Abstract
Endotoxin (bacterial lipopolysaccharide [LPS]) causes fatal septic shock via the Toll-like receptor 4 (TLR-4) protein present on innate immunity effector cells, which activates nuclear factor kappa B (NF-κB), inducing proinflammatory cytokines, including tumor necrosis factor alpha (TNF-α). An early step in this process involves nuclear sequestration of the p65-RelA NF-κB subunit, enabling transcriptional activation of target inflammatory cytokine genes. Here, we analyzed the role of the nuclear zinc finger protein Gfi1 in the TLR response using primary bone marrow-derived macrophages. We show that upon LPS stimulation, expression of Gfi1 is induced with kinetics similar to those of nuclear translocation of p65 and that Gfi1 interacts with p65 and inhibits p65-mediated transcriptional transactivation by interfering with p65 binding to target gene promoter DNA. Gfi1-deficient macrophages show abnormally high mRNA levels of the TNF-α gene and many other p65 target genes and a higher rate of TNF promoter occupancy by p65 than wild-type cells after LPS stimulation, suggesting that Gfi1 functions as an antagonist of NF-κB activity at the level of promoter binding. Our findings identify a new function of Gfi1 as a general negative regulator of the endotoxin-initiated innate immune responses, including septic shock and possibly other severe inflammatory diseases.
The inflammatory response toward microbial pathogens is orchestrated by specialized cells of the innate immune system. A critical element in the induction of this type of immune response is the activation of membrane-bound Toll-like receptor (TLR) molecules by their ligands, which can be bacterial cell wall components, such as the endotoxin lipopolysaccharide (LPS) of Gram-negative bacteria, membrane lipoproteins of Gram-positive bacteria, and nonmethylated CpG-rich DNA of both types of bacteria (1, 36). These microbial molecules, which are also referred to as “pathogen-associated molecular patterns” (PAMPs), are able to initiate a signaling cascade upon binding to their cognate TLRs that triggers the cellular response. LPS can bind to TLR4 (46) and starts several signaling cascades by recruiting the adaptor protein MyD88, members of the interleukin-1 (IL-1) receptor-associated kinase (IRAK) family, and the adapter protein TRAF-6 (tumor necrosis factor [TNF] receptor-associated factor 6) (1, 36, 51). Subsequently, this triggers several distal events, including the activation of mitogen-activated protein (MAP) kinases and also, in particular, the latent nuclear transcription factor kappa light chain enhancer of activated B cells (NF-κB), which is a critical element in TLR signaling (1, 36, 51, 56). Once activated, NF-κB regulates a series of cellular responses, including the production of proinflammatory cytokines, such as TNF-α and IL-1β (20, 38).
The members of the NF-κB/Rel family of dimeric “rapid-acting” primary transcription factors have important functions in many aspects of cell growth, survival, development, and innate as well as adaptive immunity (20). This family comprises five members, RelA, RelB, p50, p52, and cRel, which can form combinations of hetero- and homodimers (18). The most abundant form of NF-κB is the p65-p50 heterodimer, which is held inactive in the cytosol by a family of IκB-inhibitory proteins that mask the nuclear localization signals of NF-κB (11, 18). TLR signaling causes rapid activation of IκB kinase (IKK), which is composed of two catalytic subunits, IKK-α and IKK-β, and a regulatory component, IKK-γ (16, 18). Activated IKK phosphorylates IκB, which leads to proteolysis of the inhibitor through the ubiquitin-proteasome pathway. This enables nuclear import of NF-κB and subsequent transcriptional activation of NF-κB target genes, such as the gene encoding the inflammatory cytokine TNF (11, 19). In macrophages, this ultimately leads to secretion of TNF-α, IL-1β, and other inflammatory cytokines and response mediators (1, 3, 9, 36). These cytokines and mediators are very potent molecules that induce multiple reactions that counteract the growth and dissemination of pathogens, enhancing the entire adaptive immune response (24, 25).
Although inflammatory cytokines are critical for controlling the growth of pathogenic microorganisms, a persistent inflammatory response is harmful and even lethal to the host. To avoid inappropriate inflammatory responses, negative regulatory mechanisms exist to attenuate TLR signaling and to maintain a balance between activation and inhibition of this pathway (37, 43). Many of the negative TLR regulators act at different levels in the active TLR signaling pathways through negative feedback. For instance, dominant-negative forms of IRAK (IRAK-M) or MyD88 (MyD88s) block the dissociation or formation of complexes between TLRs and IRAK or MyD88 (28, 29, 35), whereas the Toll-interacting protein (Tollip) interferes with IRAK1 autophosphorylation (5), and the A20 protein affects TRAF6 ubiquitination (53). It is well accepted that inhibition of the proximal MyD88-IRAK-TRAF6 part of the TLR signaling pathway results in inactivation of more-distal post-TLR signaling components, such as decreased phosphorylation of MAPK family members and decreased DNA binding activity of NF-κB (37). Other studies have revealed that inhibition of NF-κB activity is not necessarily caused by a block in the proximal part of the TLR signaling pathway and may be regulated by nuclear factors that act through hitherto unknown molecular mechanisms (7).
One of the genes that are induced after stimulation of macrophages with TLR4 ligand LPS is that encoding the transcriptional repressor Gfi1 (growth factor independence 1) (30, 32). Intriguingly, LPS stimulation of Gfi1-deficient mouse macrophages (bone marrow-derived macrophages [BMDMs] and lung alveolar macrophages) results in an exaggerated production of proinflammatory cytokines such as TNF-α and IL-1β (30, 32, 40). Gfi1 is a 55-kDa nuclear protein with six C-terminal zinc finger domains and an N-terminal 20-amino-acid (aa) “SNAG” domain, conserved between Gfi1 and the proteins Snail and Slug (8, 12, 23, 33, 40). Gfi1 represses transcription by binding to DNA recognition sequences in target gene promoters, which requires intact SNAG and zinc finger domains (15, 48, 52, 57). Gfi1 is differentially expressed in specific immunohematopoietic cells, in the central nervous system, and in the intestine (8, 23, 26, 33, 40, 49). Studies with Gfi1−/− mice revealed a series of hematopoietic defects ranging from increased hematopoietic stem cell (HSC) proliferation with decreased self-renewal capacity to early blockage in T-cell differentiation and granulopoiesis (21, 22, 31, 54, 55), suggesting that Gfi1 is a key regulator of hematopoiesis.
Consistent with this overproduction of proinflammatory cytokines, Gfi1-deficient mice are highly susceptible to LPS-induced septic shock (30, 32), suggesting a negative regulatory role for Gfi1 in TLR-initiated signaling pathways. How such a negative regulatory role is executed by Gfi1 on the molecular level is yet unknown. In the present study, we addressed the function of Gfi1 in this process. Our data indicate that Gfi1, in contrast to most known negative regulators of TLRs, does not affect the proximal cytoplasmic components of TLR signaling pathways but rather acts at the downstream end of the pathway in the nucleus. We present evidence that Gfi1 directly binds to the p65 subunit of NF-κB and controls its DNA binding activity, thereby regulating the expression of a large number of NF-κB target genes.
MATERIALS AND METHODS
Mice.
Gfi1−/− mice were generated by homologous recombination in R1 embryonic stem cells as described previously (32). Mice were housed at the IFZ animal facility, University of Essen Medical School, under specific-pathogen-free conditions in accordance with German animal legislation or at the animal facility at the Institut de Recherches Cliniques de Montreal in accordance with the guidelines of the Canadian Council on Animal Care (CCAC).
Reagents.
Protein-free, phenol-water-extracted Salmonella enterica serovar friedenau LPS H909 was kindly provided by H. Brade (Borstel, Germany). All cytokines used in the present study were purchased from PeproTech. The following antibodies were utilized in the coimmunoprecipitation and Western blotting analyses according to the protocols recommended by the manufacturers: anti-phospho-p42/44 (Thr202/Tyr204), anti-phospho-stress-activated protein kinase (SAPK)/c-Jun N-terminal kinase (JNK) (Thr183/Tyr185), anti-phospho-p38 MAP kinase (Thr180/Tyr182), anti-phospho-Akt (Ser473), anti-IκBα, anti-phospho-IκBα, and anti-phospho-p65 (Ser536) (purchased from Cell Signaling Technology, Inc.); anti-FLAG (M2) (purchased from Sigma); and anti-p65 monoclonal antibody (C-20), anti-p50 (NLS) antibody, and anti-c-Myc antibody (C19) (from Santa Cruz Biotechnology [Santa Cruz, CA]).
Stimulation of BMDMs.
BMDMs were differentiated from marrow cells of 4- to 8-week-old C57BL/6 Gfi1-deficient mice and their wild-type (WT) littermates by use of classic methods as previously described (30, 32). Bone marrow cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, 1% penicillin-streptomycin, and 50 ng/ml macrophage colony-stimulating factor (M-CSF) for 7 to 8 days. The purity of BMDMs was determined by analysis of CD11b and F4/80 expression using flow cytometry techniques. Flow cytometry analysis showed no difference in irrelevant-antibody (IAb) and CD86 expression between wild-type and Gfi1−/− BMDMs. Before being used for various experiments, BMDMs were replated in the presence or absence of LPS at the indicated doses and time points and then harvested for analysis.
RT-PCR and real-time PCR.
Total RNA from BMDMs was isolated with TRIzol reagent (Gibco BRL). For real-time PCR (RT-PCR), the following forward and reverse primers were used: 5′-AGGCTTCAAGCCCTTTGGCTG-3′ and 5′-GTTCCTTCCCTAAACCAGAGTC-3′ (for wild-type Gfi1 transcripts), 5′-AGGCTTCAAGCCCTTTGGCTG-3′ and 5′-CATCAATGTACTTTATCATGTCTGC-3′ (for transgenic Gfi1 transcripts), 5′-AGGCAAAGCCATCCAATACTT-3′ and 5′-GTGGGTCTTAACTTGGCCTTC-3′ (for mIRAK-M), and 5′-ACGATCAGTTTCCCAGACTCA-3′ and 5′-GTGACCCACCTGCAGTACC-3′ for mSHIP1. For quantitative RT-PCR, the ABI Prism 7300 sequence detection system (Applied Biosystems, Foster City, CA) was used to determine the mRNA levels of TNF-α per the manufacturer's instructions.
Confocal microscopy.
