthcare). Mass spectra of the protein fragments were measured by matrix-assisted laser desorption ionization-time of flight tandem mass spectrometry (MALDI-TOF-MS-MS) using a proteome analyzer 4800 (Applied Biosystems). The parameters for the measurements were set as described previously (76), except that the signal-to-noise ratio for the TOF-TOF measurements was raised to 10. Proteins were identified by searching an M. gryphiswaldense databank with the Mascot search engine (search parameters are described in reference 76). Differentially expressed proteins on the 2D gels were analyzed with Delta 2D software (Decodon, Greifswald, Germany) (76). For analysis of membrane proteins, 1D gel lanes were manually cut into 10 equal slices and the slices were digested with trypsin. Liquid chromatography (LC)-coupled mass spectrometry was performed as described previously (78). Ratios of identified peptide ion abundances higher than +2 or smaller than −2 were set as a threshold indicating significant changes.
Bioinformatics.
The protein sequences of E. coli Fur (NCBI accession no. AP_001321.1), Corynebacterium diphtheriae DtxR (NCBI accession no. AAA23302.1), and Rhizobium leguminosarum RirA (NCBI accession no. YP_766387.1) were used as a query in BLAST searches of the genomes of M. gryphiswaldense WT, M. magneticum AMB-1, and M. magnetotacticum MS-1 using the BLASTP algorithm 2.2.16 (4) with a cutoff E value of 1e−05 or an amino acid similarity of >30%. Sequence alignments and construction of similarity trees were performed using MEGA4 software (66). Sequences were aligned by ClustalW (default settings), and similarity trees were constructed using the minimal evolution (ME) method (53). Fur-like proteins with NCBI accession numbers ZP_00208795.1, ZP_00052390.2, and ZP_00209401.1 present in the genome assembly of M. magnetotacticum MS-1 (NCBI accession no. AAAP00000000) were omitted from analyses, since the whole-genome sequence contigs on which the three proteins are found share no homology to other Magnetospirillum sequences but have almost 100% identity to Methylobacterium species or 80% identity to Xylanimonas cellulosilytica DSM 15894 and thus are likely to represent contaminations.
RESULTS
Identification of a putative fur gene.
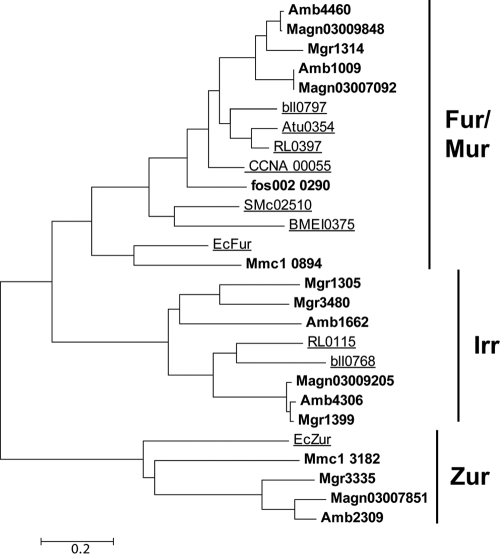
Using Corynebacterium diphtheriae DtxR (NCBI accession no. AAA23302.1) and R. leguminosarum RirA (NCBI accession no. YP_766387.1) as queries in BLASTP analysis, we failed to detect homologs of the diphtheria toxin repressor family (DtxR) or the rhizobial iron regulator RirA in the genome of M. gryphiswaldense. However, BLASTP searches with E. coli Fur (NCBI accession no. AP_001321.1) yielded five hits with significant similarities (>30%) (see Table S2 in the supplemental material). Closer inspection of the five candidate Fur proteins revealed that they fall into three different subfamilies of the Fur superfamily (Fig. 1). Three proteins (Mgr1305, Mgr1399, and Mgr3480) are more closely related to the Irr subfamily (63), whereas one protein (Mgr3335) belongs to the Zur family of putative Zn regulators (3). One single protein, Mgr1314, referred to herein as MgFur, belongs to the Fur/Mur subfamily, which comprises genuine iron- and manganese-responsive regulators (50). Mgfur is part of a putative polycistronic operon and is flanked upstream by a gene encoding a putative transcriptional regulator of the Ros/MucR family (Mgr1313) and downstream by a gene encoding a putative hemolysin-like protein (Mgr1315), as well as 13 additional genes that are transcribed in the same direction. Mgfur encodes a protein of 143 amino acid residues containing the highly conserved putative regulatory Fe-sensing site (i.e., S1) (48), consisting of amino acid residues H91, D93, E112, and H129, and the highly conserved structural Zn-binding site (i.e., S2), consisting of residues H37, E85, H94, and E105. RT-PCR analysis revealed that Mgfur was transcribed under all tested (i.e., iron-replete and -depleted) conditions (data not shown).
FIG. 1.
Similarity tree of alphaproteobacterial and E. coli Fur-like proteins. Sequences are designated by locus tags. Fur-like proteins of MTB are shown in bold. Proteins that have been characterized experimentally are underlined. Locus tags refer to M. gryphiswaldense (Mgr1305, Mgr1314, Mgr1399, Mgr3335, and Mgr3480), M. magneticum (Amb1009, Amb1662, Amb2309, Amb4306, and Amb4460), M. magnetotacticum (Magn03007092, Magn03007851, Magn03009848, and Magn03009205), Bradyrhizobium japonicum (bll0768 and bll0797), Agrobacterium tumefaciens (Atu0354), Rhizobium leguminosarum (RL0115 and RL0397), Caulobacter crescentus (CCNA_00055), uncultured MTB (fos002_0290), Sinorhizobium meliloti (SMc02510), Brucella melitensis (BMEI0375), E. coli (EcFur and EcZur), and Magnetococcus species (Mmc1_0894 and Mmc1_3182). A more extended tree showing characterized and noncharacterized Fur-like proteins is shown in Fig. S2 in the supplemental material.
We found that, within the genus Magnetospirillum, Fur is well conserved with respect to sequence (see Table S3 in the supplemental material) as well as its genomic localization within a highly conserved 11-kb region. However, while Mgfur is present in a single copy in M. gryphiswaldense, a second copy (with 79% identity to Amb4460 and 73% identity to MgFur at the amino acid level) is present in the genomes of the closely related species M. magneticum and M. magnetotacticum. Notably, in the latter strains the second ortholog is associated with a partial duplication of the mamAB operon (region R9 [43]), a 7-kb region which is absent from the genome of M. gryphiswaldense.
