Abstract
The yeast PHO5 promoter is a classical model for studying the role of chromatin in gene regulation. To enable biochemical dissection of the mechanism leading to PHO5 activation, we reconstituted the process in vitro. Positioned nucleosomes corresponding to the repressed PHO5 promoter state were assembled using a yeast extract-based in vitro system. Addition of the transactivator Pho4 yielded an extensive DNase I-hypersensitive site resembling induced PHO5 promoter chromatin. Importantly, this remodeling was energy dependent. In contrast, little or no chromatin remodeling was detected at the PHO8 or PHO84 promoter in this in vitro system. Only the PHO5 promoter harbors a high-affinity intranucleosomal Pho4 binding site (UASp) where Pho4 binding can compete with nucleosome formation, prompting us to test the importance of such competition for chromatin remodeling by analysis of UASp mutants in vivo. Indeed, the intranucleosomal location of the UASp element was critical, but not essential, for complete remodeling at the PHO5 promoter in vivo. Further, binding of just the Gal4 DNA binding domain to an intranucleosomal site could increase PHO5 promoter opening. These data establish an auxiliary role for DNA binding competition between Pho4 and histones in PHO5 promoter chromatin remodeling in vivo.
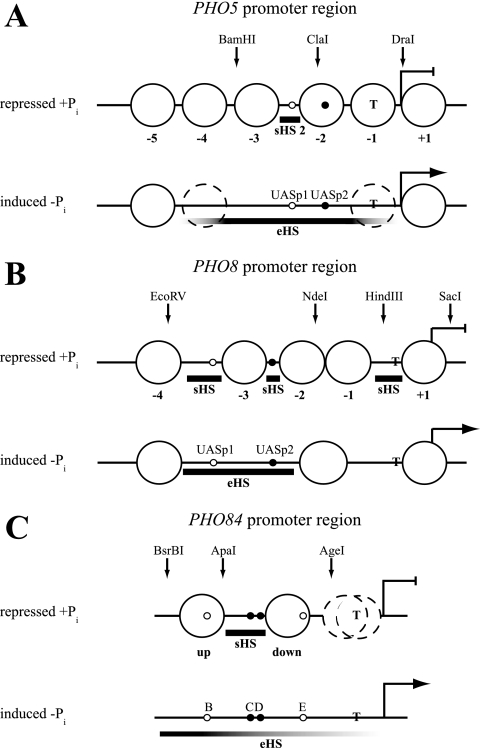
The PHO5 promoter in Saccharomyces cerevisiae represents one of the first model systems where the regulatory role of chromatin was recognized (60). Importantly, many mechanistic features that were elucidated in this well-defined and comparatively simple system turned out to be generally applicable to chromatin biology and transcriptional activation. The repressed PHO5 promoter is organized into four positioned nucleosomes (Fig. 1 A) (2). A low-affinity binding site for its essential transactivator Pho4, UASp1, is accessible in a short hypersensitive region (sHS2), and a high-affinity site, UASp2, resides in nucleosome −2. This nucleosome prevents Pho4 from binding UASp2, thereby constituting a chromatin switch for PHO5 regulation (64). Upon induction by phosphate starvation, the PHO5 promoter nucleosomes become remodeled. Nucleosomes −2 and −3 are remodeled to a greater extent than nucleosomes −1 and −4 (2, 10, 29, 50, 51), and even nucleosome −5 can become accessible in a Pho4-dependent way (29), which collectively generates an extensive nuclease-hypersensitive site (eHS).
FIG. 1.
Schematics of the nucleosomal organization at the PHO5, PHO8, and PHO84 promoter regions in the repressed (+Pi) and induced (−Pi) states. Large circles denote positioned nucleosomes, which are numbered relative to the ATG at the PHO5 promoter and relative to the short hypersensitive region just upstream of the ATG at the PHO8 promoter. The positioned nucleosomes flanking the short hypersensitive region at the PHO84 promoter are labeled “up” and “down” according to reference 67. Stippled circles stand for nucleosomes that are partially remodeled (PHO5, −Pi) (2, 10, 11, 34) or show ambiguous positioning (PHO84, +Pi) (67). Bold horizontal bars demarcate short (sHS) or extensive (eHS) DNase I-hypersensitive sites. Fading of the bar symbolizes less hypersensitivity. The positions of high-affinity Pho4 binding sites (UASp2 at PHO5 and PHO8, C and D at PHO84) are indicated by small filled circles, and the positions of low-affinity sites (UASp1 at PHO5 and PHO8, B and E at PHO84) are indicated by small open circles. The positions of the TATA box (T), the ATG (broken blunt and broken pointed arrows for repressed and induced states, respectively), and the restriction sites used for generating marker fragments are shown. The schematics are mainly based on previous publications on PHO5 (2, 29), PHO8 (3), and PHO84 (67). For a comparison of these schematics with data from other sources, see reference 67.
