Abstract
The reactive α-oxoaldehydes such as glyoxal (GO) and methylglyoxal (MG) are generated in vivo from sugars through oxidative stress. GO and MG are believed to be removed from cells by glutathione-dependent glyoxalases and other aldehyde reductases. We isolated a number of GO-resistant (GOr) mutants from Escherichia coli strain MG1655 on LB plates containing 10 mM GO. By tagging the mutations with the transposon TnphoA-132 and determining their cotransductional linkages, we were able to identify a locus to which most of the GOr mutations were mapped. DNA sequencing of the locus revealed that it contains the yqhC gene, which is predicted to encode an AraC-type transcriptional regulator of unknown function. The GOr mutations we identified result in missense changes in yqhC and were concentrated in the predicted regulatory domain of the protein, thereby constitutively activating the product of the adjacent gene yqhD. The transcriptional activation of yqhD by wild-type YqhC and its mutant forms was established by an assay with a β-galactosidase reporter fusion, as well as with real-time quantitative reverse transcription-PCR. We demonstrated that YqhC binds to the promoter region of yqhD and that this binding is abolished by a mutation in the potential target site, which is similar to the consensus sequence of its homolog SoxS. YqhD facilitates the removal of GO through its NADPH-dependent enzymatic reduction activity by converting it to ethadiol via glycolaldehyde, as detected by nuclear magnetic resonance, as well as by spectroscopic measurements. Therefore, we propose that YqhC is a transcriptional activator of YqhD, which acts as an aldehyde reductase with specificity for certain aldehydes, including GO.
Glyoxal (GO) is a reactive α-oxoaldehyde formed by the oxidative degradation of glucose, by lipid peroxidation, and by the autoxidation of ascorbic acid (21, 38). Due to its two reactive carbonyl groups, GO is capable of inducing cellular damage through multiple mechanisms. For example, GO can modify proteins and nucleic acids, causing protein malfunction and mutagenesis, respectively (12, 30). It has been reported that the advanced glycation (nonenzymatic glycosylation) end products formed by GO are associated with pathophysiological changes found in aging and neurodegenerative disease (1, 2, 18). Like GO, methylglyoxal (MG) and 3-deoxyglucosone also cause protein glycation and other types of cellular damage (12, 30).
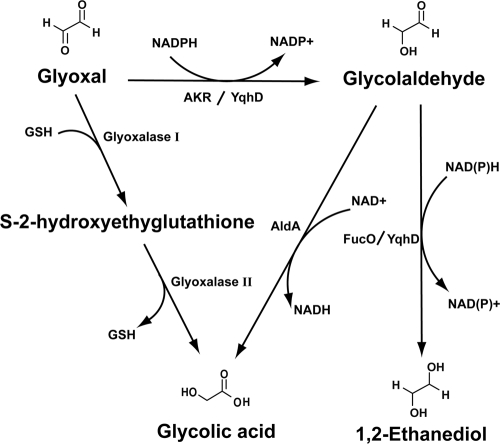
GO can be converted to glycolaldehyde, presumably by an uncharacterized aldo-keto reductase (AKR) or aldehyde reductase (Fig. 1). Glycolaldehyde can be further oxidized or reduced to glycolic acid or 1,2-ethandiol, respectively. Glycolaldehyde oxidation is mediated by aldehyde dehydrogenase and NAD+ (5, 14), while its reduction is accomplished by 1,2-propanediol oxidoreductase using NADH as a cofactor (7, 23). Glycolic acid is also believed to be generated directly from GO by glutathione (GSH)-dependent glyoxalases (31). Unlike GO, detoxification of MG is quite well characterized and is known to involve the glyoxalase system and AKRs (11, 13, 32, 34, 35). The glyoxalase system consists of glyoxalases I and II (gloA and gloB, respectively), which require GSH as a cofactor (16, 36), and which produce lactate from MG. The AKRs, including YafB, YqhE, and YghZ, are also reported to detoxify MG by generating hydroxyacetone using NADPH (13).
FIG. 1.
Proposed intracellular conversion of GO. GO can be reduced to glycolaldehyde and further to 1,2-ethandiol through involvement of the AKRs, including YafB, YqhE, and YghZ. The present study showed that YqhD mediates two-step reduction of GO to 1,2-ethandiol using NADPH. FucO can also mediate glycolaldehyde reduction (7, 23). Glycolic acid can be inefficiently produced from GO by the glyoxalase system (31), and it is also produced by the oxidation of glycolaldehyde involving AldA (5, 14, 31).
Although the YqhD protein was previously predicted to be a group III (iron-activated) NADP-dependent alcohol dehydrogenase based on sequence analysis, YqhD was recently proposed to function as an aldehyde reductase, detoxifying aldehyde compounds, including propanal, butanal, and acrolein, which are likely derived from lipid peroxidation (26). The dimeric structure of YqhD has been determined to contain NADPH and zinc at its active site (29). YqhD is structurally analogous to other enzymes, such as FucO and AdhE, that catalyze l-lactaldehyde and acetaldehyde, respectively (9, 22). YqhD has been characterized as an enzyme involved in GO metabolism, which is enzymatically similar to but structurally different from other members of the AKR family.
In the present study, we isolated a number of GO-resistant (GOr) mutants that overproduce the aldehyde reductase YqhD. Based on analysis of GOr mutations, we were able to characterize YqhC as a transcriptional activator of the yqhD gene that binds directly to the putative promoter region of yqhD. Interestingly, the GOr mutations we characterized are located in the regulatory region of YqhC and not in the DNA-binding region.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the strains used were derivatives of Escherichia coli K-12 (Table 1). MG1655 was used as the wild-type strain for gene disruption. Disruption of the yqhD and yqhE genes was achieved by using a previously described method for chromosomal gene inactivation (8). Other AKR mutants were obtained from the E. coli genome project (University of Wisconsin, Madison) and transferred to MG1655 by P1 transduction (15). The BL21(DE3) strain (Novagen) was used for overproduction and purification of protein.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | F−, λ− | Lab collection |
| CH3 | MG1655 yqhC(P63S) | This study |
| CH5 | MG1655 yqhC(A109D) | This study |
| CH6 | MG1655 yqhC(R61H) | This study |
| CH9 | MG1655 yqhC(P63Q) | This study |
| CH13 | MG1655 yqhC(V99E) | This study |
| CH20 | MG1655 yqhC(stop) (E112 nonsense)b | This study |
| CH21 | MG1655 ΔyqhD::kan | This study |
| CH26 | MG1655 ΔgloA::kan | Lab collection |
| CH27 | MG1655 ΔyqhD::tet ΔgloA::kan | This study |
| CH28 | MG1655 ΔyqhD::tet ΔgloA::kan | This study |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| CH29 | MG1655 ΔlacZ | This study |
| CH30 | CH3 ΔlacZ | This study |
| CH31 | CH20 ΔlacZ | This study |
| Plasmids | ||
| pACYC184 | ori(p15A); Tcr Cmr | Lab collection |
| pCH1 | pACYC184 yqhD | This study |
| pCH2 | pET15b yqhD | This study |
| pET-YafB-His | pET21b yafB | 13 |
| pET-YqhE-His | pET21b yqhE | 13 |
| pET-YghZ-His | pET21b yghZ | 13 |
| pON1 | pET21a yqhC | This study |
| pRS415 | Apr, lacZ operon fusion vector | 28 |
| pON2 | pRS415 yqhD operator | This study |
Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance.