MgFur transcomplements an E. coli fur mutation.
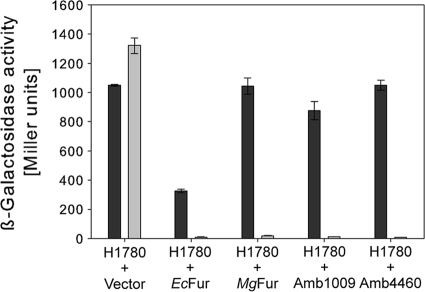
To test whether MgFur is a genuine iron-responsive regulator, the reporter strain E. coli H1780 was transcomplemented with pBBR1MCS-5Ptet, pBBR1MCS-5Ptet/amb1009, pBBR1MCS- 5Ptet/amb4460, pBBR1MCS-5Ptet/mgr1314, and pBBR1MCS-5Ptet/Ecfur, containing fur orthologs of M. magneticum, M. gryphiswaldense, and E. coli as a positive control. This strain harbors a lacZ reporter gene under the control of the promoter of the iron-regulated outer membrane protein gene fiu. Due to an undefined mutation of native fur, ß-galactosidase is constitutively expressed unless strain H1780 is transformed with plasmids containing a functional fur gene. Plasmids were transferred into E. coli H1780, and transformed strains were grown under iron-replete and iron-depleted conditions. ß-Galactosidase activities showed that MgFur and Fur proteins of M. magneticum AMB-1 and E. coli (EcFur) were able to bind to the fiu promoter and repress lacZ expression to similar extents under iron-replete conditions (Fig. 2). ß-Galactosidase activity was highest under iron-depleted conditions in all tested strains, indicating that binding of Fur to the fiu promoter was iron dependent. Strain H1780(pBBR1MCS-5Ptet/Ecfur) showed intermediate ß-galactosidase activities in the absence of iron, indicating that EcFur is able to repress lacZ expression without iron, similarly to results observed previously in several studies using high-copy-number plasmids for the expression of EcFur (38, 45). Regulation of lacZ in strain H1780 carrying an empty vector was the same irrespective of iron concentration, as ß-galactosidase activities were equally high under iron-replete and iron-depleted conditions. These results suggested that MgFur is functional in E. coli.
FIG. 2.
ß-Galactosidase activities of the fiu-lacZ fusion strain H1780 harboring the indicated plasmids and grown in LB under iron-limiting (200 μM 2,2-dipyridyl) (black bars) and iron-sufficient (100 μM FeCl3) (gray bars) conditions. The assays were performed in triplicate, and values are expressed as the means, with standard deviations displayed as error bars.
Generation and analysis of an M. gryphiswaldense fur mutant.
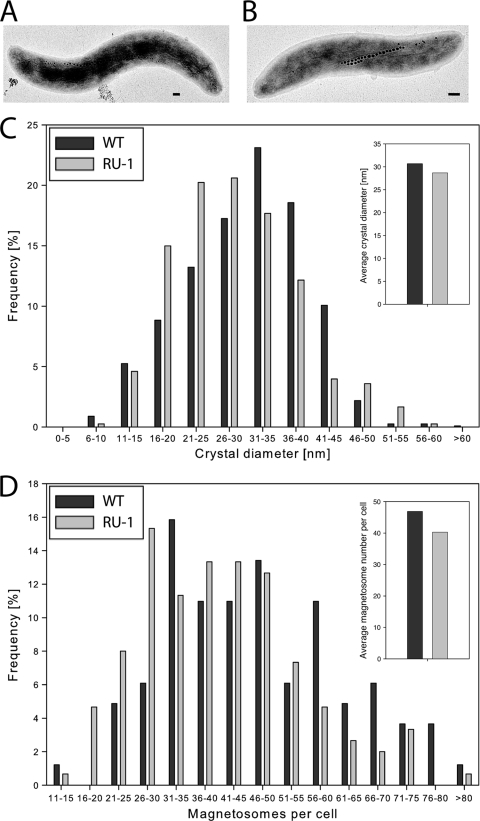
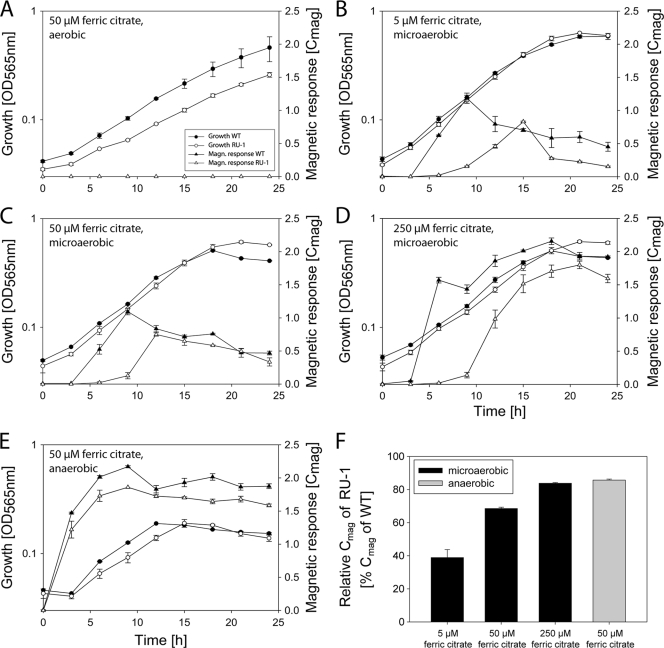
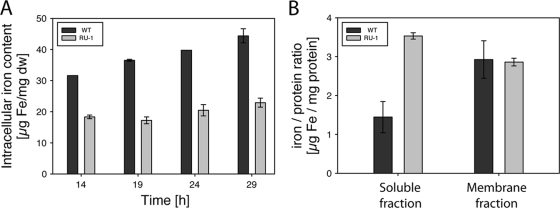
To analyze whether MgFur has iron-responsive regulatory functions in M. gryphiswaldense as well and to clarify the role of Fur in the biomineralization of magnetosomes, an unmarked fur mutant strain of M. gryphiswaldense was constructed by a cre-lox-based method (39), resulting in an unmarked in-frame deletion of fur. TEM analysis showed that the fur mutant strain RU-1 was still able to produce magnetosomes, although with diameters (28.6 ± 9.1 nm) and in numbers (40 ± 14.3 per cell) significantly reduced compared to those for the WT (46 ± 16.1 magnetosomes per cell and 30.6 nm ± 9.0 nm in diameter) (Mann-Whitney test; P ≤ 0.003) (Fig. 3). For further characterization, both strains were iron deprived by three passages in LIM supplemented with 10 μM 2,2′-dipyridyl, a medium supporting growth but not magnetite synthesis, until cellular magnetism was no longer detectable and subsequently inoculated into fresh LIM containing different iron concentrations to reinduce magnetite biomineralization. Under aerobic and anaerobic growth conditions, growth rates of the fur mutant were slightly lower than those of the WT, whereas under microaerobic conditions, RU-1 showed growth rates similar to those of the WT at all tested iron concentrations (Table 2). However, magnetosome formation in RU-1 became detectable by Cmag only about 3 h after that in the WT (Fig. 4 B). This delay was also observed with increased extracellular concentrations of ferric citrate (250 μM) (Fig. 4D). When RU-1 was cultured under anaerobic conditions, no difference in time course of magnetite formation was observed (Fig. 4E). In addition, the maximal Cmag values that were reached by RU-1 were significantly smaller than those reached by the WT. These differences were most pronounced (RU-1 Cmag reached only 40% of WT Cmag) at low iron concentrations (5 μM). Although Cmag values of the fur mutant increased with extracellular iron concentration to up to 85% of the WT value with 250 μM ferric citrate and under anaerobic conditions, they never did reach WT levels under any condition tested (Fig. 4F). Consistent with TEM and Cmag data, the total intracellular iron content after microaerobic growth with 50 μM iron was also reduced in the fur mutant by 50% compared to the level for the WT (Fig. 5 A). Transcomplementation of the fur mutant by a WT fur allele on pBBR1MCS-2fur resulted in partial restoration of the WT iron content (see Fig. S3 in the supplemental material).