The molecular mechanism of PHO5 promoter chromatin remodeling has been studied in great detail. Importantly, the opening of PHO5 promoter chromatin is not a consequence of, but rather a prerequisite for, activated transcription, as it occurs independently of transcription in a TATA box deletion mutant (19). Conversely, high levels of transcription have never been observed without chromatin remodeling. PHO5 promoter chromatin remodeling represented the first described in vivo example of histone eviction in trans (11, 34, 50, 51), a mechanism that was soon recognized to operate genomewide (8, 36, 70).
There has been a long-standing search for factors involved in PHO5 promoter chromatin remodeling. Both Pho4 and the pleiotropic homeobox protein Pho2 are essential for wild-type PHO5 promoter opening (18). Recently, the Mcm1 and forkhead proteins were shown to be involved in mitotic PHO5 induction (47). Pho2 mainly increases the binding affinity and transactivation potential of Pho4 (5), and its absence can be compensated for by overexpression of PHO4, but not vice versa (18). Likewise, though Pho2-targeted histone acetylation by Esa1 is important for Pho4 binding, it is dispensable when Pho4 is overexpressed (46). PHO5 promoter chromatin remodeling was shown to be independent of replication (53), which triggered at that time a search for chromatin remodeling factors that might be responsible for the changes in chromatin structure observed upon PHO5 induction. Many different ATP-dependent remodelers have been described and characterized (14), and 16 of the 17 remodelers in yeast (20) have been tested for a role in PHO5 promoter opening (4, 11). Of these, only Swi/Snf (4, 16, 21, 43) and Ino80 (4, 59) are involved in opening of the PHO5 promoter, to an extent where their absence causes a measurable delay. The same is true for the histone acetyltransferases Gcn5 (7, 24) and Rtt109 (66, 67) and the histone chaperones Asf1 (1, 32) and Nap1 (15). The degree of cofactor dependency varies with the degree of PHO5 induction (16, 32). Thus, while suboptimal induction conditions helped uncover a role for these cofactors, the final degree of promoter opening is not affected significantly in the absence of any of them under full induction conditions. Therefore, the search for the essential PHO5 promoter chromatin opening cofactor has so far been in vain, and Pho4 remains the only factor generally recognized as essential for PHO5 induction in vivo. This argues for a pronounced redundancy of pathways leading to PHO5 promoter chromatin remodeling (4) and somewhat encumbers further dissection of the remodeling mechanism in vivo.
Therefore, we set out to reconstitute PHO5 promoter chromatin remodeling in vitro. There were previous attempts to do this. Minichromosomes bearing the PHO5 locus were isolated ex vivo, and Pho4 was added in vitro. This induced some chromatin remodeling, as judged by smeared micrococcal nuclease ladders, but a nuclease-hypersensitive site at the proper position was not evident (26). De novo chromatin assembly of the PHO5 locus using the histone chaperone Nap1 supported some Pho4-induced remodeling and transcription. However, the resulting chromatin patterns indicated simultaneous stable binding of Pho4 and a nucleosome in the region of UASp2 (63), which is in contrast to the in vivo data (64). We established a different approach by using yeast extracts supplemented with exogenous histones and energy, which enabled the generation of a PHO5 promoter chromatin structure remarkably similar to that observed in vivo (33). This in vitro system also works well for the PHO8 (27) and PHO84 (67) promoters. These three PHO promoters are intriguing reference models, as they are coregulated by Pho4 and show prominent chromatin transitions upon induction (Fig. 1), but with somewhat differently stringent cofactor requirements (4, 7, 24, 25, 42, 67).