A nonsense mutation (334G→T, counted from the first ATG).
Isolation and localization of GOr mutations.
GOr mutants, including yqhC(P63S), were isolated from the E. coli MG1655 strain on LB plates containing 10 mM GO. For mapping, a λTnphoA-132 hopping phage carrying a tet gene encoding tetracycline resistance was used for insertional mutagenesis (39). After infection of the GOr mutant [yqhC(P63S)] cells with the phage, more than 10,000 independent clones with transposon insertions were obtained, thereby generating a yqhC(P63S)::TnphoA-132 library. By infecting the library of cells with P1 phage, we generated a P1 pool containing different genomic fragments of TnphoA-132. These phages were used to identify insertions near yqhC(P63S) after transfection into wild-type MG1655 cells and selection for both GO- and tetracycline-resistant strains.
Transposon insertion sites were identified by inverse PCR. Chromosomal DNA was prepared and completely digested with TaqI restriction enzyme, which cuts near the 5′ end of TnphoA-132, generating a fragment containing the 5′ end of the transposon with a flanking sequence. The digested chromosomal DNA was then ligated to form circular DNA. PCR amplification was performed with a pair of outwardly directed primers matched to the 5′ end of the transposon: the complement of bases 31 to 55 (TNPO, directed to the 5′ end) and the sequence of bases 67 to 91 (TNPI, directed to the 3′ end). The amplified DNA was purified after agarose gel electrophoresis and sequenced using TNPO as a primer. The sequences flanking the transposon insertion were searched for similarity against the E. coli genome database using the BLASTN program (3). After determining the locations of tags through inverse PCR and sequencing, the sites of GOr mutations were mapped by using cotransduction frequencies obtained with the insertion tags. Finally, genes responsible for GOr mutations were identified by sequencing genes in the candidate region.
Sample preparation and metabolite analysis by NMR.
A Bruker AVANCE-400 nuclear magnetic resonance (NMR) spectrometer equipped with a temperature controller was used for NMR experiments. Samples were placed in a 5-mm NMR tube and kept at 28°C during measurement. Quantitative 1H-NMR measurements were made by using a 300 pulse with a long relaxation delay. The duration of acquisition was approximately 5 min for each NMR spectrum. All measurements were made in 600 μl of solution with 10% D2O as the locking substance. To detect GO and MG reducing activity, purified YqhD protein was dialyzed for 15 h against three successive changes of 50 mM potassium phosphate buffer (pH 7.0). The enzymatic reaction for GO or MG contained purified YqhD (40 μg), GO (10 mM), or MG (3 mM), and coenzyme (2 mM NADPH) in buffer (50 mM potassium phosphate [pH 7.0]) with D2O. NMR data were collected at 0 and 6 h after mixing.
Purification of proteins.
The yqhD gene was inserted into the pET15b vector (Novagen) and transformed into the BL21(DE3) pLysS strain. The transformed cells were grown at 37°C to the mid-exponential phase (optical density at 600 nm [OD600] of 0.5) in LB broth with ampicillin (100 μg ml−1). Proteins were induced by adding 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were further grown for 4 h, harvested, and resuspended in binding buffer (20 mM Tris-Cl [pH 7.9], 5 mM imidazole, 500 mM NaCl). After disruption by sonication, cell debris was removed by centrifugation at 15,000 × g for 15 min, and protein was purified according to standard procedures for Ni-NTA columns (Novagen). Concentration and purity were determined by using the Bradford reagent and also by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
EMSA.
DNA probes containing the YqhC binding site were generated by PCR and end labeled using [γ-32P]ATP (GE Healthcare) and polynucleotide kinase (Takara). Unincorporated [γ-32P]ATP was removed by using a nucleotide removal column (Intron). For the electrophoretic mobility shift assay (EMSA), reactions were carried out using 20 μl of DNA probe (100 fmol), 1 μg of poly(dI-dC), and 0 to 200 fmol of purified protein. The probes were synthesized by PCR using 10 pmol of the following primers: 5′-GCAATTTTGTAGCATTTCTC-3′ and 5′-GTTTTTCATTGTGATCGCC-3′. To generate the mutant probes, the following site-specific mutant oligonucleotides were used: 5′-CTCGGGATCCAGATGTAGGGAGAAATGCTACAAAATTGCGC-3′ and 5′-CCCTACATCTGGATCCCGAGGCAAGACATTGGCAGAAATG-3′ for mutant 1 and 5′-TTTTACTAAATGATACGGGAGAGCCAGGGCGCTGGCGATC-3′ and 5′-TCCCGTATCATTTAGTAAAATGTCTTGCCTATTTCTCCAG-3′ for mutant 2. After synthesis of the mutant templates, the mutant probes were synthesized by PCR using the same primers used for preparation of the wild-type probe. The binding buffer contained 50 mM KCl, 20 mM Tris-Cl (pH 7.9), 1 mM dithiothreitol, 10% glycerol, and 125 μg of bovine serum albumin/ml. The mixture was kept on ice for 45 min, loaded onto an 8% low-ionic-strength polyacrylamide gel, and electrophoresed at 100 V in Tris-borate-EDTA buffer for 9 h at 4°C.
Determination of the yqhD transcriptional start site.