After the indicated incubation periods, medium was removed and cells were washed with phosphate-buffered saline (PBS), fixed for 10 min with cold methanol, washed twice with PBS, and equilibrated for 30 min in solution A (10 mM Tris, pH 7.5, 100 mM NaCl, 0.05% Tween 20, 1% bovine serum albumin [BSA]). Afterwards, cells were stained with anti-NF-κB p65 (C-20) or anti-Gfi1 (N-20 or GP33) as the primary antibody (diluted 1:50 in solution A). After 1 h of incubation at room temperature, fluorochrome-conjugated secondary antibody was added (Jackson Immunoresearch, West Grove, PA). Nuclear staining was done using Topro3 (Invitrogen). After a wash with PBS, cells were analyzed using a confocal microscope (laser scanning microscope [LSM]; Zeiss, Jena, Germany) and LSM Browser 5.0 software.
Transient-transfection and reporter gene assay.
RK13 epithelial cells and NIH 3T3 cells were maintained in DMEM supplemented with 10% fetal calf serum (FCS), penicillin-streptomycin, and l-glutamine. The cells were transfected with different constructs, as described in the figure legends, using Roti-Fect transfection reagent (Roth) according to the manufacturer's instructions. The amount of DNA was kept constant in each transfection by adding empty cytomegalovirus (CMV)-Flag vector where necessary. After 24 to 48 h, cells were lysed at 4°C in 25 mM Tris (pH 7.8), 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1% Triton X-100, 2 mM dithiothreitol (DTT), 0.3 mM phenylmethylsulfonyl fluoride (PMSF), and 2 μg/ml aprotinin. The luciferase activity was measured by luminometry. Luciferase values were normalized by cotransfecting a β-galactosidase expression vector.
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts were prepared from BMDMs after different treatment periods as described in the figure legends. Double-stranded [γ-32P]ATP NF-κB oligonucleotides (5′-AGT TGA GGG GAC TTT CCC AGC-3′ and 3′-TCA ACT CCC CTG AAA GGG TCG-5′) were used as probes. Nuclear extracts (2 μg of protein) were incubated for 20 min at 20°C in a total volume of 15 μl buffer containing 48 mM KCl, 38 mM HEPES, 1.2 mM MgCl2, 6% glycerol, 3.3% Ficoll, and 3.6 mM DTT in the presence of 2 μg of poly(dI:dC) as a nonspecific competitor. DNA-protein complexes were resolved on native (5%) polyacrylamide gels, which were subsequently dried and visualized by autoradiography.
Coimmunoprecipitation and immunoblotting.
Whole-cell extracts were incubated with specific antibodies, as indicated in the figure legends, for 2 h, followed by overnight incubation with protein A-agarose beads (Roche). For immunoprecipitation (IP) with FLAG monoclonal antibody, cell lysates were incubated with anti-FLAG (M2) affinity gel (Sigma). Immunoprecipitates and lysates were resolved by SDS-PAGE and were electrotransferred onto Hybond-C Extra polyvinyldifluoride membranes (Amersham Bioscience). After the blocking, proteins were detected by probing with primary and secondary antibodies as indicated in the figure legends, according to the manufacturers' protocols.
ChIP.
The Affymetrix chromatin immunoprecipitation (ChIP) assay protocol (Affymetrix) was used to perform the ChIP experiments according to the manufacturer's protocols, using lysates of wild-type or Gfi1−/− BMDMs (15 × 106 cells/condition) either untreated or stimulated with 10 ng/ml LPS for the indicated time periods. MatInspector software (Genomatix) was used to analyze the results. Lysates were immunoprecipitated with anti-NF-κB p65 antibody (C-20; Santa Cruz). Quantitative PCR (Q-PCR) was performed with the primer pairs indicated below, utilizing Platinum SYBR green qPCR SuperMix-UDG (Invitrogen) in accordance with the manufacturer's protocols. The enrichment ratio was calculated using the ΔΔCT method, in which the enrichment ratio is defined as 2 raised to the power [(target gene CT for input − control region CT for input) − (target gene CT for IP − control region CT for IP)], where CT is the threshold cycle (45). The primer sets used for the ChIP on the TNF promoter were P1-F (5′-TTATAGCCCTTGGGGAAGAG-3′), P1-R (5′-TTCTCCACCAAGGAAGTTTTC-3′), P2-F (5′-CAGGATTCTGTGGCAATCTG-3′), P2-R (5′-GGTTTCAGTTCTCAGGGTCCTA-3′), P3-F (5′-TCTGAAAGCTGGGTGCAT-3′), P3-R (5′-CCACTTCCTCCAAGAACTCA-3′), P4-F (5′-GACCTCACAAGCCTTCTCCT-3′), P4-R (5′-GAAAACTCACTTGGGAGCAG-3′), P5-F (5′-CCTCCCACTCCTAAACACTCTC-3′), and P5-R (5′-ATGAGGGTTCTGGGGAGA-3′).
RESULTS
Expression of Gfi1 is induced by LPS in a TNF-independent manner.
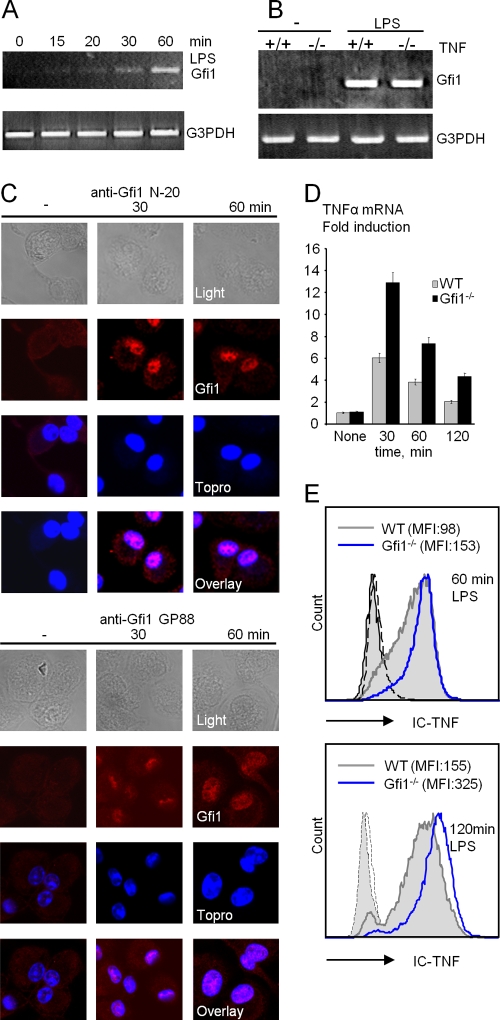
We previously showed that Gfi1 expression can be induced by the TLR4 ligand LPS in primary bone marrow-derived macrophages (BMDMs) obtained from wild-type C57BL/6 mice (30, 32). When the kinetics of this induction in BMDMs was examined, we were able to detect Gfi1 mRNA as early as 30 min after LPS stimulation (Fig. 1A). Interestingly, LPS also induced Gfi1 expression in TNF-α-deficient macrophages, indicating that Gfi1 induction is direct and not mediated by autocrine secretion of TNF-α after LPS stimulation (Fig. 1B). We further investigated whether Gfi1 expression was directly induced by LPS or indirectly through LPS-dependent cytokines. However, we found no induction of Gfi1 mRNA in BMDMs cultured after exposure to TNF-α, IL-1β, IL-6, IL-10, alpha interferon (IFN-α), IFN-γ, granulocyte-macrophage colony-stimulating factor (GM-CSF), or M-CSF (data not shown). Although it has been shown that hypoxia can also activate macrophages to produce proinflammatory cytokines (4), Gfi1 expression was undetectable in hypoxia-activated BMDMs (data not shown), further emphasizing the specificity of Gfi1 expression for TLR signaling in macrophages. When Gfi1 protein expression in BMDMs was investigated, Gfi1 was detected 30 min after LPS exposure using two different anti-Gfi1 antibodies that both revealed similar nuclear staining patterns (Fig. 1C).
FIG. 1.
Induction of Gfi1 expression by TLR4 ligand LPS in wild-type (WT) and Gfi1-deficient (Gfi1−/−) macrophages. (A) Gfi1 mRNA expression is induced in BMDMs as early as 15 to 30 min after the onset of LPS stimulation (10 ng/ml). (B) RT-PCR analysis of LPS-induced Gfi1 expression in wild-type (+/+) and TNF-deficient (−/−) BMDMs. (C) Wild-type BMDMs were treated with 10 ng/ml of LPS for the indicated times, stained with anti-Gfi1 (left panel, N-20; right panel, GP33), and analyzed using confocal microscopy. (D) TNF-α mRNA is increased in Gfi1-deficient macrophages. Quantification by Q-PCR of Gfi1 mRNA levels using RNA from wild-type (WT) and Gfi1−/− BMDMs stimulated with LPS (10 ng/ml) for the indicated times. Representative results from three independent experiments with three independent sets of mice are shown. (E) Flow cytometric analysis of intracellular TNF-α in WT and Gfi1−/− BMDMs after LPS treatment for the indicated times. Numbers in brackets represent mean fluorescence intensities. IC-TNF, intracellular TNF-α.
Enhanced TNF-α expression in Gfi1-deficient macrophages.
We have reported in several independent studies that Gfi1−/− BMDMs respond to LPS with increased TNF-α secretion measured in cell supernatants several hours after LPS stimulation, compared to wild-type BMDMs (14, 30, 32, 41). Consistent with this, we also found higher levels of TNF-α mRNA and intracellular TNF protein in Gfi1−/− BMDMs than in wild-type cells as early as 30 and 60 min after LPS stimulation, respectively (Fig. 1D and E). This suggests that the overproduction of TNF-α seen in Gfi1-deficient macrophages is due to an overproduction of TNF mRNA.