In this study, we used chromatin assembly in yeast extracts to study Pho4-induced chromatin remodeling at the PHO5, PHO8, and PHO84 promoters in vitro. At the PHO5 promoter, we observed a Pho4-dependent generation of a hypersensitive site and show explicitly that this remodeling is ATP dependent. In contrast, there was almost no Pho4- and energy-dependent remodeling at the PHO8 and PHO84 promoters, prompting us to question if the intranucleosomal location of a high-affinity UASp element, unique to the PHO5 promoter, was the key difference. Our experiments indicate that, indeed, the intranucleosomal location of the UASp2 site in the −2 nucleosome of the PHO5 promoter plays a hitherto overlooked role in PHO5 promoter opening in vivo. This argues that competition between a transcriptional activator and histones for binding to the DNA can make an important mechanistic contribution to promoter chromatin remodeling.
MATERIALS AND METHODS
Strains and media.
The following S. cerevisiae strains are described elsewhere: CY337 (52), CY339 (67), YS70 (64), YS18 (54), YS22 (5), BY4741 (12), and AH2341 [a ura3(ΔApaI to NcoI)::YIP5-Var.31 his3 leu2-3,112 pho5 pho3 gal4::TRP1] (17). Strain YS27 carries null mutations in pho4 and pho2 (5, 55) and had an additional deletion of cbf1 created with disruption plasmid pMF33 (39) to yield strain YS27 cbf1::URA3. Strain CY337 EB1615 carries the mutant H1 PHO5 promoter, as described by Lam et al. (35), and was generated by transformation of CY337 with PflMI-linearized pRS306-based plasmid EB1615, kindly provided by Erin O'Shea. Strain CY337 FE1600 carries a mutant version of EB1615 where the CACGTGG sequence in sHS2 was changed back to the UASp1 E box CACGTTT and a CACGTGG sequence was introduced close to the BstEII site (see Fig. 7). The BstEII site is a proxy for the position of the linker between nucleosomes −1 and −2 (2, 10). Both mutations were generated using the QuikChange kit (Stratagene); primers UASp1for (5′-ATTAAATTAGCACGTTTTCGCATAGAACGCAAC-3′), UASp1rev (5′-GTTGCGTTCTATGCGAAAACGTGCTAATTTAAT-3′), Bst-UASfor (5′-TATCAAATTGGTCACGTGGCTTGGCAAGGCATATAC-3′), and Bst-UASrev (5′-GTATATGCCTTGCCAAGCCACGTGACCAATTTGATA-3′); and EB1615 as the template. Strain CY339 ura3 was derived from CY339 after selection on 5-fluoroorotic acid-containing medium and confirmed for uracil auxotrophy. Strain CY341 was derived from CY337 (for a description, see Fig. 7). Plasmid pCB-UASp2-5 Δ2 was derived from pCB(LEU2) (33) and carries the BamHI-ClaI fragment of the PHO5 promoter region of strain YS70 and the ClaI-Bsu36I fragment of pCB-ΔUASp2, where the CACGTG sequence of UASp2 in plasmid pCB(LEU2) was replaced with the AAGCTT HindIII site (64). Plasmid pCB-UASp2-8 Δ2 was also derived from pCB(LEU2) and carries the ApaI-PmlI fragment of the PCR product obtained by using primers 5′-TGGCTGGATAAATGGGCCCC-3′ and 5′-ATCGCTGCACGTGGCCCGACGTAGATGACCCTTTTGTGCAGACAAAGAAAAAGCGC-3′ with pCB(LEU2) as the template and the PmlI-Bsu36I fragment of the PCR product obtained by using primers 5′-TCGGGCCACGTGCAGCGATCGAACGCAACTGCACAATGC-3′ and 5′-GTCGACATCGGCTAGTTTGC-3′ with pCB-ΔUASp2 as the template. The plasmid for overexpression of PHO4 was YEpP4 (61). The expression plasmids for the Gal4 DNA binding domain, YCpGal4(1-147) and YCpGal4(1-94), carry a CEN element and a HIS3 marker, were derived from pRJR266 (L. Gaudreau), have the Gal4 construct under the control of the GAL4 promoter and terminator, and are described in reference 17. Yeast strains were grown as described previously (4), i.e., under repressive conditions (high phosphate) in yeast extract-peptone-dextrose with 0.1 g/liter adenine and 1 g/liter KH2PO4 or in yeast nitrogen base selection medium supplemented with the required amino acids for plasmid bearing-strains and in corresponding synthetic phosphate-free medium for induction (no phosphate).