FIG. 3.
Transmission electron micrographs of the WT (A) and RU-1 (B). Bar, 100 nm. (C) Magnetite crystal size distribution determined from 200 cells by TEM. (D) Distribution of magnetosome number per cell.
TABLE 2.
Growth rates (μ) and doubling times (tD) of the WT strain and RU-1 grown under different oxygen and iron concentrations
| Strain | Parameter | Resulta under the following culture conditions |
||||
|---|---|---|---|---|---|---|
| 2% O2 and: |
0% O2 and 50 μM Fe citrate | 21% O2 and 50 μM Fe citrate | ||||
| 5 μM Fe citrate | 50 μM Fe citrate | 250 μM Fe citrate | ||||
| WT | μ (h−1) | 0.169 (±0.010) | 0.162 (±0.001) | 0.165 (±0.010) | 0.148 (±0.015) | 0.121 (±0.013) |
| tD (h) | 4.10 (±0.24) | 4.29 (±0.02) | 4.19 (±0.26) | 4.68 (±0.68) | 5.73 (±0.60) | |
| RU-1 | μ (h−1) | 0.167 (±0.003) | 0.158 (±0.001) | 0.149 (±0.019) | 0.124 (±0.003) | 0.090 (±0.002) |
| tD (h) | 4.16 (±0.07) | 4.40 (±0.00) | 4.64 (±0.59) | 5.60 (±0.14) | 7.70 (±0.17) | |
Values are the sample means of at least replicate cultures. Sample standard deviations are given in parentheses.
FIG. 4.
(A to E) Levels of growth (optical density [OD] at 565 nm) and magnetic response (Cmag) of the WT and RU-1 grown under different conditions. (F) Relative maximal Cmag of RU-1 grown in LIM, determined from results shown in panels B to E. Data are from representative experiments done in duplicate. The entire experiment was repeated three times, with comparable results. Values are given as means ± standard deviations.
FIG. 5.
(A) Time courses of total intracellular iron content of the WT and RU-1 during growth in FSM under microaerobic conditions. Values are given as means ± standard deviations (SD) from three independent replicates. (B) Iron-to-protein ratios of WT and RU-1 nonmagnetic cytoplasmic and membrane-enriched protein fractions. Values are given as means ± SD from two independent replicates.
Unlike in other Magnetospirillum species (15, 44), no siderophores could be detected in M. gryphiswaldense under any tested condition in previous studies. However, although there is no clear genomic indication of siderophore synthesis, we cannot entirely exclude the possibility of the synthesis and use of primary catecholate-like metabolites as siderophores under some unspecified conditions, as described for M. magneticum (16). Since fur mutants of several other bacteria (e.g., Pseudomonas aeruginosa and Shewanella oneidensis) showed increased or constitutive production of siderophores (49, 68), we reassessed siderophore production using the CAS decoloration assay with supernatants from cultures grown under different iron concentrations and a modified plate growth CAS assay. However, again we were unable to detect siderophore production either in the fur mutant or in the WT (data not shown).
As fur mutants of E. coli are known to be susceptible to higher iron concentrations than the WT (70), we also checked for increased sensitivity of M. gryphiswaldense RU-1 to various metals (Fe, Mn, and Co) at different concentrations. Dose-response assays under microaerobic conditions revealed identical growth yields for the WT and RU-1 between 0 and 500 μM iron as well as 0 and 2 mM manganese (see Fig. S4A and B in the supplemental material). Only at very high concentrations of iron (>1,000 μM) and manganese (>3 mM) was growth of RU-1 increasingly inhibited relative to that of the WT. No differences with respect to growth yield could be observed in the presence of Co (5 to 100 μM) (data not shown). These data indicate a different role for MgFur than for EcFur.
Since growth of M. gryphiswaldense RU-1 was impaired under aerobic conditions, we also compared the sensitivities of the WT and RU-1 strains against the superoxide-producing agent paraquat (13). Growth in the presence of 5 μM paraquat resulted in growth yields of the WT being reduced by 40%, whereas growth of RU-1 was inhibited by 60% (see Fig. S4C in the supplemental material). At higher paraquat concentrations, both strains were equally inhibited. These results suggest that MgFur might be involved in the oxidative stress response of M. gryphiswaldense.
Mössbauer spectroscopic analysis of RU-1 reveals a large pool of iron bound to a ferritin-like component.