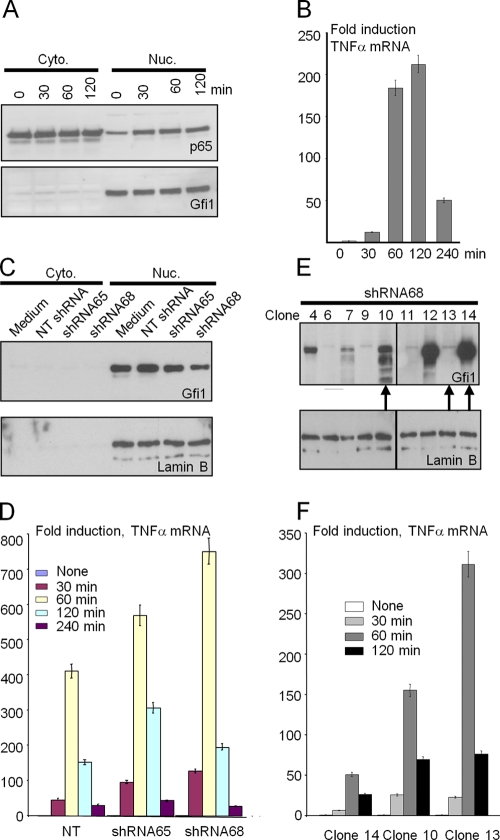
Regulation of TNF-α transcription by Gfi1 in the human cell line THP-1.
To further investigate the potential role of Gfi1 in regulating TNF-α expression, we next used the human monocytic cell line THP-1 in combination with short hairpin RNA (shRNA) against human Gfi1. As expected, LPS stimulation of THP-1 cells resulted in nuclear translocation of p65 NF-κB (Fig. 2A) and rapid induction of TNF-α expression in a time-dependent manner (Fig. 2B). Gfi1 was constitutively expressed and was found exclusively in the nuclear fraction (Fig. 2A). To silence Gfi1 expression, THP-1 cells were infected with two lentiviruses expressing specific shRNAs directed against Gfi1 sequences (shRNA65 and shRNA68) (39). A nontargeting shRNA (NT shRNA) lentivirus was used as a control (39). Cytoplasmic and nuclear fractions of infected cell populations were analyzed for Gfi1 protein levels after puromycin selection. Both shRNA65 and shRNA68 were able to decrease the level of endogenous Gfi1 protein expression, albeit to different levels (Fig. 2C). When these cells were stimulated with LPS for different time periods, we observed that lower Gfi1 expression levels correlated with higher TNF-α expression (Fig. 2D). Since shRNA68 was more efficient in silencing the Gfi1 gene, we used cells infected with this virus to prepare single-cell clones by limiting dilution that expressed different levels of Gfi1 protein (Fig. 2E). On the basis of the level of Gfi1 expression, we selected three different clones (clones 14, 10, and 13 [Fig. 2E, arrows], with high, medium, and low levels of Gfi1 expression, respectively) and stimulated the cells with LPS. We observed a similar inverse correlation between the expression levels of Gfi1 and TNF-α in the three clones as with the bulk culture (Fig. 2F), which supported our hypothesis that Gfi1 is able to dampen TNF-α gene expression upon TLR4 stimulation.
FIG. 2.
Regulation of TNF-α transcription by Gfi1 in THP-1 cells. (A) THP-1 cells stimulated with or without LPS (10 ng/ml) for the indicated times. Cytoplasmic (Cyto.) and nuclear (Nuc.) fractions were then blotted against NF-κB p65 and Gfi1 proteins. (B) THP-1 cells were treated as described for panel A, and the induction of TNF-α mRNA was determined by Q-PCR analysis. (C) THP-1 cells were infected with a lentivirus expressing shRNA against Gfi1. Nontargeting (NT) lentivirus was used as a control. Cytoplasmic (Cyto.) and nuclear (Nuc.) fractions were blotted against Gfi1 proteins. (D) Infected cells were stimulated with or without LPS (100 ng/ml) for the indicated times, and the induction of TNF-α mRNA was determined by Q-PCR analysis. (E) Cells infected with lentivirus expressing shRNA68 against Gfi1 were used to single clone THP-1 cells by a limiting dilution assay. Individual clones, indicated by clone number, were used to analyze Gfi1 expression in the nuclear fraction. The arrows at the bottom of the panel indicate the 3 clones chosen for the Q-PCR analysis shown in panel F. (F) Three individual clones (clones 14, 10, and 13) were stimulated with or without LPS (100 ng/ml) for the indicated periods of time, and the induction of TNF-α mRNA was determined by Q-PCR analysis.
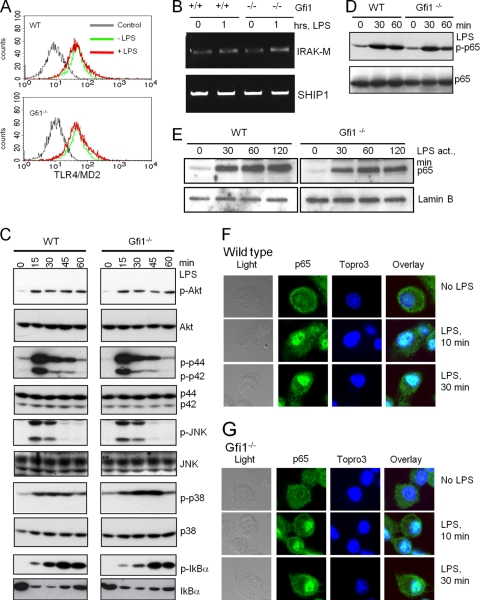
Gfi1 deficiency does not cause alterations in the LPS-induced TLR signaling pathway.
These results prompted us to examine whether Gfi1 deficiency had an effect on TLR4 itself or on its proximal signaling cascade initiated by LPS stimulation. No difference in the expression level of the LPS receptor (TLR4-MD2 complex), the SH2 domain containing inositol phosphatase (SHIP), or the serine/threonine kinase IRAK-M was detected between wild-type and Gfi1-deficient macrophages (Fig. 3 A and B), thus excluding that altered LPS receptor levels could be responsible for the enhanced TNF production in the absence of Gfi1. In addition, we did not detect any deviation of the expression and phosphorylation levels of the main constituents of the LPS signaling pathways that include Erk1/2, c-Jun N-terminal kinase (JNK), p38, and Akt (Fig. 3C). Moreover, Gfi1 deficiency did not alter the levels or the kinetics of IκBα phosphorylation, degradation, or resynthesis (Fig. 3C) or the overall cellular abundance of the p65 NF-κB subunit or its LPS-induced phosphorylation in BMDMs in a very significant manner, although slight differences cannot be entirely ruled out (Fig. 3D). Finally, cellular fractionation confirmed that LPS stimulation caused a rapid nuclear translocation of p65 NF-κB regardless of the presence or absence of Gfi1 (Fig. 3E). The kinetics of the translocation was further examined utilizing confocal immunofluorescence staining and showed that LPS stimulation resulted in nuclear translocation of the p65 NF-κB subunit, with similar kinetics in wild-type (Fig. 3F) and Gfi1−/− (Fig. 3G) BMDMs. Hence, it is very likely that the proximal and distal signaling components of the TLR4 and NF-κB pathways are not strongly affected by the absence of Gfi1.
FIG. 3.
TLR4 signaling is not affected in Gfi1−/− BMDMs. Wild-type (WT) and Gfi1−/− BMDMs were treated with medium or 10 ng/ml of LPS, and cells were harvested at the indicated time points. (A) Flow cytometry of TLR4-MD2 expression of medium-treated (−LPS) and LPS-stimulated (+LPS) wild-type (WT) and Gfi1−/− BMDMs. Staining with irrelevant antibody was used as a control. (B) Expression levels of IRAK-M and SHIP1 determined using RT-PCR. (C) The activation levels of p-Akt, p-Erk, p-JNK, p-p38, and p-IκBα were assessed using immunoblotting. Immunoblot analysis of total endogenous proteins of each signaling molecule was used to ensure equal sample loading. (D) Extracts from WT and Gfi1−/− BMDMs treated as indicated were probed with antibodies against NF-κB subunit p65 or the phosphorylated form of p65 (p-p65). (E) Nuclear extracts of wild-type and Gfi1−/− BMDMs treated as indicated were probed with antibodies against the NF-κB subunit p65. (F) WT and (G) Gfi1−/− BMDMs were treated with 10 ng/ml LPS for the indicated times, and the localization of endogenous NF-κB p65 was assessed by confocal microscopy. Nuclei were visualized by Topro3 staining.
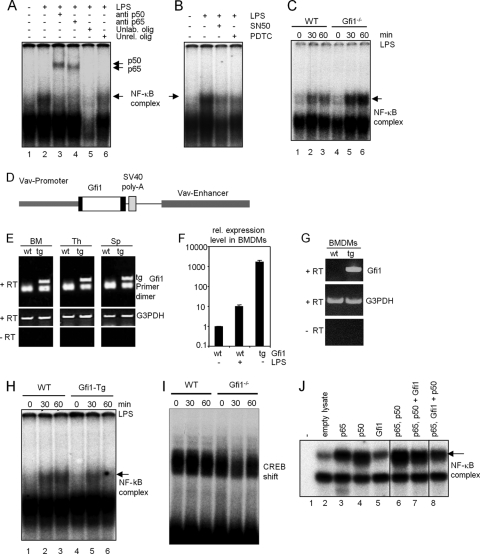
Gfi1 antagonizes NF-κB DNA binding activity.
Next, we explored the effect of Gfi1 on NF-κB DNA binding using electrophoretic mobility shift assays (EMSAs) with nuclear extracts from LPS-stimulated wild-type or Gfi1−/− BMDMs. As expected, we observed a clear induction of NF-κB-DNA complexes in response to LPS stimulation in the nuclear extract from wild-type BMDMs (Fig. 4A, lane 2) and Gfi1−/− BMDMs (not shown). The NF-κB-DNA complex formed after LPS stimulation of BMDMs was composed of the p50 and p65 NF-κB subunits (Fig. 4A, lanes 3 and 4) and was inhibited in the presence of specific inhibitors of NF-κB (Fig. 4B). When wild-type and Gfi1−/− BMDMs were stimulated with LPS under similar conditions, the amounts of the NF-κB-DNA complexes present in nuclear extracts from Gfi1−/− BMDMs were significantly larger (Fig. 4C, lanes 4, 5, and 6) than those detected in extracts from wild-type cells (Fig. 4C, lanes 1, 2, and 3) at the indicated time points. In contrast, BMDM extracts from Gfi1 transgenic mice, in which a constitutive overexpression of Gfi1 was targeted to all hematopoietic cells by the vav promoter/enhancer (Fig. 4D to G), showed decreased levels of NF-κB-DNA complexes (Fig. 4H). EMSAs with a CREB binding site showed no significant difference in CREB binding activity between nuclear extracts from wild-type and Gfi1−/− BMDMs (Fig. 4I), indicating the specificity of the assay. Similarly, when in vitro-translated proteins were used for EMSA, Gfi1 was found to interfere with the binding of the p65-p50 complex to the NF-κB binding site (Fig. 4J). The effect was detectable when p65 and Gfi1 were cotranslated and p50 was added to the complex (Fig. 4J, lane 8).