FIG. 7.
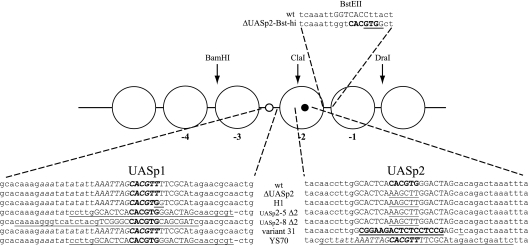
PHO5 promoter UASp mutants. Schematic of PHO5 promoter mutants with altered UASp elements. The bases of the core hexanucleotide E box of the UASp elements are shown in bold uppercase letters. The UASp2 E box was deleted by conversion to a HindIII site. Mutated DNA regions are underlined. Uppercase bases at the endogenous UASp1 and UASp2 positions correspond to the Pho4 dimethyl sulfate footprint region, and italic bases correspond to the Pho2 dimethyl sulfate footprint region (3, 6, 65). All underlined bases at the UASp1 position of the UASp2-8 Δ2 mutant stem from the UASp2 region of the PHO8 promoter. The bold underlined uppercase bases at the UASp2 position in variant 31 correspond to the introduced Gal4 binding site. The A-to-T point mutation downstream introduced a SacI site. The BstEII site in the wild-type sequence of the linker between the −1 and −2 nucleosomes is indicated in uppercase letters.
DNase I indirect end labeling and restriction enzyme accessibility assay of in vivo chromatin.
Preparation of yeast nuclei and chromatin analysis by restriction nuclease and DNase I digestion with indirect end labeling were done as described previously (2, 22, 23, 62). For the chromosomal PHO5 locus, ApaI (DNase I analysis) or HaeIII (restriction enzyme analysis), for the variant 31 PHO5 locus in strain AH2341, HindIII (DNase I analysis) or HindIII/SalI (restriction enzyme analysis), for the PHO8 locus, BglII, and for the PHO84 locus, SspI were used for secondary cleavage. For the PHO5 locus on plasmids pCB-UASp2-5 Δ2 and pCB-UASp2-8 Δ2, secondary cleavage was done with NciI. Hybridization probes were PCR products corresponding to the following genomic regions: probe-DNase I-PHO5, bases −760 to −1296; probe-RE-PHO5, bases −276 to −537 from the ATG of the PHO5 open reading frame (ORF); the probe for the variant 31 PHO5 promoter mutant in strain AH2341, the HindIII-BamHI fragment of pBR322 (19); probe PHO5-plasmid, bases 172 to 379 of pBR322; probe-PHO8, bases +78 to +568 from the ATG of the PHO8 ORF; probe-PHO84, bases −1083 to −1428 from the ATG of the PHO84 gene. Probes were labeled with [α-32P]dCTP using the kit PrimeIt II (Stratagene). Blots were exposed to X-ray films (Fuji Super RX) using intensifier screens (DuPont Lightening Plus) and scanned in CMYK mode (MikroTek ScanMaker i900). Scanned images were imported into Adobe Photoshop CS2, and the total image was further manipulated by conversion into grayscale format and linear level adjustment. Sometimes parts of the image were rearranged as indicated in the figures and figure legends. Quantification of restriction enzyme accessibility assays was done using a PhosphorImager (Fuji FLA3000, AIDA software version 3.52.046).
Yeast whole-cell extract preparation.
Yeast whole-cell extract was prepared as previously described (27, 67) but from strain YS27 cbf1::URA3 and with Tris-HCl instead of HEPES-KOH in the extraction buffer.
Chromatin assembly.