Previous Mössbauer experiments revealed that the intracellular iron pool of the WT grown under microaerobic conditions comprises mainly magnetite, a ferritin-like component, and a ferrous high-spin component (20). Here, we performed a comparative determination of intracellular iron metabolites and their contributions in M. gryphiswaldense WT and RU-1 by using transmission Mössbauer spectroscopy (TMS) analyses. Prior to TMS analyses, cultures of M. gryphiswaldense WT and RU-1 were passaged three times under microaerobic, iron-replete conditions. To study iron metabolites in iron-induced cells, an additional culture of RU-1 was passaged three times under microaerobic, iron-depleted conditions (RU-1 −Fe) until no magnetism was observed by Cmag measurements. After three passages, all cultures were transferred to fresh FSM supplemented with 40 μM 57Fe(citrate)2 (WT) or a mixture of 20 μM 57Fe(citrate)2 and 20 μM 56Fe(citrate)2 (RU-1) and cultivated under microaerobic conditions.
Mössbauer spectra are characterized by three different parameters: the isomer shift, δ; the quadrupole splitting, ΔEQ; and the magnetic hyperfine field, BHF. The isomer shift, δ, which originates from the electric monopole interaction between the nucleus and the electronic shell, is a measure of the degree of covalent bonding of the iron atom with a ligand and is also an attribute for the oxidation state of the iron atom. The quadrupole splitting, ΔEQ, originates from the electric quadrupole interaction between the nucleus and the electronic shell and is a measure for the symmetry of the metal chelate and for the covalent distribution of ligand-metal bonding. The magnetic hyperfine field, BHF, is a result of magnetic dipole interaction between the nucleus and electrons and generates six-line or even more complicated spectra. Whole-cell, late-log Mössbauer analyses revealed the presence of metabolites showing characteristics of a ferrous iron high-spin metabolite, ferric iron bound to a ferritin-like metabolite, and magnetite in both strains under all tested conditions (see Fig. S5 in the supplemental material). However, the relative contributions of the metabolites differed between the WT and the fur mutant strain (Table 3). Whereas ferritin-like metabolite (50.4%) and magnetite (49.2%) contributed almost equally to the intracellular iron pool of the WT, the most abundant iron species found in RU-1 (75.8% for RU-1 +Fe and 89.5% for RU-1 −Fe) exhibited Mössbauer parameters similar to those of ferritins. Magnetite accounted for only 22.2% (+Fe) and 8.9% (−Fe) of the intracellular iron pool of RU-1, confirming the reduced magnetite biomineralization in the fur mutant observed by TEM. In all samples, only a small relative contribution came from a ferrous iron high-spin, ferrochelatin-like iron species also found in many bacterial and fungal systems (11). However, while in the WT the ferrous iron high-spin metabolite contributed only 0.35% to the intracellular iron pool, in RU-1 this metabolite was increased to 2% (RU-1 +Fe) and 1.6% (RU-1 −Fe).
TABLE 3.
Mössbauer parameters of M. gryphiswaldense WT and RU-1
| Metabolite | Parameter | Result for strain |
||
|---|---|---|---|---|
| WT | RU-1 +Fe | RU-1 −Fe | ||
| Ferritin-like metabolite | δ (mms−1) | 0.45 | 0.45 | 0.46 |
| ΔEQ (mms−1) | 0.76 | 0.67 | 0.70 | |
| Contribution (%) | 50.4 | 75.8 | 89.5 | |
| Contribution (% × g−1)a | 23.3 | 38.3 | 32.6 | |
| Ferrous high-spin metabolite | δ (mms−1) | 1.27 | 1.28 | 1.28 |
| ΔEQ (mms−1) | 2.81 | 2.85 | 2.90 | |
| Contribution (%) | 0.35 | 2 | 1.6 | |
| Contribution (% × g−1) | 0.16 | 1 | 0.6 | |
| Magnetite, site A | δ (mms−1) | 0.37 | 0.32 | 0.32 |
| ΔEQ (mms−1) | 0 | 0 | 0 | |
| Contribution (%) | 16.4 | 7.4 | 3 | |
| Contribution (% × g−1) | 7.6 | 3.7 | 1.1 | |
| BHF (T) | 49.1 | 48.4 | 48.9 | |
| Magnetite, site B | δ (mms−1) | 0.75 | 0.82 | 0.82 |
| ΔEQ (mms−1) | 0 | 0 | 0 | |
| Contribution (%) | 32.8 | 14.8 | 5.9 | |
| Contribution (% × g−1) | 15.2 | 7.5 | 2.1 | |
| BHF (T) | 47.1 | 46.9 | 45 | |
| Ferritin-like metabolite/magnetite | FMR | 1.02 | 3.41 | 10.1 |
Relative contribution normalized by sample mass.
Since the contribution ratio of ferritin-like iron to magnetite (FMR) was changed from 1.02 in the WT to 3.4 and 10.1 in the fur mutant, these data suggested that an increased amount of iron was bound to proteins in RU-1. To test this assumption, we analyzed the iron-to-protein ratios of nonmagnetic cell fractions of the WT and RU-1 grown to late log phase under microaerobic conditions. The iron-protein ratio of the membrane fraction was 3 μg Fe/mg protein for both strains. However, in the soluble fraction, the iron-protein ratio of the WT was only 1.44 μg Fe/mg protein, whereas in the fur mutant, the iron-protein ratio was increased more than 2-fold (3.53 μg Fe/mg protein) (Fig. 5B), which was consistent with the estimations derived from TMS analysis.
Analyis of the putative Fur regulon.
To analyze putative targets of Fur, we performed a Fur titration assay (FURTA) (64). In a FURTA, high-copy-number plasmids carrying promoters with putative Fur-binding elements are introduced into E. coli H1717, a strain carrying a lacZ reporter construct under the control of the Fur-regulated promoter fhuF. If plasmids contain sequences capable of binding Fur, a titration of the repressor will occur, leading to expression of the lacZ reporter. E. coli strain H1717 was transformed with the high-copy-number plasmid pGEM-T Easy or pGEM-T Easy promoter construct from M. gryphiswaldense, including putative promoter regions of mamDC, mamAB, mms16 (apdA), mgr4079, and rplK as well as the promoter PfhuF of E. coli. PfhuF and PrplK served as controls, whereas PmamDC and PmamAB have been shown to be iron regulated (56). In addition, PmamDC of M. magneticum and M. magnetotacticum was previously predicted in silico to be Fur regulated (50). All strains formed pink colonies on iron-depleted MacConkey lactose agar (see Fig. S6 in the supplemental material), showing high ß-galactosidase activities, since EcFur does not repress the fhuF promoter in the absence of available iron. Strains harboring the plasmids pGEMPfhuF (positive control) and pGEMPmamDC also formed pink colonies on agar plates supplemented with 30 μM FeCl3, suggesting that Fur was titrated out by Fur binding sequences within the tested promoters. All other strains formed white or only slightly pinkish colonies on iron-replete agar plates, indicating that Fur represses the expression of lacZ in the presence of iron. These data also demonstrated that Fur is involved in the transcriptional regulation of at least some magnetosome genes.