FIG. 4.
Enhanced NF-κB DNA binding in Gfi1−/− macrophages. Wild-type (WT) and Gfi1−/− BMDMs were treated with medium or 10 ng/ml of LPS. Nuclear extracts were collected at the indicated time points for an electrophoretic mobility shift assay (EMSA) using a radiolabeled DNA probe containing an NF-κB (p65-p50) binding site sequence. (A) EMSA with nuclear extracts from WT BMDMs before and after LPS stimulation. An arrow indicates the position of the p65-p50-DNA complex, generated after 1 h of LPS stimulation (arrowheads represent supershift with antibodies recognizing the p65 and p50 subunits of NF-κB). Addition of excess unlabeled NF-κB oligonucleotide or unlabeled irrelevant oligonucleotide (lanes 5 and 6) or NF-κB inhibitors (SN-50 and PDTC) (B) disrupted the formation of the p65-p50-DNA complexes and demonstrated the specificity of the binding reaction. (C) EMSA with nuclear extracts from WT and Gfi1−/− BMDMs before and after LPS stimulation. An arrow indicates the induction of the p65-p50-DNA complex after LPS stimulation in wild-type (lanes 1 to 3) and Gfi1−/− (lanes 4 to 6) BMDMs. (D) Schematic representation of the vav-Gfi1 construct used to generate transgenic (tg) mice. (E) Expression of the vav-Gfi1 transgene detected by RT-PCR in bone marrow (BM), thymus (Th), and spleen (Sp). (F and G) Expression of the vav-Gfi1 transgene detected in BMDMs by Q-PCR (F) and by RT-PCR (G). (H) EMSA with nuclear BMDM extracts from WT or vav-Gfi1 tg mice before and after LPS stimulation. An arrow indicates the position of the p65-p50-DNA complex, generated after 1 h of LPS stimulation. (I) Control shift experiments with a labeled CREB binding site revealed similar amounts of protein in all samples of nuclear extracts from wild-type (WT) and Gfi1−/− BMDMs. (J) EMSA with in vitro-translated proteins as indicated. Lane 7, p65 and p50 were cotranslated, and then Gfi1 in vitro-translated protein was added; lane 8, p65 and Gfi1 were cotranslated, and then p50 in vitro-translated protein was added.
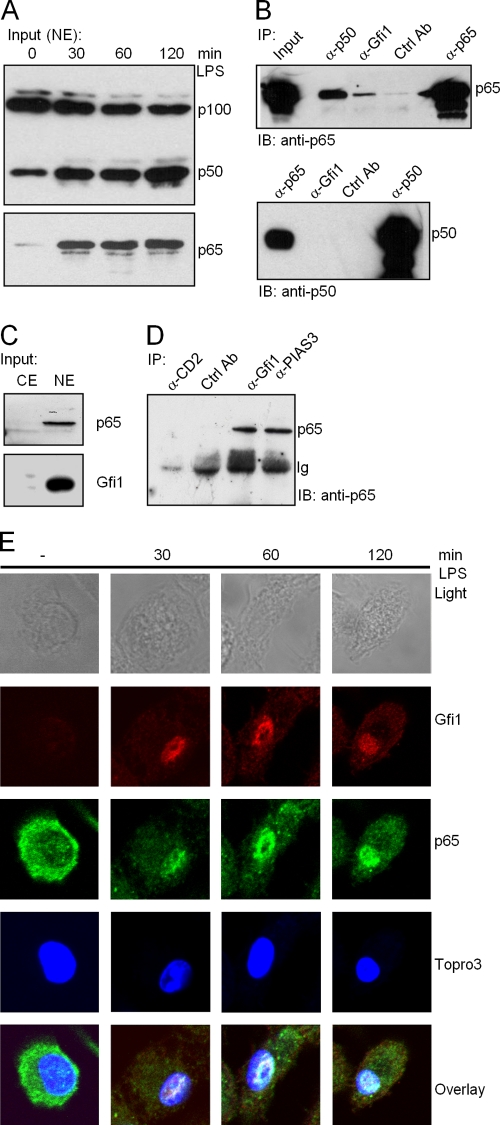
Gfi1 interacts with the p65 subunit of NF-κB.
Given the results gathered from the NF-κB DNA binding studies, we next tested whether endogenous Gfi1 and NF-κB directly interact. To this end, we performed immune precipitation assays with extracts from both THP-1 and Jurkat cells, which express both proteins endogenously (Fig. 2A and 5C). Stimulation of THP-1 cells with LPS at the indicated time points significantly increased the amounts of both the p50 and the p65 NF-κB subunits in the nuclear fraction (Fig. 5A). As shown in Fig. 5B, the NF-κB p65 subunit was coprecipitated with anti-Gfi1- or anti-p50-specific antibodies. Irrelevant control antibody failed to pull down the p65 subunit, excluding immunoprecipitation due to nonspecific binding. Immunoprecipitation with anti-p65 was used as a positive control. These results indicated that p65 forms complexes with both the Gfi1 and p50 proteins.
FIG. 5.
Endogenous NF-κB subunit p65, but not p50, interacts with Gfi1. (A) Nuclear translocation of NF-κB subunits p65 and p50 after LPS stimulation of THP-1 cells for the indicated times. NE, nuclear extracts. (B) Extracts from THP-1 cells were immunoprecipitated with the indicated antibodies, and the precipitates were analyzed by immunoblotting using anti-p65 antibody (upper panel) or anti-p50 antibody (lower panel). (C) Input levels of endogenous p65 and Gfi1 proteins in Jurkat cells; both are readily detected in Jurkat nuclear extracts (NE) but are absent in cytoplasmic extracts (CE). (D) Extracts from Jurkat cells were immunoprecipitated with the indicated antibodies, and the precipitates were analyzed by immunoblotting using anti-p65 antibodies. Anti-PIAS3 was used as a positive control. Anti-CD2 and anti- atrophrin were used as negative controls and show the specificity of p65 immunoprecipitation. (E) Wild-type BMDMs were treated with 10 ng/ml LPS for the indicated periods of time, and the localization of endogenous NF-κB and Gfi1 proteins was assessed by confocal microscopy. Nuclei were visualized by Topro3 staining.
Since the NF-κB-DNA complexes formed after LPS stimulation of BMDMs contained both the p50 and the p65 NF-κB subunits (Fig. 4A), we checked for a Gfi1-p50 interaction under similar conditions. The results revealed that the NF-κB p50 subunit was not coprecipitated with anti-Gfi1 antibody (Fig. 5B, lower panel). Immunoprecipitation with anti-p65 was used as a positive control. Similarly, when extracts from Jurkat cells were used, p65 was coprecipitated with an anti-Gfi1-specific antibody (Fig. 5C and D) or, as a positive control, with an antibody against PIAS3, which has been shown to bind to p65 (27). Irrelevant antibodies against CD2 or atrophin failed to pull down the NF-κB p65 subunit, validating the specificity of the conditions used for immunoprecipitation (Fig. 5D). The results indicated that Gfi1 forms complexes only with the NF-κB p65 subunit and not with the p50 subunit.
We next examined the Gfi1-p65 interaction at the endogenous level in BMDMs in which the NF-κB pathway was activated by LPS stimulation for the indicated periods of time. Cells were analyzed by confocal microscopy using specific antibodies against Gfi1 and the NF-κB p65 subunit, and the nucleus was observed using DNA-specific staining with the dye Topro3. Whereas Gfi1 protein was absent in unstimulated BMDMs, a noticeable induction of Gfi1 was observed in the nucleus after 30 min of LPS treatment (Fig. 5E). As expected, p65 was detectable in the cytoplasm of unstimulated cells; however, p65 was found to rapidly translocate from the cytoplasm to the nucleus 30 min after LPS stimulation (Fig. 5E). Consistent with our immunoprecipitation experiments, confocal microscopy clearly showed that Gfi1 and p65 were colocalized in specific regions of the nucleus after LPS induced the p65 nuclear translocation (Fig. 5E).
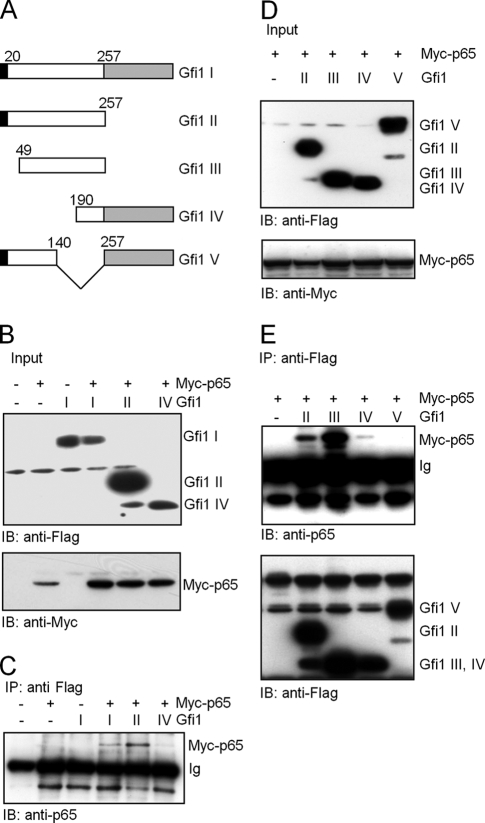
To narrow down a specific Gfi1 protein domain responsible for the Gfi1-p65 interaction, similar immunoprecipitation experiments were performed with NIH 3T3 cells transfected with constructs encoding Myc-tagged p65, Flag-tagged full-length Gfi1, or mutants of Gfi1 (Fig. 6A). Twenty-four hours posttransfection, whole-cell lysates were prepared and used for coimmunoprecipitation assays using an anti-Flag antibody, followed by immunoblotting with an anti-p65-specific antibody. Full-length Gfi1 and Gfi1 mutants III and IV, which lack the SNAG domain, and Gfi1 mutants II and III, which lack the zinc finger DNA binding motifs, were still able to bind p65. In contrast, deletions present in Gfi1 mutant V located in the intermediary domain of Gfi1 abolished the Gfi1-p65 interaction. This points to the possibility that a region between aa positions 140 and 257 may play a role in the Gfi1-p65 interaction.