De novo assemblies and salt gradient dialysis assemblies were done as described previously (27, 33, 67). Drosophila embryo histone octamers and recombinant yeast histones were prepared as described previously (37, 38, 56). The DNA template for the de novo chromatin assembly reaction was plasmid pCB(LEU2), and for all salt gradient dialysis chromatin reconstitution reactions, they were circular, supercoiled, pUC19-based plasmids where the multiple cloning site contained the PHO5, PHO8, or PHO84 ORF plus upstream regions as follows. Plasmid pUC19-PHO5 was generated by inserting the 3,149-bp HindIII-PstI fragment of the PHO5 locus into pUC19. Plasmids pUC19-PHO8 and pUC19-PHO84 (67) were prepared, respectively, by ligating a 3.5-kb PCR product, generated using primers 5′-CCATGTGCATAGGATCCGGACGTTTGCCATAGTGTTG-3′ and 5′-CAGTCAGACGCTGCAGGGGAGAGTTAGATAGGATCAGT-3′ or 5′-CCGGAATTCTCGAGTCATGATTTGGAACAGCTCC-3′ and 5′-CGCGGATCCGCAGAGAGATGTGAGGAAAT-3′ and genomic DNA from strain BY4741 as the template, via PstI and BamHI or via EcoRI and BamHI, respectively, into pUC19.
Shifting nucleosome positions in vitro.
A 100-μl shifting reaction mixture contained 1 μg DNA in total, either pUC19-PHO5 and -PHO8 or pUC19-PHO84 and -PHO8 in equimolar amounts, preassembled into chromatin by salt gradient dialysis. The reaction mixture was incubated with or without yeast extract (∼250 μg protein, judged from Coomassie-stained gel lanes in comparison to a protein standard) and with or without a regenerative energy system (3 mM ATP, 3 mM MgCl2, 30 mM creatine phosphate [Sigma], and 50 ng/μl creatine kinase [Roche Applied Science]) in assembly buffer (20 mM HEPES-KOH [pH 7.5], 10% glycerol, 80 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 2.5 mM dithiothreitol [DTT]) for 2 h at 30°C. If called for, Pho4 and/or Pho2 were added to 3 μg/100 μl (Pho4-SPA6), 3.4 μg/100 μl (wild-type Pho4), and 4.2 μg/100 μl (Pho2), respectively, and incubated for 1 h more at 30°C if addition was not in parallel with the yeast extract. Preparations of the chromatin remodeling complexes RSC and SWI/SNF were kind gifts from Tom Owen-Hughes. The amounts added to the shifting reactions were sufficient to remodel the equivalent chromatin mass of mononucleosomal templates (T. Owen-Hughes, personal communication). In addition, we showed that salt gradient dialysis chromatin templates with the PHO5 and PHO8 loci were effectively remodeled upon addition of the corresponding amount of the RSC preparation, as judged by ATP-dependent changes in the DNase I pattern and ClaI restriction enzyme accessibility (F. Ertel, C. J. Wippo, and P. Korber, unpublished data). ATP was depleted from reconstitution reaction mixtures by a factor of at least 106 by adding apyrase (M0393L; NEB) to a concentration of 3 to 4 U/100 μl and incubation for 30 min at 30°C.
DNase I indirect end labeling and restriction enzyme accessibility assay of in vitro reconstituted chromatin.
Ten-microliter aliquots of a reconstitution reaction mixture was mixed with an equal volume of digestion buffer (20 mM HEPES-KOH [pH 7.5], 12% glycerol, 4 mM MgCl2, 5.5 mM CaCl2, 2.5 mM DTT, 80 mM NaCl, 0.1 mg/ml bovine serum albumin) containing DNase I (04716728001; Roche Applied Science) at concentrations in the range of 0.02 to 0.1 U/ml (salt gradient dialysis chromatin) or 2 to 15 U/ml (salt gradient dialysis chromatin with extract) and incubated at room temperature for 5 min. In all DNase I mapping experiments, chromatin samples were digested with a range of DNase I concentrations. However, due to space limitations, only one lane or a few representative lanes are shown in the figures. The digestion reactions were stopped by adding 4 μl of STOP buffer (10 mM EDTA, 4% sodium dodecyl sulfate [SDS]), and the DNA was purified by digestion with proteinase K and ethanol precipitation.
Prior to restriction enzyme digestions, ATP was removed by apyrase. Two-microliter aliquots of an apyrase-treated reconstitution reaction mixture was mixed with 15 μl of restriction enzyme digestion buffer (20 mM HEPES KOH [pH 7.5], 4.5 mM MgCl2, 2.5 mM DTT, 80 mM NaCl, 0.5 mM EGTA) and digested with two different concentrations of each restriction enzyme. The reactions were stopped by adding 4 μl STOP buffer, and the DNA was purified as described above. Blotting, hybridization, and secondary cleavage were done as for the analysis of in vivo chromatin.