Total RNA was prepared from yqhC(P63S) cells using Tri-Reagent solution (MRC). For 5′-RACE (rapid amplification of cDNA ends) analyses, 5-μg aliquots of total RNA were reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) with 0.5 pmol of the primer 5′-GCTGCGGCGATAAATTTGG-3′ and 10 μl of 150 mM NaOH. The mixture was incubated at 65°C for 1 h for the removal of RNA. A PCR purification kit (Labopass, Korea) was used to purify the DNA. After the homopolymeric A-tail was added to the 3′ end of the cDNA using terminal transferase (Roche), the sample was purified again by using a PCR purification kit (Labopass). The tailed cDNA served as a template for PCR amplification using the following primers: 5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTTTTT-3′ and 5′-GCTGCGGCGATAAATTTGG-3′. The PCR product was sequenced by using an ABI 3100 sequencer.
Construction of lac reporter plasmid and β-galactosidase assay.
The pRS415 plasmid was used to construct a transcriptional fusion for reporter assay (28). The DNA fragment containing the putative yqhD operator sequences (425 bp, including 5′ 60 bp of the open reading frame) were inserted into EcoRI-BamHI site of lacZ 5′-untranslated region of pRS415, which was introduced into recipient cells. The β-galactosidase assay was performed according to the method of Miller (20). Cells were grown in LB to an OD600 of 1 and treated with 0 or 1 mM GO for 70 min. Then, 0.2-ml portions of the cell cultures were suspended in Z-buffer to a final 1 ml. Next, 100 μl of chloroform and 50 μl of 0.1% SDS were added, followed by vortexing for 10 s. A 0.2-ml portion of o-nitrophenyl-β-d-galactoside (ONPG; 4 mg/ml) was added to mixture, and the sample was vortexed again. The enzyme reaction was stopped by adding 0.5 ml of 1 M Na2CO3 when the yellow color was developed. After we measuring the OD420 and the OD550, the LacZ activity was calculated according to the method of Miller (20).
qRT-PCR.
The MG1655 and yqhC(stop) strains were cultured in LB medium at 37°C with agitation to an OD600 of 1. The cultures were then treated with 0 and 1 mM GO for 30 min, which were sedimented and used for RNA purification. Then, 1 μg of total RNA from different samples was used for reverse transcription (RT). cDNA synthesis was carried out by using ImPromII reverse transcriptase (Promega). Portions (2 μl) of the cDNAs synthesized were used for real-time quantitative RT-PCR (qRT-PCR). Reaction mixtures (20 μl) included SYBR green I, 1 U of HS Taq polymerase, 5 mM MgCl2, 2 mM deoxynucleoside triphosphates (Prime Q-Mastermix), and 0.5 pmol of specific primers for yqhD and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; a reference). The primers for yqhD were as follows: forward, 5′-ACGCGAACAAATTCCTCACGATGC-3′, and reverse, 5′-GCTCAATACCGCCAAATTCCAGCA-3′. The primers for GAPDH were as follows: forward, 5′-ACTTACGAGCAGATCAAAGC-3′, and reverse, 5′-AGTTTCACGAAGTTGTCGTT-3′. Bio-Rad CFX-96 was used for PCR and for detecting the fluorescence change. The ratios of the cycle threshold (CT) values, as determined from the Bio-Rad CFX manager software, were compared between samples (27).
Enzymatic assay.
The enzymatic activity of purified YqhD was measured at 25°C by using a Beckman Coulter DU800 spectrophotometer by monitoring the initial rate of oxidation of NADPH at 340 nm. Substrate specificity was assessed by using 1 ml of 50 mM potassium phosphate buffer (pH 7.0) with 2 mM NADPH as a coenzyme. In all reactions, nonenzymatic conversions were subtracted from the observed initial reaction rates. The apparent Km and kcat values were determined by measuring the initial rate over a range of substrate concentrations.
Cell viability assay for determining lethal concentrations of aldehyde compounds.
The cell viability after exposure to aldehyde compounds was determined by growing cells on LB plates containing different concentrations of aldehyde compounds. Fresh colonies of wild-type and mutant strains were grown overnight in LB broth, diluted 100-fold in the same medium, and incubated with shaking until the OD600 reached 1.0. The cells were diluted from 10−1 to 10−6 and spotted (4 μl) onto LB plates containing different concentrations of aldehyde compounds. Growth was assessed after 12 to 14 h of incubation at 37°C.
Measuring the inhibitory concentrations of aldehyde compounds.
To compare the 50% inhibitory concentrations (IC50s) among strains, we measured the ODs of cells growing in media containing different concentrations of GO. A 96-well culture plate containing 180 μl of LB medium per well was treated with different concentrations of GO. Portions (20 μl) of inoculum containing cells at an OD600 of 1 were then added to each well. The 96-well plate was sandwiched between two acrylic plates (preheated) for efficient temperature equilibration, followed by incubation at 37°C with shaking (220 rpm) for 6 h. The OD595 was measured by using a microplate reader (model 680; Bio-Rad). The IC50 was estimated by using Sigmaplot (SPSS, Inc., Chicago, IL) by fitting it to the sigmoidal dose-response equation.
RESULTS
Isolation and localization of GOr E. coli mutants.
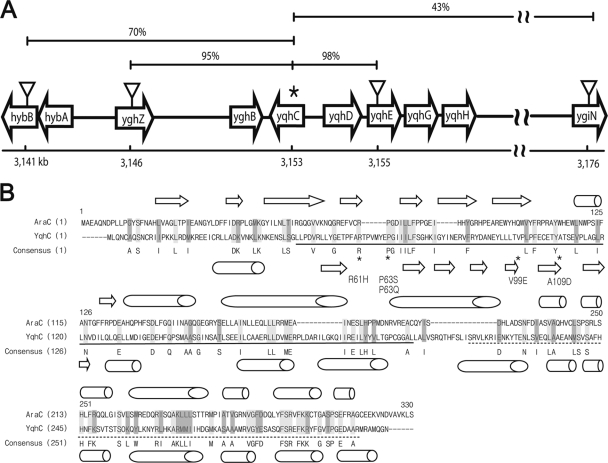
We isolated GOr mutants from the E. coli MG1655 strain on LB plates containing 10 mM GO. The mutations were mapped by random tagging with TnphoA-132 insertions (39) and were further analyzed by cotransduction with phage P1. The insertion tags were localized to two different chromosomal loci, one of which was found to be in the region ∼70 min (ca. 3,152 kb of the E. coli genome). The tags at 70 min were found to be in the hybB (3,141-kb) and ygiN (3,176-kb) genes (Fig. 2A), which were linked to the GOr locus with cotransduction frequencies of 70 and 43%, respectively, indicating that the candidate locus was somewhere between the two tags (Fig. 2A). We included two more markers, yqhE::cat (13) and yghZ::kan, for fine mapping and found that the GOr locus was very closely linked to yqhE::cat and yghZ::kan, with cotransduction frequencies of 98 and 95%, respectively. Mapping of the other GOr locus is currently under way (C. Lee et al., unpublished).