Proteomic analysis of the Fur regulon.
To identify other putative constituents of the Fur regulon in addition to the tested targets, we performed a proteome-wide analysis of cytoplasmic and membrane-enriched protein fractions of the WT and the fur mutant grown under iron-replete and under iron-depleted conditions. In total, 719 proteins were identified in the membrane-enriched fractions by 1D LC-MS-MS analysis, and 735 spots were detected on 2D gels of the cytoplasmic fractions (see Fig. S7 in the supplemental material). By use of 1D LC-MS-MS proteomic analysis, 23 proteins whose genes are part of a 130-kb genomic magnetosome island (MAI), harboring most magnetosome genes (71), were identified. Eighteen of these identified genes are part of the mam and mms operons encoding magnetosome proteins. Analysis of the expression data revealed almost no differences in expression level of these identified MAI proteins between the WT and RU-1 (see Table S4 in the supplemental material). The only exception was Mms6, a protein that was reported to affect magnetite crystal formation in vitro (6), which showed an expression level reduced by 55% in RU-1 under iron-replete conditions compared to that for the WT. Mgr4109, a putative type I secretion system ATPase encoded within the MAI, was detected in RU-1 but not in the WT.
Significant changes in expression levels were observed for 14 proteins encoded outside the MAI (Table 4), of which 12 were upregulated in RU-1 and two were upregulated in the WT under iron-depleted and iron-replete conditions. Five of the proteins with altered expression levels are components of putative iron uptake systems, comprising all iron uptake systems predicted from the genome of M. gryphiswaldense (R. Uebe, unpublished data).
TABLE 4.
Proteins that are differentially expressed between the WT and RU-1 under different growth conditions (−Fe and +Fe), as identified by LC-MS-MSa
| Expression level in RU-1 vs WT | NCBI accession no. or locus tag | Protein identification/function | Molecular size (kDa) | No. of peptide ions |
Relative expression level in RU-1 |
||||
|---|---|---|---|---|---|---|---|---|---|
| WT |
RU-1 |
RU-1 −Fe/ WT −Fe | RU-1 +Fe/ WT +Fe | ||||||
| −Fe | +Fe | −Fe | +Fe | ||||||
| Upregulated | MGR0081 | TonB-dependent outer membrane receptor | 72 | NDb | ND | 17 | 23 | Unique | Unique |
| MGR0236 | Bacterial extracellular solute-binding protein | 36 | 12 | ND | 25 | 13 | 2.08 | Unique | |
| MGR0237 | Putative diguanylate cyclase/phosphodiesterase with PAS sensor domain | 76 | ND | ND | 10 | 13 | Unique | Unique | |
| MGR0662 | Hypothetical protein | 32 | ND | ND | 10 | 12 | Unique | Unique | |
| MGR0705 | Conserved hypothetical protein | 11 | ND | ND | 19 | 14 | Unique | Unique | |
| MGR0706 | Conserved hypothetical protein | 12 | 11 | ND | 26 | 28 | 2.36 | Unique | |
| MGR0707 | Conserved hypothetical protein | 12 | ND | ND | 17 | 14 | Unique | Unique | |
| MGR1021 | Periplasmic trypsin-like serine protease | 56 | ND | ND | 11 | 12 | Unique | Unique | |
| MGR1446 | FeoB2 | 83 | 10 | ND | 53 | 50 | 5.30 | Unique | |
| MGR1447 | FeoA2 | 9 | 13 | 11 | 31 | 34 | 2.38 | 3.09 | |
| MGR1593 | Transcriptional regulator ArsR family | 20 | ND | ND | 14 | 13 | Unique | Unique | |
| MGR4109 | HlyB type I secretion system ATPase | 81 | ND | ND | 11 | 11 | Unique | Unique | |
| Downregulated | MGR0698 | CydA cytochrome d ubiquinol oxidase subunit 1 | 58 | 10 | 11 | ND | ND | Unique (WT) | Unique (WT) |
| ABL14106 | FeoB1 | 76 | 40 | 50 | ND | 10 | Unique (WT) | −5.00 | |
Characteristics of proteins with putative relation to iron metabolism are shown in bold.
ND, not detected.
Mgr0236, a putative bacterial extracellular binding protein that displays 59.8% similarity to the periplasmic ferric iron binding protein FutA2 (7), showed a 2-fold-increased expression in RU-1 versus in the WT under iron-depleted conditions. Under iron-replete conditions, Mgr0236 was repressed in the WT but was still detectable in the fur mutant, as was also observed by 2D gel analysis (data not shown). Interestingly, mgr0236 is highly similar to mgr4079, a pseudogene copy of mgr0236, which encodes a protein truncated by the first 60 amino acids (82.7% similarity to Mgr0236) and which is located within the MAI of M. gryphiswaldense. Mgr0081, a TonB-dependent outer membrane receptor putatively involved in the uptake of iron-siderophore complexes, was detected in RU-1 but not in the WT. The ferrous iron transport system FeoAB2 (Mgr1447 and Mgr1446) was expressed in a 2-fold-larger amount in RU-1 than in the WT. In addition to these iron transport systems, other proteins putatively involved in metal metabolism showed different expression levels between RU-1 and the WT. This included a transcriptional regulator of the metal-binding ArsR family that was detectable in RU-1 but not in the WT. Three highly basic proteins, whose genes are colocalized with a putative heavy-metal-transporting P-type ATPase, also showed an increased expression level in the fur mutant. Other proteins that showed differential expression had no obvious relation to metal metabolism (Mgr0237, Mgr0662, Mgr1021, and Mgr4109).
The genome of M. gryphiswaldense encodes a second copy of the Feo iron uptake system, designated feoAB1, which was previously shown to have an accessory role in magnetite biomineralization (51). Under iron-depleted and -replete conditions, the second Feo system is expressed in smaller amounts in the fur mutant than in the WT. The iron-containing protein CydA, a cytochrome oxidase, was also expressed at higher levels in the WT than in RU-1, since it was detected only in the WT.