Expression and purification of recombinant 6×His-tagged Pho4 and Pho2.
Expression and purification of recombinant Pho4 and Pho2 were previously described (6). A BbrPI/NcoI fragment of the PHO4 locus was cloned into pET21d (Stratagene), which was cut with EagI, blunt ended, and cut with NcoI. This adds 10 amino acids to the C terminus of Pho4 [AALE(H)6], the last six being the histidine tag. The pET21d-based Pho2 expression plasmid was a kind gift from D. Stillman. The PHO4-SPA6 mutation (57) was introduced into the pET21d-PHO4 expression plasmid by transfer of a 280-bp SfuI/ClaI restriction fragment from plasmid EB1043 (kind gift of E. O'Shea). Escherichia coli strain BL21(DE3)/pLysS (Stratagene) was transformed with either construct and grown at 37°C (Pho4 expression) or 28 to 30°C (Pho2 expression) in logarithmic phase in LB medium with chloramphenicol (34 μg/ml) and ampicillin (300 μg/ml). For induction, the culture was treated with 1 mM isopropyl-β-d-thiogalactopyranoside and incubated for an additional 3 h. The cells were harvested by centrifugation, washed, and resuspended in sonication buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl). Complete protease inhibitor without EDTA (Roche Applied Science) and 0.1% NP-40 were added. Cells lysis was achieved by a freeze-thaw cycle (liquid nitrogen) and sonication (50% amplitude, four separate 20-s bursts, >1 min of cooling on ice between bursts; SONOPULS 2200 ultrasonic homogenizer equipped with sonotrode MS73; Bandelin Electronic, Berlin, Germany). The cell lysate was cleared by centrifugation (12,000 rpm, relative centrifugal force [rcf] of 17,210, 4°C, 30 min; SS34 [Sorvall]), and the supernatant was applied in batch to 1 ml of Ni2+-nitrilotriacetic acid agarose (Qiagen; equilibrated in sonication buffer). After incubation for 1 h with agitation in a cold room, the resin was washed twice in batch with ice-cold sonication buffer, transferred into a column, and washed with 100 ml sonication buffer-60 mM imidazole (pH 8.0)-1× Complete protease inhibitor without EDTA (Roche Applied Science). Pho4-6xHis or Pho2-6xHis was eluted from the column with 10 ml sonication buffer-1 M imidazole (pH 8.0)-1× Complete protease inhibitor without EDTA. Fractions were tested by SDS-polyacrylamide gel electrophoresis and Coomassie staining for the band of the proper size. Positive fractions were pooled and dialyzed (molecular mass cutoff, 6,000 to 8,000 Da) overnight at 4°C against 20 mM Tris-HCl (pH 8.0)-100 mM NaCl-10% glycerol-0.5 mM DTT-0.5 mM EDTA-0.2 mM phenylmethylsulfonyl fluoride-1 mM benzamidine.
Luciferase ATP assay.
ATP concentrations were measured using the Enliten Luciferase reagent (Promega) and a luminometer (Lumat; Berchtold, Tuttlingen, Germany) according to the manufacturer's directions. As this assay is very sensitive and no longer linear if the ATP concentration is too high, samples were usually diluted by a factor of 10,000 to 100,000 with water.
Separation of salt gradient dialysis chromatin populations in sucrose gradients.