FIG. 2.
Mapping of GOr mutations to yqhC using transposon insertions. (A) Using nearby TnphoA-132 insertions isolated as described in Materials and Methods, the locations of yqhC mutations were determined based on their cotransductional linkages. The candidate locus was found at ∼70 min on the E. coli linkage map, with the physical coordinates shown underneath. Using insertions located close to the mutations, i.e., hybB (70% cotransduction frequency), ygiN (43%), yghZ (95%), and yqhE (98%), the candidate locus was further narrowed down to the region containing the yghB, yqhC, and yqhD genes. Eventually, the mutation was revealed by sequencing the yqhC gene (asterisk), a member of the AraC transcriptional activator family. (B) YqhC is a homolog of the AraC transcriptional activator with 13.6% amino acid identity based on alignment with AlignX (program provided in Vector NTI). The point mutations in Table 2 are marked by asterisks, with α-helices (cylinders) and β-sheets (arrows) shown as predicted by Jpred3 (http://www.compbio.dundee.ac.uk/www-jpred/). The predicted regulatory and DNA-binding domains are shown with solid and dashed lines, respectively (http://www.genome.jp/kegg/).
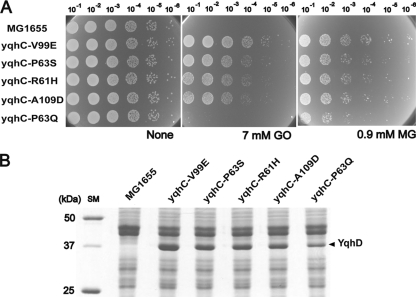
Based on the fact that YqhD was proposed to function as an alcohol dehydrogenase/aldehyde reductase (29) and the fact that it is flanked by the divergently transcribed yqhC gene, which is predicted to be an AraC-type transcriptional regulator, we chose to sequence the yqhC gene first and found mutations associated with GOr. The YqhC protein shows 13.6% amino acid identity with AraC (Fig. 2B). The GOr mutants identified had single changes in the yqhC gene, as shown in Table 2 and Fig. 2B. All of the mutations were in frame and were clustered in the putative regulatory domain of YqhC (Fig. 2B). There was variation in the effectiveness of GO resistance in different mutants, as shown in Fig. 3A, which was quantitated by measuring inhibition of cell growth after treatment with different concentrations of GO in order to determine the IC50s (see Materials and Methods).
TABLE 2.
GOr mutations found in the yqhC gene
| Allele | Mutational change |
IC50b for GO (mM) | |
|---|---|---|---|
| Nucleotide (bp)a | Amino acid | ||
| Wild type | None | 2.87 | |
| yqhC(P63S) | 187C→T | P63S | 5.99 |
| yqhC(A109D) | 326C→A | A109D | 5.79 |
| yqhC(R61H) | 182G→A | R61H | 5.91 |
| yqhC(V99E) | 296T→A | V99E | 6.65 |
| yqhC(P63Q) | 188C→A | P63Q | 5.49 |
That is, the position from the beginning of the yqhC open reading frame.
The value (IC50) for the yqhD insertion mutant is 1.88 mM.
FIG. 3.
yqhC mutations confer resistance to 2-oxoaldehydes and alter protein expression patterns. (A) Point mutations of yqhC exhibit a spectrum of resistance to GO and MG. The most resistant mutant, yqhC(V99E), shows normal growth in media containing up to 7 mM GO and 0.9 mM MG, conditions that are lethal for wild-type MG1655. The yqhD(P63Q) mutation had the lowest level of resistance to both substrates. (B) yqhC mutations result in the overexpression of a 40-kDa protein, as visualized on a SDS-12% PAGE gel, which was identified as YqhD by MALDI-TOF MS analysis. The levels of YqhD expression are roughly proportional to the degrees of oxoaldehyde resistance among the yqhC mutants.
YqhC mutations result in increased expression of YqhD protein.
When we examined total protein expression in GOr mutants and compared it with that of wild-type cells, we found overproduction of a protein shown on a SDS-12% PAGE gel to be 43 kDa, which was characterized as YqhD after matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) fingerprinting (Fig. 3B). The overexpression of YqhD was also confirmed by two-dimensional gel electrophoresis as a spot with a predicted pI value of 6.0, which is close to the predicted pI for YqhD of 5.72 (data not shown). No overproduction of the yqhE gene product was detected, suggesting that only expression of YqhD, and not the downstream gene yqhE, is likely to be regulated by YqhC. This result implies that enhanced expression of YqhD specifically results from missense mutations in yqhC, which is consistent with the fact that expression of YqhD protein was relatively low in wild-type cells (Fig. 3B). Our results also showed that levels of YqhD expression in mutant cells are roughly proportional to their degree of GO resistance (Fig. 3B), supporting the notion that YqhD is responsible for resistance to GO.
YqhC acts as a transcriptional activator for the yqhD gene.
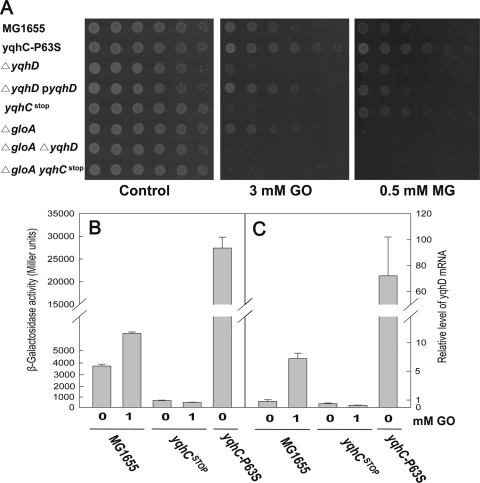
In order to assess the role of yqhC in yqhD regulation, we obtained a yqhC(stop) mutant with a nonsense change (E112) in the yqhC gene that produced a peptide with a deletion of two thirds of its coding region (Table 1). This peptide lacked the helix-turn-helix that was predicted to serve as the DNA-binding domain, and we observed that the mutant strain confers sensitivity to GO (Fig. 4A). Thus, we concluded that YqhC functions as an activator for yqhD, supporting the notion that GOr mutations in yqhC are specific changes activating yqhD. We further demonstrated that the yqhD activation by YqhC indeed occurs at the transcription level by constructing a β-galactosidase reporter and also by analyzing transcription with real-time qRT-PCR (Fig. 4BC). The promoter region of yqhC was cloned into the transcriptional fusion plasmid pRS415 containing lacZ (see Materials and Methods for details). When cells were grown in LB medium, the wild type exhibited some level of yqhD transcription (Fig. 4B), as measured by a β-galactosidase assay, which was increased about 2-fold in the presence of GO at 1 mM. The removal of YqhC by introducing the yqhC(stop) mutation essentially abolished the transcription of yqhD, which is unchanged upon addition of GO. As expected, the yqhD transcription was dramatically enhanced by the presence of constitutively activating mutation of yqhC(P63S). The levels of yqhD transcripts measured by real-time qRT-PCR were consistent with the data of the reporter assay, again confirming the transcriptional activation of yqhD gene by YqhC. It is of interest that the yqhD transcription is enhanced by the addition of GO.