DISCUSSION
We identified and analyzed the genuine Fur-like iron regulator MgFur (Mgr1314), which is one of five predicted proteins of the Fur superfamily in M. gryphiswaldense, with four of them (including MgFur) being putative iron-responsive transcriptional regulators. This number is notably high compared to those for the genomes of other bacteria of the alphaproteobacterial clade, which contain between zero (e.g., Rickettsia and Ehrlichia species) and three (e.g., Bradyrhizobium japonicum) genes encoding iron-responsive regulators (50). The multitude of putative iron regulators may either reflect the need for very strict iron homeostasis in M. gryphiswaldense and indicate particularly fine-tuned and versatile iron regulation or simply represent functional redundancy.
The colocalization of fur homologues with magnetosome genes in several cultivated and uncultivated MTB (30), as well as the identification of putative Fur binding sites within the promoter regions of two magnetosome operons (50), suggested a close link between Fur and the regulation of biomineralization. However, our data indicate that MgFur is not essential for magnetite synthesis, as the fur mutant was still able to produce functional magnetite crystals, albeit fewer and smaller than those produced by the WT. The reduction of total iron accumulation by 50% in RU-1 was due to the lower content of magnetite as shown by Mössbauer (TMS) analysis and iron measurements. On the other hand, these analyses also revealed that, compared to the WT, in which iron is deposited mainly as magnetite, in RU-1 the largest proportion of bulk intracellular iron is bound to proteins in the nonmagnetic fraction, as indicated by a 2.4-fold-increased iron-to-protein ratio of the nonmagnetic fraction. In particular, the intracellular iron in RU-1 was bound mostly to an as-yet-unidentified ferritin-like metabolite, which appears to be upregulated in the mutant compared to the level in the wild type when incubated under identical conditions, based on previous analyses (20). A similar effect was observed in a fur mutant of Helicobacter pylori (9), where a deregulation of the iron storage protein ferritin (Pfr) was observed, leading to higher expression rates of Pfr. In contrast, in an E. coli fur mutant, ferritin-bound iron was decreased, resulting in 2.5-fold-lower intracellular iron contents (1), whereas relative intracellular levels of ferrous iron were substantially increased, as detected by TMS (B. Matzanke, unpublished data). Thus, although MgFur was able to substitute for EcFur in E. coli in an iron-dependent manner, our observations argue for a somewhat distinct regulatory role in M. gryphiswaldense.
Although levels of intracellular ferrous iron detectable by TMS were relatively low in both M. gryphiswaldense WT and RU-1 compared to observations in E. coli, we found that ferrous iron signals were significantly increased in TMS of M. gryphiswaldense RU-1. The increase in protein-bound iron, and in particular the increased proportion of free ferrous iron, might explain the observed sensitivity of RU-1 against O2- and paraquat-induced oxidative stress, since ferrous iron promotes the generation of radical oxygen species via the Fenton reaction (77), and we did not observe any changes in expression levels of proteins of the oxidative stress response. Unexpectedly, the fur mutant was also growth impaired under anaerobic conditions, indicating that the increased intracellular iron concentration interferes with enzymes of the anaerobic metabolism.
Although putative Fur binding sites within the promoter regions of magnetosome operons were predicted in silico (50) and in part also experimentally confirmed in this study, proteomic analyses revealed that the expression levels of most detected magnetosomal proteins were unaffected by the deletion of fur. The only exception is the magnetosome protein Mms6, which showed a decreased expression in RU-1 grown under iron-sufficient conditions. Therefore, we cannot entirely exclude the possibility that the magnetosomal mamDC promoter exhibits some unspecific affinity to EcFur. Differential expression patterns were observed for several proteins putatively involved in iron uptake for general iron metabolism, indicating an important role of Fur in iron homeostasis of M. gryphiswaldense. Remarkably, Mgr0532 and Mgr0533, representing putative bacterioferritins, did not exhibit differential expression in RU-1 versus the WT, suggesting that they are not identical with the ferritin-like constituent that caused an increased signal in TMS. However, it cannot be excluded that the increased ferritin-like pool is a result of increased iron binding to bacterioferritin due to higher intracellular iron concentrations. In summary, the proteomic approach revealed only 14 proteins whose expression was significantly altered between the WT and RU-1. This is consistent with observations of several fur mutants, where Fur plays only a minor role in iron homeostasis (17, 45, 72), compared to the large Fur regulon of E. coli, in which up to 100 genes are down- or upregulated by the direct or indirect effect of Fur (26, 40). In several alphaproteobacteria, proteins other than Fur, such as RirA (69, 73) and Irr (63), have taken over the function of a global iron-responsive regulator. By genome analysis of M. gryphiswaldense, we also identified three proteins (Mgr1305, Mgr1399, and Mgr3480) within the Fur superfamily which are more closely related to the Irr subfamily (63) (Fig. 1). It seems possible that these Irr-like proteins are also involved in more complex regulatory networks overlapping the MgFur regulon. A recent study addressed the function of a presumptive Fur-like protein of M. gryphiswaldense (Mgr1399) which, however, was assigned as Irr-like in our study. Supposedly, deletion of Mgr1399 resulted in a nonnmagnetic phenotype (29). However, in the absence of transcomplementation experiments these results are not conclusive, given the high genetic instability of the magnetic phenotype that gives rise to frequent spontaneous mutations within the MAI of M. gryphiswaldense (71). Therefore, future work is required to study the contribution of the Irr-like proteins to iron homeostasis in more detail.
In conclusion, our data demonstrate that MgFur is involved in iron homeostasis and, to a lesser extent, also affects magnetite biomineralization. Since most magnetosome proteins exhibited similar levels of expression between the WT and RU-1, the reduced magnetite synthesis most likely is caused by indirect consequences of the deregulated phenotype. For example, it could be that the increased uptake of iron into the cytoplasm and its subsequent sequestration by cytoplasmic proteins lead to a decreased pool of iron available for magnetite biomineralization in the fur mutants, whereas in the WT, MgFur might be involved in balancing the competing demands for biochemical iron supply and magnetite biomineralization (20). A possible explanation for the observed delay of magnetite synthesis in iron-induced cells grown under microaerobic, but not anaerobic, conditions might be that the increased pool of cytoplasmic iron-binding proteins has to be saturated before iron becomes available for magnetite biomineralization. However, further studies including the identification and biochemical characterization of individual iron-sequestering cellular constituents are required to provide deeper insights into the regulation of biomineralization.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (Schu 1080/6-2).