FIG. 6.
Interaction of Gfi1 with the p65 subunit of NF-κB in NIH 3T3 transfected cells. (A) Schematic representation of full-length Gfi1 (I) and four Gfi1 mutants (II, III, IV, and V) used for coimmunoprecipitation after transfection of constructs into NIH 3T3 cells. Gfi1 deletion mutants Gfi1 I (positions 1 to 257) and Gfi1 III (positions 49 to 257) lack the C-terminal zinc finger domain (gray box), and Gfi1 mutants III and IV lack the N-terminal SNAG repressor domain (black box). (B) NIH 3T3 cells were transiently transfected with Flag-tagged full-length Gfi1 (I) or the Gfi1 mutants (II and IV) and Myc-tagged p65, as indicated, and input levels were controlled by immunoblotting (IB) with anti-Myc or anti-Flag antibodies. (C) Whole-cell lysates of the transfected cells were subjected to coimmunoprecipitation (IP) with anti-Flag antibodies, followed by immunoblotting (IB) with anti-p65 antibodies. (D) NIH 3T3 cells were transiently transfected with the indicated Flag-tagged Gfi1 mutants (II, III, IV, and V) and Myc-tagged p65, as indicated, and input levels were controlled by immunoblotting (IB) with anti-Myc or anti-Flag antibodies. (E) Whole-cell lysates of transfected cells were subjected to coimmunoprecipitation (IP) with anti-Flag antibodies, followed by immunoblotting (IB) with anti-p65 or anti-Flag antibodies.
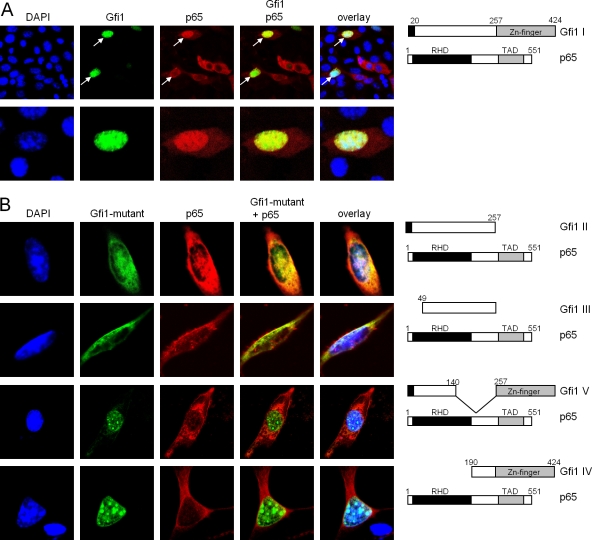
To further validate the interaction between Gfi1 and the p65 subunit of NF-κB, we used immunofluorescence analyses of cells transiently transfected with constructs able to direct the expression of either Gfi1-GFP fusion proteins, Flag-tagged Gfi1 (Gfi1 I) or Gfi1 mutants (Gfi1 II to Gfi1 V), or full-length p65 in different combinations. When both full-length Gfi1 (Gfi1 I) and p65 were coexpressed, a complete merge of both fluorescence signals was observed, indicating an interaction of both in the nucleus (Fig. 7A). Deletion of the zinc finger domain of Gfi1 (Gfi1 II) or both the SNAG repressor domain and the zinc finger domains (Gfi1 III) resulted in a partial loss of colocalization of p65 and Gfi1 (Fig. 7B). Deletions in the intermediary domain of Gfi1 (mutants Gfi1 IV and V) resulted in a complete loss of p65-Gfi1 interaction (Fig. 7B).
FIG. 7.
Gfi1 colocalizes with p65 in the nucleus. (A) NIH 3T3 fibroblasts were transfected with constructs directing the expression of full-length Gfi1 as a fusion protein with GFP or a full-length Myc-tagged p65 protein. Nuclei were visualized by DAPI (4′,6-diamidino-2-phenylindole) staining, and p65 was visualized by staining with anti-Myc antibodies and rhodamine-labeled secondary antibodies. Cells were analyzed with a laser scanning microscope (LSM). The merged pictures demonstrate colocalization of Gfi1 (green) and p65 (red) in cells that coexpress both proteins (white arrows). (B) Cotransfection of the indicated expression constructs, as described for panel A, that allow the production of either full-length Myc-tagged p65 or the Flag-tagged Gfi1 II, III, V, and IV mutants. Gfi1 mutants II and III show a partial colocalization with p65 (first and second rows), but Gfi1 mutants IV and V showed no colocalization (third and fourth rows).
Gfi1 interferes with p65 DNA binding and not p65 transcriptional transactivation.
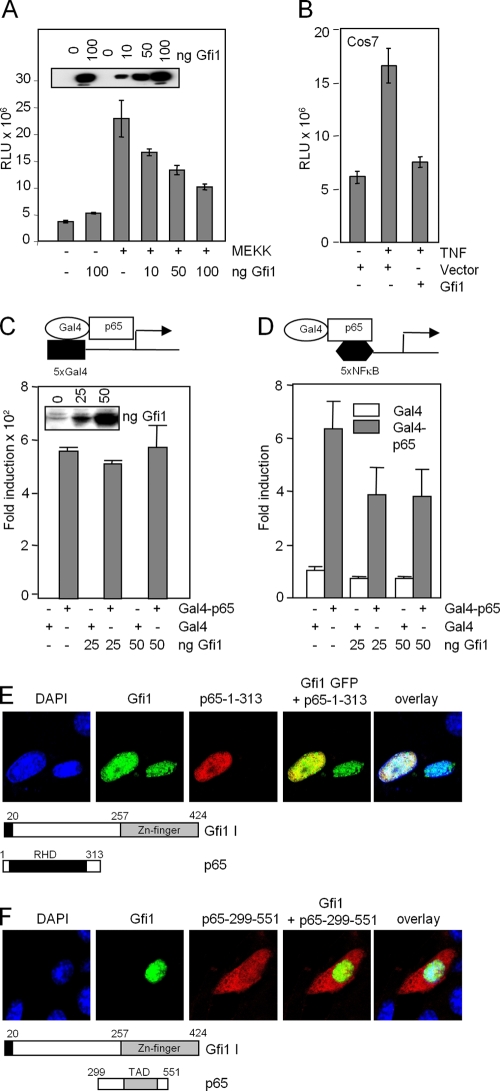
The NF-κB p65 subunit has been shown to possess both a DNA binding domain for its cognate NF-κB binding site on target gene promoter DNA and a transactivation domain for binding to other proteins (10, 47). To distinguish which of these p65 functions was affected by Gfi1, we first cotransfected a luciferase reporter construct containing five synthetic NF-κB binding sites (5xNF-κB) with increasing amounts of Gfi1 expression vector. MEK kinase (MEKK) was used to activate the NF-κB signaling pathway in transfected RK13 cells (17), and we observed that overexpression of Gfi1 inhibited NF-κB-mediated gene activation in a dose-dependent manner (Fig. 8A). A similar effect of Gfi1 could be observed with the same reporter in Cos7 cells treated with TNF to activate the NF-κB pathway (Fig. 8B).
FIG. 8.
Gfi1 represses NF-κB transcriptional activity and binds to the p65 Rel homology domain. (A and B) Reporter gene assays with RK13 cells (A) or COS7 cells (B) with a luciferase vector containing 5xNF-κB binding sites. Activation of NF-κB was achieved by cotransfecting the expression constructs for MAP kinase MEKK (A) or by treating the cells with TNF that activates p65 (B). Expression levels of Gfi1 are shown in the inset in panel A. RLU, relative light units. (C and D) Gfi1 interferes with DNA binding and not transcriptional transactivation of p65. Reporter gene assays were performed with NIH 3T3 cells transfected with combinations of expression constructs encoding Gal4 (white bar) or Gal4-p65 (gray bar) fusion proteins, increasing amounts of Flag-Gfi1, and either a 5xGal4 reporter (C) or a 5xNF-κB reporter (D). All data are representative of three independent experiments. (E) Coexpression of the indicated p65 mutants lack-ing either the TAD domain or the Rel homology domain (RHD) with full-length Gfi1. Interaction between Gfi1 and the RHD part of p65 is clearly observed. (F) The p65 mutant containing only the TAD remains in the cytoplasm, indicating that it lacks the ability to interact with Gfi1.
This effect of Gfi1 on NF-κB-mediated gene activation could be due to either Gfi1 blocking DNA binding of the NF-κB p65-p50 heterodimer to its target NF-κB binding sites or, alternatively, Gfi1 inhibiting p65-mediated gene transactivation. To address these possibilities, we used a Gal4-p65 fusion protein, which can activate reporter constructs carrying either Gal4 binding sites or NF-κB binding sites. This Gal4-p65 expression construct was transiently transfected into NIH 3T3 cells with reporter constructs containing either five copies of the Gal4 binding site (5xGal4) or five copies of the NF-κB binding site (5xNF-κB) (Fig. 8C and D). Gfi1 showed an inhibitory effect on Gal4-p65-induced transcriptional activity only when the 5xNF-κB reporter construct was used (Fig. 8D). In contrast, no inhibitory effect of Gfi1 on transcriptional activity was noted when the 5xGal4 reporter construct was used (Fig. 8C). These results suggest that Gfi1 may act by interfering with DNA binding of p65 to NF-κB binding sites in promoter DNA and that Gfi1 does not act by blocking transcriptional transactivation by p65.
Immunofluorescence experiments with transfected cells showed that when p65 deletion mutants were coexpressed with Gfi1, a colocalization indicating an interaction was observed only with the Rel homology domain (RHD), not with the trans-activation domain of p65 (Fig. 8E and F), confirming that Gfi1 interacts with the DNA binding domain and not with the transactivating domain of p65.
Gfi1 controls TNF-α promoter occupancy by NF-κB.