FIG. 4.
Transcriptional regulation of yqhD by YqhC and their roles in GO detoxification. (A) Phenotypes of yqhC, yqhD, and gloA deletion mutants and complementation with a cloned yqhD plasmid. The yqhD deletion mutant showed an enhanced susceptibility to GO and MG relative to wild-type cells. The oxoaldehyde sensitivity of yqhD deletion was rescued by pACYC184-cloned yqhD expressed under its own promoter. The yqhC and yqhD deletion mutants show apparent GO sensitivity but not MG sensitivity. In contrast, the gloA deletion mutant exhibits a sensitivity to only MG, but not to GO. The transcriptional activity of yqhD was measured by the fusion of β-galactosidase reporter (B) and by real-time qRT-PCR analysis (C). Compared to the wild type, yqhD expression was dramatically enhanced in yqhC(P63S), whereas the yqhC(stop) mutant exhibited almost no expression of yqhD. In wild-type MG1655, yqhD expression was increased by an addition of GO. The cells at an OD600 of 1 were treated 1 mM GO for 70 and 30 min in β-galactosidase assay and real-time qRT-PCR, respectively.
The yqhD-defective strain exhibits a GOs phenotype, which can be complemented by a yqhD clone (pACYC184 yqhD) that produces the protein under its own promoter (Fig. 4A). Since YqhD protein was not highly expressed from this plasmid (data not shown), the resistance of the plasmid to GO was only slightly enhanced relative to that of wild-type MG1655. In order to assess the contributions of the glyoxalases to the intracellular removal of MG and GO, we tested yqhC and yqhD mutants on a gloA-negative background (Fig. 4A). Although the gloA mutant was susceptible to MG, its sensitivity to GO was essentially the same as that of the wild type (MG1655). In contrast, yqhD mutants showed greater sensitivity to GO but no difference in MG sensitivity. The sensitivity of yqhC(stop) mutants to these compounds was similar to that of the yqhD mutants. However, the effects of yqhD and yqhC mutations on MG sensitivity became visible in a gloA-deficient background (data not shown) or in a yqhC(P63S) mutant overexpressing YqhD, indicating that YqhD plays a role in MG detoxification, which is consistent with its catalytic activity, as shown in Table 3.
TABLE 3.
Substrate specificities and kinetics of YqhD
| Substrate | Product | Sp acta (nmol min−1 mg−1) | Km (mM) | kcat (min−1) | kcat/Km (min−1 M−1) |
|---|---|---|---|---|---|
| Glyoxal | Glycolaldehyde | 36,250 | 11.53 | 618 | 5.36 × 104 |
| Glycolaldehyde | 1,2-Ethandiol | 61,902 | 28.28 | 3,258 | 1.09 × 105 |
| Butanaldehyde | Butanol | 58,587 | 0.40 | 1,983 | 4.99 × 106 |
| Methylglyoxal | Hydroxyacetone | 26,528 | 2.60 | 284 | 1.09 × 105 |
| Glyceraldehyde | Glycerol | 26,363 | 1.36 | 204 | 1.49 × 105 |
| Hydroxyacetone | 1,2-Propanediol | 3,050 | 76.90 | 144 | 1.87 × 103 |
Enzyme activities were measured at room temperature with 2 mM NADPH, unless otherwise stated (0.1 mM for hydroxyacetone). The concentrations of the substrates were 200 mM glyoxal and glycolaldehyde, 10 mM butanal and glyceraldehyde, 20 mM MG, and 500 mM hydroxyacetone. A 10-μg portion of YqhD was used.
YqhC binds to the operator region of yqhD gene.
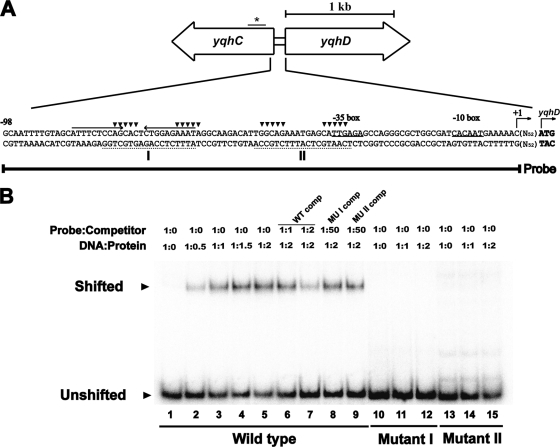
Phylogenetic analysis of YqhC revealed its close similarity to AraC and showed that it is somewhat distant from the other AraC cluster containing GadX, GadY, and AppY (data not shown). The consensus sequence of the AraC binding site is not yet clearly defined; however, the consensus binding sequence for SoxS, another member of AraC family, is well characterized (40). Using sequence analysis of the 5′ region of yqhD gene to search for a promoter with the SoxS-binding sequence, several candidates were found, one of which is located in the open reading frame (ORF) region of yqhC as shown previously (26). The predicted binding site (shown with an asterisk in Fig. 5A) in the yqhC ORF appeared not to bind to YqhC (data not shown). Others are clustered ∼50 bp upstream of the yqhD ORF (Fig. 5A), and these sequences overlap with a 24-bp palindrome consisting of two 10-bp repeating units. We carried out a gel mobility shift assay on DNA fragments amplified by PCR containing the putative operator regions, which were tested for their ability to bind to purified YqhC protein. As shown in Fig. 5B, YqhC binds to DNA containing the operator region (the SoxS-binding consensus sequence with the palindrome) in concentration-dependent manner (lanes 1 to 5). In order to verify that YqhC indeed binds to this region, a gel mobility shift assay was carried out with two mutated probes in which 10 residues in the two SoxS consensus sequences were altered (shown with an inverted triangle in Fig. 5A). As shown in Fig. 5B, YqhC did not bind to the mutated probes (lanes 10 to 15), indicating that the mutated residues are indeed part of the YqhC-binding sequence. Furthermore, the mutated probes were unable to compete with the wild-type probe (lanes 6 to 9, Fig. 5B), confirming that the altered residues in the yqhD operator region are directly involved in YqhC binding. The location of this putative promoter sequence for the yqhD gene is consistent with the transcription initiation site revealed by 5′-RACE analysis, as shown in Fig. 5A.