Fifty micrograms of DNA after salt gradient dialysis assembly into chromatin (equimolar amounts of pUC19-PHO5 and -PHO8 in 500 μl) was prepared as previously described (27, 67). Four hundred nanograms of DNA packaged in chromatin was left on ice as an input control. The rest of the chromatin was dialyzed against polyethylene glycol (PEG; Sigma-Aldrich) to reduce the volume to 150 μl. Seven hundred nanograms of DNA packaged in chromatin was put aside as a second input control. The concentrated chromatin was loaded onto 11 ml of a 15 to 40% sucrose gradient (Gradient Master; BIOCOMP) in salt dialysis buffer (10 mM Tris HCl [pH 7.6], 50 mM NaCl, 1 mM EDTA, 0.05% Igepal) and centrifuged for 16 h at 30,000 rpm (rcf, 111,132) at 4°C (Beckman ultracentrifuge, SW41 Ti rotor, polyallomer centrifuge tubes [14 by 89 mm]; Beckman). Five-hundred-microliter fractions were collected in tubes containing 100 μg bovine serum albumin using a syringe microfractionator (Brandel). Four hundred microliters of each fraction was stored at 4°C, while100 μl was treated with 4% SDS-0.1 M EDTA, digested with 140 μg proteinase K (Roche) for 3 h at 37°C, and precipitated with ethanol. The DNA content was analyzed spectrophotometrically (NanoDrop Spectrophotometer ND-1000; Peqlab) and by agarose gel electrophoresis in order to choose representative chromatin-containing fractions. The remaining 400-μl aliquot was dialyzed against salt dialysis buffer (low salt) and concentrated with PEG to counteract the volume increase during dialysis. Portions (1.5 μg) of chromatinized DNA of the selected fractions and of the two input controls were used for shifting nucleosome positions in vitro and afterwards analyzed by DNase I indirect end labeling.
Separation of extract-treated chromatin populations by differential MgCl2 precipitation.
Chromatin reconstituted by salt gradient dialysis and treated with extract with or without addition of Pho4 was precipitated stepwise by successive additions of MgCl2 (0.2, 1, 2.5, 6, 8, 10, and 15 mM MgCl2 contributions to the final concentration), always using the supernatant of the preceding precipitation reaction mixture. After each MgCl2 addition, the sample was incubated on ice for 15 min and centrifuged for 15 min at 13,000 rpm (rcf, 15,800) and 4°C in a tabletop centrifuge (Eppendorf). This resulted in a precipitated chromatin fraction in the pellet and a soluble chromatin fraction in the supernatant. The precipitated chromatin fractions were resuspended in assembly buffer, analyzed for DNA content (Nanodrop and agarose gel electrophoresis), and assayed by DNase I indirect end labeling.
Histone acetyltransferase (HAT) filter binding assay using Gcn5 and chromatin templates.
The HAT filter binding assay was done as previously described (28), with the following changes. Comparable histone amounts of Drosophila histone octamers or salt gradient dialysis chromatin were incubated with or without recombinant Gcn5 and 0.5 μl [3H]acetyl coenzyme A (CoA; 3.6 Ci/mmol; Amersham) in a total volume of 14 μl for 1 h at 30°C. Half of the reaction mixture was stopped by adding 1 μl 100 mM acetic acid. In order to assess the effects of HATs and histone deacetylases endogenous to the yeast extract, the second half of the reaction mixture was incubated with 44 μl of 20 mM HEPES-KOH (pH 7.5)-10% glycerol-80 mM KCl-0.5 mM EGTA-2.5 mM DTT-3 mM ATP-4.5 mM MgCl2-30 mM creatine phosphate (Sigma)-50 ng/μl creatine kinase (Roche Applied Science)-yeast extract-3.4 μg/100 μl Pho4-4.2 μg/100 μl Pho2-50 mM trichostatin A (Sigma) for 2 h at 30°C and quenched with 1 μl 100 mM acetic acid. Each reaction mixture was spotted onto a P81 paper filter (1.5 by 1.5 cm; Whatman), washed three times for 5 min at room temperature with 50 mM sodium carbonate (pH 9.2), air dried, and quantified in a scintillation counter (LS1801; Beckman).
RESULTS
The presence of Pho4 during de novo chromatin assembly in vitro leads to a DNase I-hypersensitive site at the PHO5 promoter.
We previously showed de novo generation of in vivo-like nucleosome positioning over the PHO5 promoter using a yeast extract in vitro chromatin assembly system (33). In this system, a supercoiled plasmid bearing the yeast PHO5 locus is incubated with Drosophila embryo histone octamers, yeast whole-cell extract, and an energy-regenerating system.