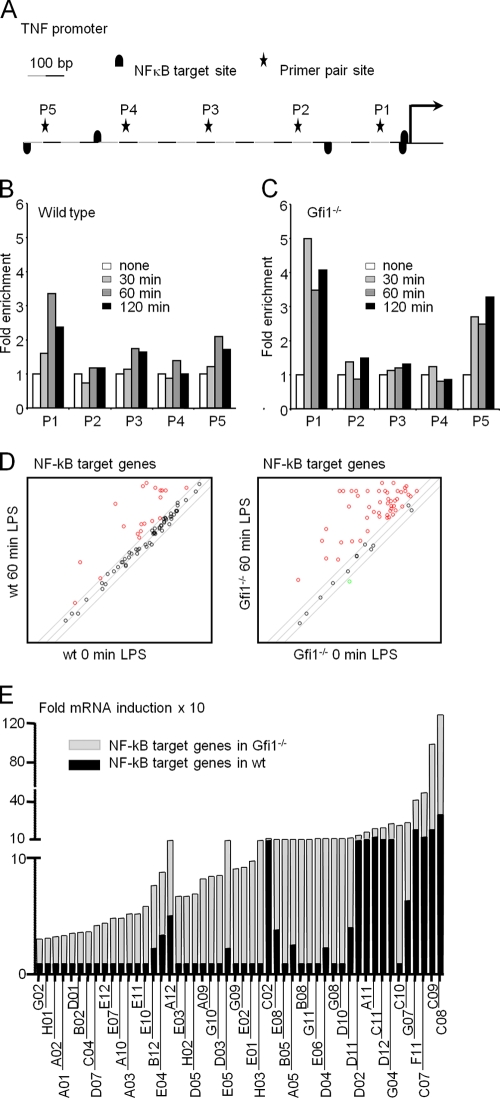
To test whether Gfi1 could interfere with p65 promoter binding in BMDMs, we stimulated wild-type and Gfi1-deficient BMDMs with LPS and performed chromatin immunoprecipitation (ChIP) with antibodies specific for the NF-κB p65 subunit and five sets of primer pairs that cover 1 kb of the TNF-α promoter region in 200-bp intervals (P1 to P5) (Fig. 9A). The first proximal site (P1) in the TNF promoter was found to be occupied by p65 as early as 30 min after LPS treatment in wild-type BMDMs (Fig. 9B). In addition, the recruitment of the p65 subunit to the P1 region of the TNF-α promoter peaked at 60 min and decreased after 120 min of LPS stimulation (Fig. 9B). Very little or no binding of p65 was detected at the other primer pairs sites of the TNF-α promoter (P2-P5). In comparison to what was observed in wild-type BMDMs, the occupancy rate of the p65 subunit at the P1 region of the TNF-α promoter was markedly increased in Gfi1−/− BMDMs as early as 30 min after LPS treatment (Fig. 9C). Importantly, the recruitment of the p65 subunit to the P1 region of the TNF-α promoter appeared to be sustained at high levels even after 120 min of LPS stimulation. A similar increased and sustained level of p65 subunit occupancy at the P5 region of the TNF-α promoter was also detected in Gfi1−/− BMDMs, compared to the level in wild-type BMDMs after LPS stimulation (Fig. 9C), suggesting that two sites for NF-κB binding exist in the TNF-α promoter region and that their rates of occupancy by p65 depend on the presence or absence of Gfi1.
FIG. 9.
Enhanced TNF-α promoter occupancy in Gfi1-deficient macrophages and regulation of NF-κB target gene expression by Gfi1. (A) Schematic representation of 1 kb of the TNF-α promoter as assessed by ChIP analysis. Primer pair sites (P1 to P5) and NF-κB target sites are indicated. Wild-type (B) and Gfi1−/− (C) BMDMs were treated with medium or 10 ng/ml of LPS for the indicated periods of time. Cells were harvested for ChIP experiments with anti-p65 antibodies, and occupancy of p65 at the indicated sites was determined using Q-PCR with five sets of primer pairs. Data represent the average relative fold inductions at each primer site with respect to the level for nontreated BMDMs (set to 1). The data are representative of two independent sets of experiments. (D) BMDMs from wild-type (wt) and Gfi1−/− mice were stimulated with LPS (10 ng/ml) for 60 min or wereleft unstimulated (0 min). Total RNA was extracted and used for the PCR array. Scatter plot of NF-κB target genes induced 60 min after LPS stimulation in BMDMs from WT mice (left panel; red circles represent 19 genes) and Gfi1−/− mice (right panel; red circles represent 50 genes). (E) Relative fold mRNA inductions of NF-κB target genes in wild-type mice (WT; black columns) and Gfi1−/− mice (KO; gray columns). The code of each NF-κB target gene is indicated based on the manufacturer's indications for the PCR array. The gene products corresponding to the codes are indicated in Table 1.
Gfi1 is a negative regulator of NF-κB target genes.
These results prompted us to investigate whether the absence of Gfi1 only affects p65-mediated expression of the TNF-α gene or whether Gfi1 might have a role as a more general regulator of many or all NF-κB target genes in BMDMs. To test this, we stimulated wild-type and Gfi1-deficient BMDMs with LPS for 60 min and used a PCR array to evaluate whether the expression of 84 known direct NF-κB target genes and other inflammatory effector genes are affected by Gfi1. We observed that 19 NF-κB target genes (22.6% of all target genes tested) were upregulated after LPS stimulation in wild-type BMDMs (Fig. 9D, left panel, and E, black bars). In contrast, we found that in Gfi1-deficient BMDMs, 50 NF-κB target genes (59.5% of all target genes tested) were upregulated after LPS stimulation (Fig. 9D, right panel, and E, gray bars). Moreover, the degree of upregulation (relative fold induction) of NF-κB target genes was significantly higher in Gfi1-deficient BMDMs than in wild-type BMDMs (Fig. 9E, gray bars, and Table 1), indicating that Gfi1 participates in the regulation of many NF-κB target genes and not just the TNF-α gene.
TABLE 1.
Relative fold inductions of NF-κB targets in wild-type and Gfi1−/− BMDMs by LPS stimulation
| Position | Gene product | Fold inductiona |
|
|---|---|---|---|
| wt | Gfi1−/− | ||
| G02 | Tnfrsf1a | 1.0 | 2.1 |
| H01 | Gusb | 1.0 | 2.2 |
| A02 | Atf1 | 1.0 | 2.3 |
| A01 | Akt1 | 1.0 | 2.4 |
| D01 | Irak2 | 1.0 | 2.6 |
| B02 | Crebbp | 1.0 | 2.7 |
| C04 | Ikbkb | 1.0 | 2.8 |
| D07 | Mapk3 | 1.0 | 3.3 |
| E12 | Tbk1 | 1.0 | 3.5 |
| E07 | Ripk1 | 1.0 | 3.9 |
| A10 | Casp8 | 1.0 | 3.9 |
| A03 | Atf2 | 1.0 | 4.3 |
| E11 | Stat1 | 1.0 | 4.3 |
| E10 | Smad3 | 1.0 | 4.9 |
| B12 | Gja1 | 2.3 | 5.4 |
| E04 | Rel | 3.5 | 5.4 |
| A12 | Cflar | 5.1 | 5.7 |
| E03 | Raf1 | 1.0 | 5.8 |
| H02 | Hprt1 | 1.0 | 5.8 |
| D05 | Ltbr | 1.0 | 6.0 |
| A09 | Casp1 | 1.0 | 7.3 |
| G10 | Traf2 | 1.0 | 7.5 |
| D03 | Jun | 1.0 | 7.6 |
| E05 | Rela | 2.3 | 7.9 |
| G09 | Tradd | 1.0 | 8.2 |
| E02 | Eif2ak2 | 1.0 | 8.3 |
| E01 | Kaf2b | 1.0 | 8.8 |
| H03 | Hsp90ab1 | 1.0 | 10.9 |
| C02 | Icam1 | 11.9 | 12.0 |
| E08 | Ripk2 | 3.9 | 14.0 |
| B05 | Lpar1 | 1.0 | 14.4 |
| A05 | Bcl3 | 2.6 | 14.7 |
| B08 | F2r | 1.0 | 16.3 |
| G11 | Traf3 | 1.0 | 18.5 |
| E06 | Relb | 1.0 | 22.2 |
| D04 | Lta | 2.4 | 25.1 |
| G08 | Tollip | 1.0 | 28.8 |
| D10 | Nfkb1 | 1.0 | 29.2 |
| D11 | Nfkb2 | 4.1 | 29.7 |
| D02 | Irf1 | 14.1 | 49.5 |
| A11 | Ccl2 | 21.3 | 71.5 |
| C11 | IL-6 | 40.8 | 88.0 |
| D12 | Nfkbia | 18.6 | 120.3 |
| G04 | Cd40 | 22.3 | 157.6 |
| C10 | IL1r1 | 1.0 | 162.0 |
| G07 | Tnfsf14 | 6.4 | 179.8 |
| F11 | Tnf | 117.8 | 306.6 |
| C07 | IL-10 | 40.2 | 455.1 |
| C09 | IL1b | 123.6 | 873.1 |
| C08 | IL1a | 272.5 | 1173.6 |
Values shown are relative fold inductions of 50 NF-κB target genes upregulated by LPS stimulation in wild type (wt) and Gfi1−/− BMDMs.
DISCUSSION
The transcription factor NF-κB is an essential element of the TLR-mediated response in cells of the innate immune system. In macrophages, one of the physiological consequences of TLR stimulation is the release of inflammatory cytokines such as TNF, IL-1β and IL-6. The latent transcription factor NF-κB functions as an important bridge connecting TLR stimulation by endotoxins and the production of proinflammatory cytokines, in particular the TNF-α gene, which is a direct NF-κB target gene (1, 18, 56). Here, we present evidence that in mouse macrophages, the zinc finger protein Gfi1 regulates the TLR signaling pathway by directly antagonizing NF-κB and preventing it from binding to its two NF-κB binding sites in target gene TNF-α promoter DNA after LPS stimulation. This finding may have important implications since a better understanding of how TLR signaling is regulated may facilitate the development of new strategies for controlling TLR-mediated inflammatory diseases.
We have reported that macrophages upregulate Gfi1 mRNA expression after stimulation with LPS (30, 32, 41) and also upon treatment with other TLR ligands, such as CpG and PGN (41). Our experiments with other cytokines and with TNF-deficient cells indicate that the induction of Gfi1 mRNA expression is elicited upon TLR4 signaling and not by inflammatory cytokines that are produced after TLR stimulation, for instance, through an autocrine loop. Our findings that TNF-deficient cells show Gfi1 mRNA induction similar to that in wild-type cells and the observation that TNF, IL-1β, and IL-6 cannot induce Gfi1 expression support this notion. Moreover, in contrast to what was found for other previously described regulators of TLR signaling, such as SHIP1 (50), SOCS1 (13, 34, 42), and IRAK-M (35), the rapid induction of Gfi1 mRNA transcription seen within minutes of LPS stimulation in the present study is consistent with a role for Gfi1 as an immediate-early negative regulator of TLR signaling.