FIG. 5.
Binding of YqhC protein to the yqhC-yqhD intergenic region. (A) Relative locations of the yqhC and yqhD genes and the predicted promoter and regulatory sequences. The −10 and −35 sequences of the yqhD promoter are underlined with the translational start site shown in bold. The transcriptional start site of yqhD is also shown and was determined by 5′-RACE (see Materials and Methods). The predicted YqhC binding sites are shown with broken lines and boldface letters (I and II) designating the consensus sequences [(N3)GCA(N9)AAA(N2)] for SoxS binding that are conserved in the target sequences of AraC-XylS transcriptional regulators (17). The palindromic sequence is indicated by arrows. The DNA probe used for the gel shift assay (shown in panel B) contained the region from −98 to +1 of the yqhD promoter. (B) Gel mobility shift assay using YqhC to identify the promoter region. The cores (▾) of the predicted binding sites were mutated to generate mutant I and II probes, each having 10 bp. Molar ratios of DNA to protein, as well as probe DNA to competitor DNA, are shown; “WT comp” represents the addition of the cold wild-type probe as a competitor, and “MUI comp” and “MUII comp” indicate cold mutant probe. Probes were labeled with [γ-32P]ATP as described in Materials and Methods.
YqhD is an aldehyde reductase converting GO to ethandiol using NADPH.
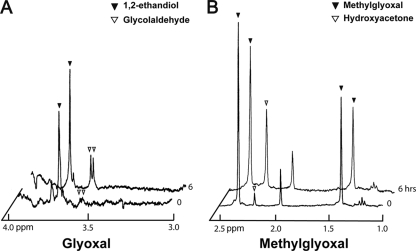
YqhD was initially characterized as an alcohol dehydrogenase based on its three-dimensional structure (29), but a recent report indicated that it is an aldehyde reductase showing specificity for certain aldehydes, including butanaldehyde, malondialdehyde, and acrolein, which are derived from lipid peroxidation (26). Thus, we suspected that the YqhD enzyme is involved in the metabolism of GO and/or glycolaldehyde. When we tested the ability of purified YqhD to metabolize various short-chain aldehydes, it exhibited fairly broad specificity with the highest specific activity for glycolaldehyde (Table 3), although its Km value was estimated to be a little bit high (28.28 mM). Furthermore, the enzyme showed activity against GO, generating glycolaldehyde using NADPH as a cofactor. Thus, we have shown that the YqhD enzyme mediates catalysis of the two consecutive reactions involved in the conversion from GO to ethandiol and thereby efficiently removes two toxic aldehydes. When we incubated GO (10 mM) with YqhD for 6 h and analyzed the resulting products by NMR, increased amounts of glycolaldehyde and ethandiol were observed (Fig. 6). YqhD also exhibited activity against MG, generating hydroxyacetone as the major reaction product (Table 3, Fig. 6), which accounted for more than 90% of the substrate converted. This estimation was based on molar quantification of hydroxyacetone generation versus MG disappearance. Commercially available MG contains only a small amount of acetate and no other compounds such as hydroxyacetone, as shown on the NMR spectrum, even after a longer period of incubation.
FIG. 6.
1H-NMR spectra showing the conversion of oxoaldehydes by YqhD. (A) Glycolaldehyde (▾) and 1,2-ethandiol (▿) were produced after 6 h of incubation with purified YqhD protein (40 μg), 10 mM GO, and 2 mM NADPH in 50 mM potassium phosphate buffer (pH 7.0) and were detected by 1H-NMR spectroscopy. The commercial GO reagent is contaminated with some 1,2-ethandiol. The GO peak is hidden by water noise at 5.0 ppm (not shown). (B) Hydroxyacetone (▿) was produced from MG (▾, 3 mM) by YqhD (40 μg). For this reaction, the same condition described for glycoaldehyde was used. It should be noted that the MG reagent is contaminated with acetate showing a peak ∼2 ppm.
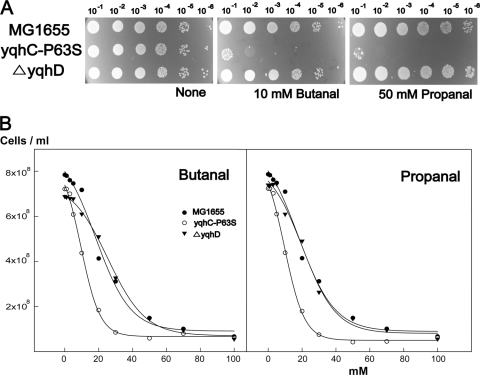
The enzymatic specificity of YqhD for both GO and glycolaldehyde is consistent with the prevalence of GOr mutants (Fig. 2). In fact, when we compared the GO and glycolaldehyde reducing activities of crude extracts from the yqhC(P63S) mutant, which expresses yqhD constitutively, to those of ΔyqhD strains, a significant difference was noted (78 ± 5 and 90 ± 2 versus 42 ± 3 and 62 ± 3 nmol min−1 mg−1, respectively, with 100 mM GO or 20 mM glycolaldehyde, plus 2 mM NADPH and 100 μg of protein). Since YqhD has been previously reported to remove aldehyde compounds derived from lipid peroxidation, we tested the YqhD-overproducing strain for its ability to remove butanal and propanal. Surprisingly, YqhD activity increased the toxic effects of these aldehydes by converting them to their corresponding alcohols, i.e., butanol and propanol (Fig. 7A). These alcohols are already known to be highly toxic to cells (4, 37). When we examined the toxic effects of butanol and propanol added externally, we found that the lethal and effective concentrations were much higher than those of the corresponding aldehydes; the IC50s of butanol and propanol in wild-type E. coli (MG1655) were more than 100 and 200 mM, respectively.
FIG. 7.