We thank Klaus Hantke, University of Tübingen, and Gregor Grass, University of Halle, for their gift of strains E. coli H1717 and E. coli H1780. We thank Anna Pollithy for help with cloning.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y. S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, B.-E., J. Cha, E.-J. Lee, A.-R. Han, C. Thompson, and J.-H. Roe. 2006. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59:1848-1858. [DOI] [PubMed] [Google Scholar]
- 3.Althaus, E. W., C. E. Outten, K. E. Olson, H. Cao, and T. V. O'Halloran. 1999. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry 38:6559-6569. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 6.Arakaki, A., J. Webbs, and T. Matsunaga. 2003. A novel protein tightly bound to bacterial magnetite particles in Magnetospirillum magnetotacticum strain AMB-1. J. Biol. Chem. 278:8745-8750. [DOI] [PubMed] [Google Scholar]
- 7.Badarau, A., S. J. Firbank, K. J. Waldron, S. Yanagisawa, N. J. Robinson, M. J. Banfield, and C. Dennison. 2008. FutA2 is a ferric binding protein from Synechocystis PCC 6803. J. Biol. Chem. 283:12520-12527. [DOI] [PubMed] [Google Scholar]
- 8.Baeuerlein, E., and D. Schüler. 1995. Biomineralisation: iron transport and magnetite crystal formation in Magnetospirillum gryphiswaldense. J. Inorg. Biochem. 59:107. [Google Scholar]
- 9.Bereswill, S., S. Greiner, A. H. M. van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Böhnke, R., and B. F. Matzanke. 1995. The mobile ferrous iron pool in Escherichia coli is bound to a phosphorylated sugar derivative. Biometals 8:223-230. [DOI] [PubMed] [Google Scholar]
- 12.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 13.Bus, J. S., S. D. Aust, and J. E. Gibson. 1974. Superoxide- and singlet oxygen-catalyzed lipid peroxidation as a possible mechanism for paraquat (methyl viologen) toxicity. Biochem. Biophys. Res. Commun. 58:749-755. [DOI] [PubMed] [Google Scholar]
- 14.Büttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mäder, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 15.Calugay, R. J., H. Miyashita, Y. Okamura, and T. Matsunaga. 2003. Siderophore production by the magnetic bacterium Magnetospirillum magneticum AMB-1. FEMS Microbiol. Lett. 218:371-375. [DOI] [PubMed] [Google Scholar]
- 16.Calugay, R. J., H. Takeyama, D. Mukoyama, Y. Fukuda, T. Suzuki, K. Kanoh, and T. Matsunaga. 2006. Catechol siderophore excretion by magnetotactic bacterium Magnetospirillum magneticum AMB-1. J. Biosci. Bioeng. 101:445-447. [DOI] [PubMed] [Google Scholar]
- 17.da Silva Neto, J. F., V. S. Braz, V. C. S. Italiani, and M. V. Marques. 2009. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res. 37:4812-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faivre, D., L. H. Böttger, B. F. Matzanke, and D. Schüler. 2007. Intracellular magnetite biomineralization in bacteria proceeds by a distinct pathway involving membrane-bound ferritin and an iron(II) species. Angew. Chem. Int. Ed. Engl. 46:8495-8499. [DOI] [PubMed] [Google Scholar]
- 21.Faivre, D., N. Menguy, M. Posfai, and D. Schüler. 2008. Environmental parameters affect the physical properties of fast-growing magnetosomes. Am. Mineral. 93:463-469. [Google Scholar]
- 22.Faivre, D., and D. Schüler. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875-4898. [DOI] [PubMed] [Google Scholar]
- 23.Grünberg, K., E. C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunnlaugsson, H. 2006. A simple model to extract hyperfine interaction distributions from Mössbauer spectra. Hyperfine Interact. 167:851-854. [Google Scholar]
- 26.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 27.Hantke, K. 1987. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol. Gen. Genet. 210:135-139. [DOI] [PubMed] [Google Scholar]
- 28.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 29.Huang, Y., W. Zhang, W. Jiang, C. Rong, and Y. Li. 2007. Disruption of a fur-like gene inhibits magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. Biochemistry (Mosc.) 72:1247-1253. [DOI] [PubMed] [Google Scholar]
- 30.Jogler, C., W. Lin, A. Meyerdierks, M. Kube, E. Katzmann, C. Flies, Y. Pan, R. Amann, R. Reinhardt, and D. Schüler. 2009. Toward cloning of the magnetotactic metagenome: identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl. Environ. Microbiol. 75:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jogler, C., and D. Schüler. 2009. Genomics, genetics, and cell biology of magnetosome formation. Annu. Rev. Microbiol. 63:501-521. [DOI] [PubMed] [Google Scholar]
- 32.Johnston, A., J. Todd, A. Curson, S. Lei, N. Nikolaidou-Katsaridou, M. Gelfand, and D. Rodionov. 2007. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other α-proteobacteria. Biometals 20:501-511. [DOI] [PubMed] [Google Scholar]
- 33.Katzmann, E., A. Scheffel, M. Gruska, J. Plitzko, and D. Schüler. 2010. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome biomineralization and chain assembly in Magnetospirillum gryphiswaldense. Mol. Microbiol. doi: 10.1111/j.1365.2958.2010.07202.x. [DOI] [PubMed]
- 34.Kawaguchi, R., J. Burgess, and T. Matsunaga. 1992. Phylogeny and 16S rRNA sequence of Magnetospirillum sp. AMB-1, an aerobic magnetic bacterium. Nucleic Acids Res. 20:1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komeili, A., Z. Li, D. K. Newman, and G. J. Jensen. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242-245. [DOI] [PubMed] [Google Scholar]
- 36.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 37.Lee, J., and J. Helmann. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485-499. [DOI] [PubMed] [Google Scholar]
- 38.Lowe, C. A., A. H. Asghar, G. Shalom, J. G. Shaw, and M. S. Thomas. 2001. The Burkholderia cepacia fur gene: co-localization with omlA and absence of regulation by iron. Microbiology 147:1303-1314. [DOI] [PubMed] [Google Scholar]
- 39.Marx, C., and M. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 40.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 41.Milagres, A. M. F., A. Machuca, and D. Napoleao. 1999. Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J. Microbiol. Methods 37:1-6. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Murat, D., A. Quinlan, H. Vali, and A. Komeili. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. U. S. A. 107:5593-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paoletti, L. C., and R. P. Blakemore. 1986. Hydroxamate production by Aquaspirillum magnetotacticum. J. Bacteriol. 167:73-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker, D., R. M. Kennan, G. S. Myers, I. T. Paulsen, and J. I. Rood. 2005. Identification of a Dichelobacter nodosus ferric uptake regulator and determination of its regulatory targets. J. Bacteriol. 187:366-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199-1210. [DOI] [PubMed] [Google Scholar]