A second very important observation that we reported earlier was that Gfi1-deficient macrophages react with increased production of TNF-α and that Gfi1-deficient mice succumb quickly, with symptoms of septic shock, after LPS treatment (32). In TNF−/−/Gfi1−/− double-deficient mice, the effect of Gfi1 deficiency is rescued (30), suggesting that the high susceptibility of Gfi1 knockout mice toward LPS-induced septic shock is indeed mediated by heightened TNF-α production. In light of these findings, we had hypothesized that Gfi1 is induced by LPS through TLR signaling and acts as an upstream negative regulator of the TNF gene. Our new finding reported here, that in Gfi1−/− macrophages the TNF-α mRNA level is increased severalfold over the wild-type level as early as 30 min after LPS stimulation, supports this conclusion and further emphasizes the role of Gfi1 in the regulation of TNF-α expression at the transcriptional level.
TLR activation is important for an infected host organism, since, on the one hand, it is essential for provoking the innate response and enhancing adaptive immunity against pathogens (2, 44), while, on the other hand, members of the TLR family are also involved in the pathogenesis of autoimmune, chronic inflammatory, and infectious diseases (6). One of the most severe diseases is sepsis caused by LPS, an agonist of TLR4 (46). A number of negative regulatory mechanisms that can dampen TLR-signaling pathways exist and have been described, suggesting multiple distinct types of safety mechanisms for controlling harmful inflammatory responses (5, 28, 29, 35, 53). Many of the negative regulators involved act directly on proximal signaling events, for instance, at the level of adaptor molecules or upstream kinases (e.g., MyD88, IRAK, and TRAF6), which then affect more-distal events, such as the activation of MAP kinases and NF-κB. Our experiments suggest that Gfi1 does not interfere with the TLR signaling pathway at the proximal level, since the activation of cytoplasmic signaling molecules of the MAP kinase and PI3K pathways, as well as the cytoplasmic and nuclear components of the NF-κB pathway, appears unaltered in Gfi1-deficient cells after LPS stimulation when compared to the level in wild-type cells, although more-subtle effects cannot be entirely ruled out.
It is conceivable that Gfi1 dampens the physiological effects of the TLR4 response by downregulating TNF-α mRNA production through two mechanisms, first, by direct interference with the action of NF-κB, for instance, by blocking its binding to DNA, and second, by inhibiting its transactivation capacity. Our experiments presented here support the first mechanism, and we propose a model in which Gfi1 interacts with the p65 subunit of NF-κB and prevents it from binding to its cognate NF-κB binding sites present in the TNF-α promoter and very likely also the promoters of more than 50 other NF-κB target genes. Several lines of evidence support this hypothesis. First, electrophoretic mobility shift assays demonstrated that increased amounts of NF-κB-DNA complexes are present in LPS-stimulated BMDMs from Gfi1 knockout mice and decreased amounts in BMDMs from overexpressing vav-Gfi1 transgenic mice, compared to the amounts in wild-type macrophages. Second, reporter gene assays showed that Gfi1 blocks NF-κB-dependent transcription of synthetic target gene promoters appended to a luciferase gene. Third, when Gal4 fusions to the DNA binding or transactivation domains of p65 were tested, Gfi1 was found to inhibit only the activity of the fusion protein that retained the DNA binding domain of p65. Fourth, immunofluorescence experiments using transfected cells indicated that Gfi1 binds to the RHD domain of the p65 subunit of NF-κB, which contains sequences that contact DNA. Fifth, chromatin immunoprecipitation experiments with primers covering the TNF-α promoter demonstrated a higher rate of occupancy of the proximal NF-κB binding site by the p65 subunit in Gfi1-deficient macrophages than in wild-type cells. Last, PCR array analysis showed derepression of many NF-κB target genes in Gfi1-deficient BMDMs, compared to the level in wild-type BMDMs, suggesting that Gfi1 may play a role as a general negative regulator of NF-κB in inflammatory responses. Although these experiments are consistent with a direct interaction between Gfi1 and p65, it cannot be ruled out that other proteins act as intermediary factors or that the effect of Gfi1 on p65 and the NF-κB complex is mediated by a more indirect mechanism. More experimentation is required to resolve this question.
Our experiments show that 30 min after LPS stimulation, expression of the Gfi1 gene is induced, and Gfi1 protein is made and forms a complex with the NF-κB subunit p65 protein, which has translocated to the nucleus by this time. Also, at this time, TNF-α mRNA is upregulated and is present in Gfi1-deficient cells at a higher level than in wild-type cells. In addition, our ChIP experiments show that the TNF-α promoter is occupied by the NF-κB p65 subunit and that TNF-α mRNA is expressed as early as 30 min after LPS stimulation. When Gfi1 is absent, the TNF-α promoter shows a markedly higher p65 occupancy rate at two sites than at one site in wild-type cells and also a higher level of expression than in wild-type cells 30 min after LPS stimulation. This could be consistent with two different models: in the first model, the interaction between p65 and Gfi1 proteins directly precludes the access of p65 to the TNF-α promoter; in the second model, Gfi1 protein may act indirectly by masking a region in the RH domain of p65, which is responsible for the heterodimerization of p65 with p50 (which is required for promoter binding). The latter model is supported by gel shift experiments with in vitro-translated p65, p50, and Gfi1. Both models could explain how the binding of p65 to NF-κB target gene promoters is prevented by an interaction of Gfi1 with p65.
It remains to be shown how the interaction of p65 with Gfi1 precisely inhibits the DNA binding of NF-κB. One possibility is that Gfi1 simply competes with NF-κB binding sites for p65 and that in the absence of Gfi1, more p65 is free to access the TNF-α promoter, as reflected by a higher p65 occupancy rate as seen in our ChIP experiments. Since no known Gfi1 binding sites are present in the 1-kb proximal region of the TNF-α promoter tested here, and we were unable to detect Gfi1 by ChIP on the 1-kb 5′ upstream region of the TNF-α promoter (not shown), it seems unlikely that Gfi1 occupies the TNF-α promoter either in a direct way or through binding to p65. In addition, a model in which Gfi1 binds to NF-κB target sites through p65 seems unlikely since this model would be inconsistent with a number of findings reported here, such as the higher promoter occupancy rate of p65 in Gfi1-deficient cells, the increased level of p65-DNA complex formation in the absence of Gfi1 as detected by EMSA, and finally also data from our luciferase reporter assays using Gal4-p65 fusion proteins.
Since Gfi1 is a transcriptional repressor, another alternative explanation for the upregulation of NF-κB target gene expression could be that Gfi1 represses a coactivator of p65 or another transcriptional activator of p65 target genes. Again, this seems unlikely in light of our findings reported here. It is difficult to picture how such a model would lead to a higher level of p65-DNA complex formation or a higher rate of p65 occupancy at target gene promoters. Considering this, a more likely model is that after LPS stimulation, Gfi1 competes with NF-κB binding sites for p65 and simply titrates out p65 molecules able to bind to NF-κB target promoters rapidly. The facts that higher levels of p65-DNA complexes can be detected in Gfi1−/− cells and that a higher rate of promoter occupancy by p65 can be found in the absence of Gfi1 also argues for this hypothesis. This model would explain why a large number of NF-κB-responsive genes become upregulated by p65 in Gfi1 deficient cells and is consistent with the kinetics of the effects observed after TLR4 stimulation. However, it does not explain why some of the genes are not superinducible by Gfi1 deficiency. This may be due to a mechanism that restricts the action of Gfi1 to a subset of p65 target genes. A number of other p65-responsive promoters have to be tested similarly to the TNF promoter to answer this question and to confirm this hypothesis.
The evidence that we present here supports the view that the transcription factor Gfi1 acts as a general negative regulator for the TLR4 signaling pathway, which is critical for the transduction of inflammatory signals, for example, after exposure to bacterial cell wall antigens such as the endotoxin LPS. A clinical manifestation where this signaling pathway rapidly derails and is not under appropriate control is septic shock, and Gfi1-deficient mice treated with LPS indeed show symptoms reminiscent of septic shock. Our results suggest that Gfi1 prevents overactivity of the LPS-TLR4 pathway by dampening the effects of NF-κB at the downstream endpoint of TLR4 signaling in the nucleus. This represents an important advance in our understanding of the mechanisms that can lead to unregulated inflammatory reactions and may provide a theoretical basis for future interventional strategies designed to prevent death from septic shock.
Acknowledgments
We are indebted to Angelika Warda, Wojciech Wegrzyn, and Inge Spratte for technical assistance, to Petra Plessow and Tomas Civela for excellent animal care, and to K. Shuai, K. Ruckdeschel, and E. Gulbins for critical reading of the manuscript. We particularly thank Helmut Brade for purified LPS and Ke Shuai for providing reporter gene constructs and p65 expression vectors. We also thank Hugo Bellen for anti-Gfi1 antibodies.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG), SPP 1110 Innate Immunity, by the Fonds der Chemischen Industrie and the IFORES Program of the University of Essen Medical School, by the Institut de Recherches Cliniques de Montreal (IRCM), by the Canadian Foundation for Innovation, by the Canadian Institutes for Health Research, and by a Canada Research Chair (tier 1) to T.M.
Footnotes
Published ahead of print on 14 June 2010.