Activity of YqhD on butanal and propanal. (A) Spot assays on LB plates containing 10 mM butanal or 50 mM propanal indicate that the yqhC(P63S) strain overexpressing YqhD became highly sensitive to these compounds, which was not observed in the strain lacking YqhD. (B) Growth inhibition with various concentrations of butanal and propanal was observed and plotted to obtain IC50s. The IC50s for the MG1655, yqhC(P63S), and ΔyqhD strains for butanal and propanal were 16.4 and 15.5 mM, 9.5 and 9.6 mM, and 24.9 and 18.5 mM, respectively.
DISCUSSION
Certain short-chain carbohydrates are known to be highly reactive because of their unprotected aldehyde groups (24). GO is a member of the 2-oxoaldehydes, a reactive subgroup, and is believed to be a product of glucose oxidation (33). However, in contrast to MG, the intracellular presence and degradation of GO have not been clearly elucidated. We have presented evidence that E. coli cells are capable of detoxifying GO with a reducing enzyme, using NADPH as a cofactor. Our genetic study to isolate GO resistance mutations revealed the identity of an enzyme YqhD that is overexpressed in constitutively active yqhC mutants. The prevalence of such mutants might be due to the rather simple mechanism of activation of yqhD by YqhC. We demonstrated that YqhC protein directly binds to the promoter region of the yqhD gene, which contains the SoxS-like binding sequence, as well as a 24-bp palindrome (Fig. 5). More importantly, the isolation of yqhC mutants allowed us to characterize the YqhD enzyme, which catalyzes the reaction of various aldehydes.
YqhD has been classified as an alcohol dehydrogenase (29), and it is also highly similar to the 1,2-propanediol oxidoreductase FucO and to the C-terminal half of AdhE, a glycolytic enzyme producing ethanol from acetaldehyde. YqhD is structurally different from the members of the AKR family, which share a characteristic motif for NADPH binding; however, growing evidence has indicated that YqhD homologs are primarily involved in the reduction of aldehyde compounds (26). It has been demonstrated that the enzyme AdhE acquired alcohol dehydrogenase activity through the introduction of a simple missense mutation, A267T, an example of the evolutionary adaptation to an oxidative enzyme (19). The aldehyde reductase activity of YqhD was first identified by its ability to convert 3-hydroxypropionaldehyde to 1,3-propanediol (10). Later, YqhD was proposed to function as a detoxifying enzyme removing short-chain aldehydes, such as butanal and propanal, generated from lipid peroxidation (26). However, the proposed role of YqhD in the removal of such compounds seems unlikely based on our in vivo evidence indicating that the enzymatic conversion of butanal and propanal results in the intracellular accumulation of even more toxic compounds: butanol and propanol, respectively (Fig. 7). The unexpected intracellular toxicity of butanol and propanol may suggest the presence of a more sensitive in vivo target for these alcohols that is not easily accessible from an exogenously added compound.
Although the intracellular detoxification of MG is known to occur through multiple pathways, including the glutathione-mediated glyoxalase I/II system, the metabolic pathways for GO have not been well established. We observed that the GO sensitivity of a gloA-deficient strain was not much different from that of the wild-type strain, suggesting that the glyoxalase system does not serve as an efficient pathway for GO detoxification. Thus, there may be other pathways for intracellular removal of GO. E. coli cells might have an advantage in detoxifying GO through their expression of the YqhD enzyme, since it is capable of converting GO to nontoxic ethandiol with a single enzyme. However, it appears that cellular strategies for removal of short-chain aldehydes are not specific, as exemplified by the presence of AKRs with rather nonspecific substrate specificities. We have previously demonstrated that several AKRs, including YghZ and YqhE, are involved in the catalysis of MG and related compounds (13).
Intracellular production of 2-oxoaldehyde is enhanced by oxidative stress (25, 33). Thus, it is possible that the regulation of aldehyde removal is associated with the regulation of the oxidative stress response. An example would be that glycolaldehyde induces the SoxR/S regulon (6). Although the YqhC-YqhD regulatory system was discovered through a specific class of regulatory mutations in YqhC that confer cellular resistance to GO, the mechanism underlying its activation still needs to be elucidated. Although we observed that the transcription of yqhD was activated by GO, the identity of intracellular signal for YqhC is yet to be discovered. Finding such signal may provide an insight into the underlying regulatory mechanism for responses to aldehyde, as well as oxidative stresses. During the present study, we isolated other GOr mutations localized at 37 centisome region that were unlinked from yqhC (Lee et al., unpublished). Further characterization of these mutants may reveal other mechanism associated with carbonyl stress and its detoxification.
Acknowledgments
This study was supported by the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Education, Science, and Technology, Republic of Korea, to C.P.
We thank Dongkyu Kim for the isolation of mutant strains.
Footnotes
Published ahead of print on 11 June 2010.
REFERENCES
- 1.Abordo, E. A., H. S. Minhas, and P. J. Thornalley. 1999. Accumulation of alpha-oxoaldehydes during oxidative stress: a role in cytotoxicity. Biochem. Pharmacol. 58:641-648. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, M. U., E. Brinkmann-Frye, T. P. Degenhardt, S. R. Thorpe, and J. W. Baynes. 1997. N-Epsilon-(carboxyethyl)lysine, a product of the chemical modification of proteins by methylglyoxal, increases with age in human lens proteins. Biochem. J. 324(Pt. 2):565-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atzori, L., M. Dore, and L. Congiu. 1989. Aspects of allyl alcohol toxicity. Drug Metab. Drug Interact. 7:295-319. [PubMed] [Google Scholar]
- 5.Badia, J., et al. 1991. l-Lyxose metabolism employs the l-rhamnose pathway in mutant cells of Escherichia coli adapted to grow on l-lyxose. J. Bacteriol. 173:5144-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benov, L., and I. Fridovich. 2002. Induction of the soxRS regulon of Escherichia coli by glycolaldehyde. Arch. Biochem. Biophys. 407:45-48. [DOI] [PubMed] [Google Scholar]
- 7.Boronat, A., E. Caballero, and J. Aguilar. 1983. Experimental evolution of a metabolic pathway for ethylene glycol utilization by Escherichia coli. J. Bacteriol. 153:134-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Costanzo, L., G. A. Gomez, and D. W. Christianson. 2007. Crystal structure of lactaldehyde dehydrogenase from Escherichia coli and inferences regarding substrate and cofactor specificity. J. Mol. Biol. 366:481-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emptage, M., et al. 2006. Process for the biological production of 1,3-propanediol with high titer. E. I. du Pont de Nemours and Company, New York, NY.