- 47.Platero, R., L. Peixoto, M. R. O'Brian, and E. Fabiano. 2004. Fur is involved in manganese-dependent regulation of mntA (sitA) expression in Sinorhizobium meliloti. Appl. Environ. Microbiol. 70:4349-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pohl, E., J. C. Haller, A. Mijovilovich, W. Meyer-Klaucke, E. Garman, and M. L. Vasil. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903-915. [DOI] [PubMed] [Google Scholar]
- 49.Prince, R. W., C. D. Cox, and M. L. Vasil. 1993. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J. Bacteriol. 175:2589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodionov, D., M. Gelfand, J. Todd, A. Curson, and A. Johnston. 2006. Computational reconstruction of iron- and manganese-responsive transcriptional networks in α-proteobacteria. PLoS Comput. Biol. 2:1568-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rong, C., Y. Huang, W. Zhang, W. Jiang, Y. Li, and J. Li. 2008. Ferrous iron transport protein B gene (feoB1) plays an accessory role in magnetosome formation in Magnetospirillum gryphiswaldense strain MSR-1. Res. Microbiol. 159:530-536. [DOI] [PubMed] [Google Scholar]
- 52.Rudolph, G., H. Hennecke, and H.-M. Fischer. 2006. Beyond the Fur paradigm: iron-controlled gene expression in rhizobia. FEMS Microbiol. Rev. 30:631-648. [DOI] [PubMed] [Google Scholar]
- 53.Rzhetsky, A., and M. Nei. 1992. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 9:945-967. [Google Scholar]
- 54.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 55.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. H. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schübbe, S., C. Würdemann, J. Peplies, U. Heyen, C. Wawer, F. O. Glöckner, and D. Schüler. 2006. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 72:5757-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schüler, D., and E. Baeuerlein. 1998. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 180:159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schüler, D., and E. Baeuerlein. 1996. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch. Microbiol. 166:301-307. [DOI] [PubMed] [Google Scholar]
- 59.Schüler, D., R. Uhl, and E. Baeuerlein. 1995. A simple light scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol. Lett. 132:139-145. [Google Scholar]
- 60.Schultheiss, D., R. Handrick, D. Jendrossek, M. Hanzlik, and D. Schüler. 2005. The presumptive magnetosome protein Mms16 is a poly(3-hydroxybutyrate) granule-bound protein (phasin) in Magnetospirillum gryphiswaldense. J. Bacteriol. 187:2416-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89-94. [DOI] [PubMed] [Google Scholar]
- 62.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 63.Small, S. K., S. Puri, I. Sangwan, and M. R. O'Brian. 2009. Positive control of ferric siderophore receptor gene expression by the Irr protein in Bradyrhizobium japonicum. J. Bacteriol. 191:1361-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stojiljkovic, I., A. J. Bäumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 65.Stookey, L. L. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779-781. [Google Scholar]
- 66.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 67.Tao, X., N. Schiering, H.-Y. Zeng, D. Ringe, and J. R. Murphy. 1994. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol. Microbiol. 14:191-197. [DOI] [PubMed] [Google Scholar]
- 68.Thompson, D. K., A. S. Beliaev, C. S. Giometti, S. L. Tollaksen, T. Khare, D. P. Lies, K. H. Nealson, H. Lim, J. Yates, C. C. Brandt, J. M. Tiedje, and J. Zhou. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (Fur) mutant of Shewanella oneidensis: possible involvement of Fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Todd, J. D., G. Sawers, and A. W. B. Johnston. 2005. Proteomic analysis reveals the wide-ranging effects of the novel, iron-responsive regulator RirA in Rhizobium leguminosarum bv. viciae. Mol. Genet. Genomics 273:197-206. [DOI] [PubMed] [Google Scholar]
- 70.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ullrich, S., M. Kube, S. Schübbe, R. Reinhardt, and D. Schüler. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 187:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Vliet, A. H. M., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180:5291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Viguier, C., P. O'Cuiv, P. Clarke, and M. O'Connell. 2005. RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol. Lett. 246:235-242. [DOI] [PubMed] [Google Scholar]
- 74.Viollier, E., P. W. Inglett, K. Hunter, A. N. Roychoudhury, and P. Van Cappellen. 2000. The ferrozine method revisited: Fe(II)/Fe(III) determination in natural waters. Appl. Geochem. 15:785-790. [Google Scholar]
- 75.Voigt, B., T. Schweder, D. Becher, A. Ehrenreich, G. Gottschalk, J. Feesche, K.-H. Maurer, and M. Hecker. 2004. A proteomic view of cell physiology of Bacillus licheniformis. Proteomics 4:1465-1490. [DOI] [PubMed] [Google Scholar]
- 76.Voigt, B., T. Schweder, M. J. J. B. Sibbald, D. Albrecht, A. Ehrenreich, J. Bernhardt, J. Feesche, K.-H. Maurer, G. Gottschalk, J. M. v. Dijl, and M. Hecker. 2006. The extracellular proteome of Bacillus licheniformis grown in different media and under different nutrient starvation conditions. Proteomics 6:268-281. [DOI] [PubMed] [Google Scholar]
- 77.Winterbourn, C. C. 1995. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol. Lett. 82-83:969-974. [DOI] [PubMed] [Google Scholar]
- 78.Wolff, S., H. Hahne, M. Hecker, and D. Becher. 2008. Complementary analysis of the vegetative membrane proteome of the human pathogen Staphylococcus aureus. Mol. Cell. Proteomics 7:1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang, J., I. Sangwan, A. Lindemann, F. Hauser, H. Hennecke, H.-M. Fischer, and M. R. O'Brian. 2006. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol. Microbiol. 60:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.