REFERENCES
- 1.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 3.Beutler, B., and E. T. Rietschel. 2003. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 3:169-176. [DOI] [PubMed] [Google Scholar]
- 4.Blouin, C. C., E. L. Page, G. M. Soucy, and D. E. Richard. 2004. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood 103:1124-1130. [DOI] [PubMed] [Google Scholar]
- 5.Burns, K., J. Clatworthy, L. Martin, F. Martinon, C. Plumpton, B. Maschera, A. Lewis, K. Ray, J. Tschopp, and F. Volpe. 2000. Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat. Cell Biol. 2:346-351. [DOI] [PubMed] [Google Scholar]
- 6.Cook, D. N., D. S. Pisetsky, and D. A. Schwartz. 2004. Toll-like receptors in the pathogenesis of human disease. Nat. Immunol. 5:975-979. [DOI] [PubMed] [Google Scholar]
- 7.Dobrovolskaia, M. A., A. E. Medvedev, K. E. Thomas, N. Cuesta, V. Toshchakov, T. Ren, M. J. Cody, S. M. Michalek, N. R. Rice, and S. N. Vogel. 2003. Induction of in vitro reprogramming by Toll-like receptor (TLR)2 and TLR4 agonists in murine macrophages: effects of TLR “homotolerance” versus “heterotolerance” on NF-kappa B signaling pathway components. J. Immunol. 170:508-519. [DOI] [PubMed] [Google Scholar]
- 8.Duan, Z., and M. Horwitz. 2003. Gfi-1 oncoproteins in hematopoiesis. Hematology 8:339-344. [DOI] [PubMed] [Google Scholar]
- 9.Fujihara, M., M. Muroi, K. Tanamoto, T. Suzuki, H. Azuma, and H. Ikeda. 2003. Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: roles of the receptor complex. Pharmacol. Ther. 100:171-194. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, S., and M. S. Hayden. 2008. New regulators of NF-kappaB in inflammation. Nat. Rev. Immunol. 8:837-848. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109(Suppl.):S81-S96. [DOI] [PubMed] [Google Scholar]
- 12.Gilks, C. B., S. E. Bear, H. L. Grimes, and P. N. Tsichlis. 1993. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol. Cell. Biol. 13:1759-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gingras, S., E. Parganas, A. de Pauw, J. N. Ihle, and P. J. Murray. 2004. Re-examination of the role of suppressor of cytokine signaling 1 (SOCS1) in the regulation of toll-like receptor signaling. J. Biol. Chem. 279:54702-54707. [DOI] [PubMed] [Google Scholar]
- 14.Grassme, H., J. Jin, B. Wilker, G. von Kurthy, W. Wick, M. Weller, T. Moroy, and E. Gulbins. 2006. Regulation of pulmonary Pseudomonas aeruginosa infection by the transcriptional repressor Gfi1. Cell. Microbiol. 8:1096-1105. [DOI] [PubMed] [Google Scholar]
- 15.Grimes, H. L., T. O. Chan, P. A. Zweidler-McKay, B. Tong, and P. N. Tsichlis. 1996. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 16:6263-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacker, H., and M. Karin. 2006. Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006:re13. [DOI] [PubMed]
- 17.Hagemann, C., and J. L. Blank. 2001. The ups and downs of MEK kinase interactions. Cell. Signal. 13:863-875. [DOI] [PubMed] [Google Scholar]
- 18.Hayden, M. S., and S. Ghosh. 2008. Shared principles in NF-kappaB signaling. Cell 132:344-362. [DOI] [PubMed] [Google Scholar]
- 19.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-kappaB. Genes Dev. 18:2195-2224. [DOI] [PubMed] [Google Scholar]
- 20.Hayden, M. S., A. P. West, and S. Ghosh. 2006. NF-kappaB and the immune response. Oncogene 25:6758-6780. [DOI] [PubMed] [Google Scholar]
- 21.Hock, H., M. J. Hamblen, H. M. Rooke, J. W. Schindler, S. Saleque, Y. Fujiwara, and S. H. Orkin. 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431:1002-1007. [DOI] [PubMed] [Google Scholar]
- 22.Hock, H., M. J. Hamblen, H. M. Rooke, D. Traver, R. T. Bronson, S. Cameron, and S. H. Orkin. 2003. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18:109-120. [DOI] [PubMed] [Google Scholar]
- 23.Hock, H., and S. H. Orkin. 2006. Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr. Opin. Hematol. 13:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Hoebe, K., E. M. Janssen, S. O. Kim, L. Alexopoulou, R. A. Flavell, J. Han, and B. Beutler. 2003. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat. Immunol. 4:1223-1229. [DOI] [PubMed] [Google Scholar]
- 25.Iwasaki, A., and R. Medzhitov. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987-995. [DOI] [PubMed] [Google Scholar]
- 26.Jafar-Nejad, H., and H. J. Bellen. 2004. Gfi/Pag-3/senseless zinc finger proteins: a unifying theme? Mol. Cell. Biol. 24:8803-8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang, H. D., K. Yoon, Y. J. Shin, J. Kim, and S. Y. Lee. 2004. PIAS3 suppresses NF-kappaB-mediated transcription by interacting with the p65/RelA subunit. J. Biol. Chem. 279:24873-24880. [DOI] [PubMed] [Google Scholar]
- 28.Janssens, S., and R. Beyaert. 2003. Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol. Cell 11:293-302. [DOI] [PubMed] [Google Scholar]
- 29.Janssens, S., K. Burns, E. Vercammen, J. Tschopp, and R. Beyaert. 2003. MyD88S, a splice variant of MyD88, differentially modulates NF-kappaB- and AP-1-dependent gene expression. FEBS Lett. 548:103-107. [DOI] [PubMed] [Google Scholar]
- 30.Jin, J., H. Zeng, K. W. Schmid, M. Toetsch, S. Uhlig, and T. Moroy. 2006. The zinc finger protein Gfi1 acts upstream of TNF to attenuate endotoxin-mediated inflammatory responses in the lung. Eur. J. Immunol. 36:421-430. [DOI] [PubMed] [Google Scholar]
- 31.Karsunky, H., I. Mende, T. Schmidt, and T. Moroy. 2002. High levels of the onco-protein Gfi-1 accelerate T-cell proliferation and inhibit activation induced T-cell death in Jurkat T-cells. Oncogene 21:1571-1579. [DOI] [PubMed] [Google Scholar]
- 32.Karsunky, H., H. Zeng, T. Schmidt, B. Zevnik, R. Kluge, K. W. Schmid, U. Duhrsen, and T. Moroy. 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30:295-300. [DOI] [PubMed] [Google Scholar]
- 33.Kazanjian, A., E. A. Gross, and H. L. Grimes. 2006. The growth factor independence-1 transcription factor: new functions and new insights. Crit. Rev. Oncol. Hematol. 59:85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinjyo, I., T. Hanada, K. Inagaki-Ohara, H. Mori, D. Aki, M. Ohishi, H. Yoshida, M. Kubo, and A. Yoshimura. 2002. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity 17:583-591. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi, K., L. D. Hernandez, J. E. Galan, C. A. Janeway, Jr., R. Medzhitov, and R. A. Flavell. 2002. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 110:191-202. [DOI] [PubMed] [Google Scholar]
- 36.Lemaitre, B. 2004. The road to Toll. Nat. Rev. Immunol. 4:521-527. [DOI] [PubMed] [Google Scholar]
- 37.Liew, F. Y., D. Xu, E. K. Brint, and L. A. O'Neill. 2005. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5:446-458. [DOI] [PubMed] [Google Scholar]
- 38.Maeda, S., and M. Omata. 2008. Inflammation and cancer: role of nuclear factor-kappaB activation. Cancer Sci. 99:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montoya-Durango, D. E., C. S. Velu, A. Kazanjian, M. E. Rojas, C. M. Jay, G. D. Longmore, and H. L. Grimes. 2008. Ajuba functions as a histone deacetylase-dependent co-repressor for autoregulation of the growth factor-independent-1 transcription factor. J. Biol. Chem. 283:32056-32065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moroy, T. 2005. The zinc finger transcription factor Growth factor independence 1 (Gfi1). Int. J. Biochem. Cell Biol. 37:541-546. [DOI] [PubMed] [Google Scholar]
- 41.Moroy, T., H. Zeng, J. Jin, K. W. Schmid, A. Carpinteiro, and E. Gulbins. 2008. The zinc finger protein and transcriptional repressor Gfi1 as a regulator of the innate immune response. Immunobiology 213:341-352. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa, R., T. Naka, H. Tsutsui, M. Fujimoto, A. Kimura, T. Abe, E. Seki, S. Sato, O. Takeuchi, K. Takeda, S. Akira, K. Yamanishi, I. Kawase, K. Nakanishi, and T. Kishimoto. 2002. SOCS-1 participates in negative regulation of LPS responses. Immunity 17:677-687. [DOI] [PubMed] [Google Scholar]
- 43.O'Neill, L. A. 2008. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity 29:12-20. [DOI] [PubMed] [Google Scholar]
- 44.Pasare, C., and R. Medzhitov. 2005. Toll-like receptors: linking innate and adaptive immunity. Adv. Exp. Med. Biol. 560:11-18. [DOI] [PubMed] [Google Scholar]
- 45.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 47.Saklatvala, J., J. Dean, and A. Clark. 2003. Control of the expression of inflammatory response genes. Biochem. Soc. Symp. 70:95-106. [DOI] [PubMed] [Google Scholar]
- 48.Saleque, S., J. Kim, H. M. Rooke, and S. H. Orkin. 2007. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell 27:562-572. [DOI] [PubMed] [Google Scholar]
- 49.Shroyer, N. F., D. Wallis, K. J. Venken, H. J. Bellen, and H. Y. Zoghbi. 2005. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 19:2412-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sly, L. M., M. J. Rauh, J. Kalesnikoff, C. H. Song, and G. Krystal. 2004. LPS-induced upregulation of SHIP is essential for endotoxin tolerance. Immunity 21:227-239. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki, N., S. Suzuki, and W. C. Yeh. 2002. IRAK-4 as the central TIR signaling mediator in innate immunity. Trends Immunol. 23:503-506. [DOI] [PubMed] [Google Scholar]
- 52.Vassen, L., K. Fiolka, S. Mahlmann, and T. Moroy. 2005. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 33:987-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wertz, I. E., K. M. O'Rourke, H. Zhou, M. Eby, L. Aravind, S. Seshagiri, P. Wu, C. Wiesmann, R. Baker, D. L. Boone, A. Ma, E. V. Koonin, and V. M. Dixit. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694-699. [DOI] [PubMed] [Google Scholar]
- 54.Yucel, R., H. Karsunky, L. Klein-Hitpass, and T. Moroy. 2003. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J. Exp. Med. 197:831-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng, H., R. Yucel, C. Kosan, L. Klein-Hitpass, and T. Moroy. 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23:4116-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, G., and S. Ghosh. 2000. Molecular mechanisms of NF-kappaB activation induced by bacterial lipopolysaccharide through Toll-like receptors. J. Endotoxin Res. 6:453-457. [DOI] [PubMed] [Google Scholar]
- 57.Zweidler-Mckay, P. A., H. L. Grimes, M. M. Flubacher, and P. N. Tsichlis. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 16:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]