- 11.Jez, J. M., M. J. Bennett, B. P. Schlegel, M. Lewis, and T. M. Penning. 1997. Comparative anatomy of the aldo-keto reductase superfamily. Biochem. J. 326(Pt. 3):625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasper, M., C. Roehlecke, M. Witt, H. Fehrenbach, A. Hofer, T. Miyata, C. Weigert, R. H. Funk, and E. D. Schleicher. 2000. Induction of apoptosis by glyoxal in human embryonic lung epithelial cell line L132. Am. J. Respir. Cell Mol. Biol. 23:485-491. [DOI] [PubMed] [Google Scholar]
- 13.Ko, J., I. Kim, S. Yoo, K. B. Min, Kim, and C. Park. 2005. Conversion of methylglyoxal to acetol by Escherichia coli aldo-keto reductases. J. Bacteriol. 187:5782-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBlanc, D. J., and R. P. Mortlock. 1971. Metabolism of d-arabinose: a new pathway in Escherichia coli. J. Bacteriol. 106:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennox, E. S. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 16.MacLean, M. J., L. S. Ness, G. P. Ferguson, and I. R. Booth. 1998. The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol. Microbiol. 27:563-571. [DOI] [PubMed] [Google Scholar]
- 17.Martin, R. G., W. K. Gillette, S. Rhee, and J. L. Rosner. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation, and spatial relationship to the core promoter. Mol. Microbiol. 34:431-441. [DOI] [PubMed] [Google Scholar]
- 18.Matkovics, B., M. Kotorman, I. S. Varga, D. Q. Hai, and C. Varga. 1997. Oxidative stress in experimental diabetes induced by streptozotocin. Acta Physiol. Hung. 85:29-38. [PubMed] [Google Scholar]
- 19.Membrillo-Hernandez, J., P. Echave, E. Cabiscol, J. Tamarit, J. Ros, and E. C. Lin. 2000. Evolution of the adhE gene product of Escherichia coli from a functional reductase to a dehydrogenase: genetic and biochemical studies of the mutant proteins. J. Biol. Chem. 275:33869-33875. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 21.Mlakar, A., A. Batna, A. Dudda, and G. Spiteller. 1996. Iron (II) ions induced oxidation of ascorbic acid and glucose. Free Radic. Res. 25:525-539. [DOI] [PubMed] [Google Scholar]
- 22.Montella, C., L. Bellsolell, R. Pérez-Luque, J. Badía, L. Baldoma, M. Coll, and J. Aguilar. 2005. Crystal structure of an iron-dependent group III dehydrogenase that interconverts l-lactaldehyde and l-1,2-propanediol in Escherichia coli. J. Bacteriol. 187:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obradors, N., E. Cabiscol, J. Aguilar, and J. Ros. 1998. Site-directed mutagenesis studies of the metal-binding center of the iron-dependent propanediol oxidoreductase from Escherichia coli. Eur. J. Biochem., 258:207-213. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien, P. J., A. G. Siraki, and N. Shangari. 2005. Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit. Rev. Toxicol. 35:609-662. [DOI] [PubMed] [Google Scholar]
- 25.Okado-Matsumoto, A., and I. Fridovich. 2000. The role of α,β-dicarbonyl compounds in the toxicity of short chain sugars. J. Biol. Chem. 275:34853-34857. [DOI] [PubMed] [Google Scholar]
- 26.Perez, J. M., et al. 2008. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 283:7346-7353. [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 29.Sulzenbacher, G., K. Alvarez, R. H. Van Den Heuvel, C. Versluis, S. Spinelli, V. Campanacci, C. Valencia, C. Cambillau, H. Eklund, and M. Tegoni. 2004. Crystal structure of Escherichia coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J. Mol. Biol. 342:489-502. [DOI] [PubMed] [Google Scholar]
- 30.Thornalley, P. J. 2002. Glycation in diabetic neuropathy: characteristics, consequences, causes, and therapeutic options. Int. Rev. Neurobiol. 50:37-57. [DOI] [PubMed] [Google Scholar]
- 31.Thornalley, P. J. 1998. Glutathione-dependent detoxification of alpha-oxoaldehydes by the glyoxalase system: involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chem. Biol. Interact. 111-112:137-151. [DOI] [PubMed] [Google Scholar]
- 32.Thornalley, P. J. 1990. The glyoxalase system: new developments toward functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thornalley, P. J., A. Langborg, and H. S. Minhas. 1999. Formation of glyoxal, methylglyoxal, and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem. J. 344(Pt. 1):109-116. [PMC free article] [PubMed] [Google Scholar]
- 34.Vander Jagt, D. L., and L. A. Hunsaker. 2003. Methylglyoxal metabolism and diabetic complications: roles of aldose reductase, glyoxalase-I, betaine aldehyde dehydrogenase, and 2-oxoaldehyde dehydrogenase. Chem. Biol. Interact 143-144:341-351. [DOI] [PubMed] [Google Scholar]
- 35.Vander Jagt, D. L., B. Robinson, K. K. Taylor, and L. A. Hunsaker. 1992. Reduction of trioses by NADPH-dependent aldo-keto reductases: aldose reductase, methylglyoxal, and diabetic complications. J. Biol. Chem. 267:4364-4369. [PubMed] [Google Scholar]
- 36.Vander Jagt, D. L. 1993. Glyoxalase II: molecular characteristics, kinetics, and mechanism. Biochem. Soc. Trans. 21:522-527. [DOI] [PubMed] [Google Scholar]
- 37.Vollherbst-Schneck, K., J. A. Sands, and B. S. Montenecourt. 1984. Effect of butanol on lipid composition and fluidity of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 47:193-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells-Knecht, K. J., D. V. Zyzak, J. E. Litchfield, S. R. Thorpe, and J. W. Baynes. 1995. Mechanism of autoxidative glycosylation: identification of glyoxal and arabinose as intermediates in the autoxidative modification of proteins by glucose. Biochemistry 34:3702-3709. [DOI] [PubMed] [Google Scholar]
- 39.Wilmes-Riesenberg, M. R., and B. L. Wanner. 1992. TnphoA and TnphoA′ elements for making and switching fusions for study of transcription, translation, and cell surface localization. J. Bacteriol. 174:4558-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood, T. I., et al. 1999. Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol. Microbiol. 34:414-430. [DOI] [PubMed] [Google Scholar]