Under repressing conditions in vivo, Pho4 is kept outside the nucleus by a process involving phosphorylation by the cyclin/cyclin-dependent kinase complex Pho80/Pho85 (31). This compartmentalization is abrogated in the yeast extract system so that otherwise cytosolic Pho4 might induce chromatin remodeling at the PHO5 promoter in vitro. Since we wished to assemble the positioned nucleosomes of the repressed state, we used yeast extracts from pho4 mutant strains. We even used extracts from a pho4 pho2 cbf1 triple mutant (27), thereby eliminating most of the factors known to bind in the PHO5 promoter region. However, these precautions proved unnecessary as even wild-type extracts assembled the repressed promoter state (F. Ertel, C. B. Hertel, C. J. Wippo, and P. Korber, unpublished data), suggesting that the concentration of endogenous Pho4 or other factors in the extract is too low to have an activating effect in vitro. It also had no importance whether extracts were prepared from phosphate-starved, i.e., induced, cells that harbor mainly unphosphorylated “active” Pho4 (Ertel and Korber, unpublished). Thus, Pho4 endogenous to the yeast extract, regardless of its phosphorylation state, failed to generate the extensive hypersensitive site that is a hallmark of induced PHO5 promoter chromatin (2), presumably due to its low abundance.
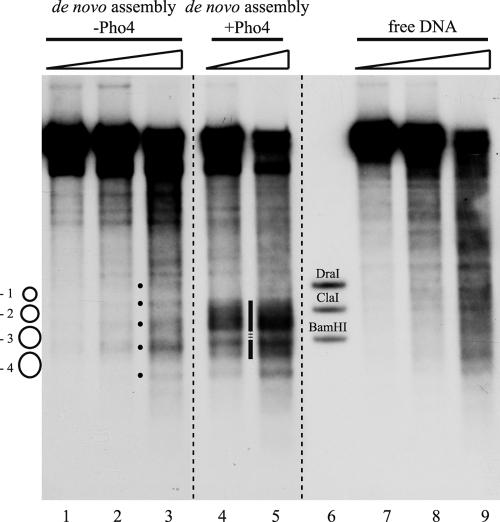
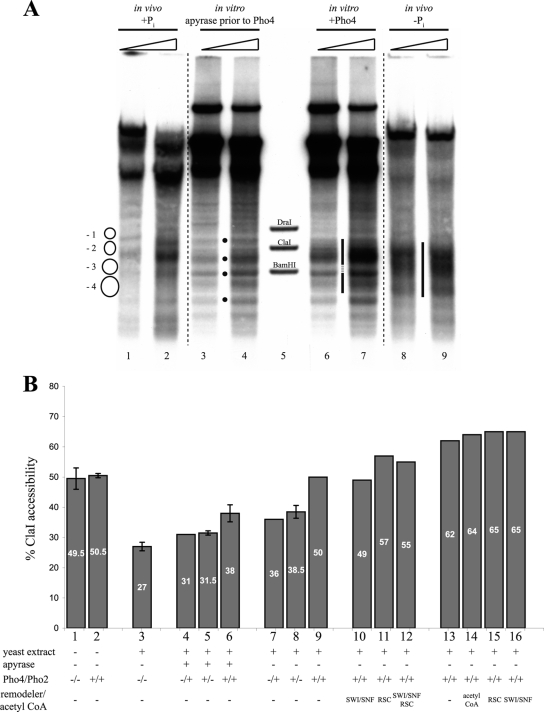
We tested if adding Pho4 exogenously during de novo chromatin assembly in vitro led to a more open chromatin organization. Indeed, the presence of recombinant Pho4 generated a prominent hypersensitive site reaching from the −2 into the −4 nucleosome (Fig. 2, lanes 4 and 5), a pattern clearly different from the positioning of nucleosomes in the absence of Pho4 (lanes 1 to 3) and also not present in a digest of free DNA (lanes 7 to 9).
FIG. 2.
Pho4-induced generation of a hypersensitive site at the PHO5 promoter during de novo assembly in vitro. DNase I indirect end-labeling analysis of the PHO5 promoter region in chromatin assembled de novo with yeast extract (from pho4 strain YS22) in vitro with or without the addition of exogenous Pho4 and in free DNA. The schematic on the left denotes positioned nucleosomes as in the schematic of the PHO5 promoter in Fig. 1A. Black dots between lanes 2 and 3 mark the bands corresponding to the linker regions between the positioned nucleosomes. The vertical bar between lanes 4 and 5 highlights the hypersensitive region generated by the addition of Pho4. The stippled region interrupting the vertical bar points to a region of maintained protection from DNase I. Ramps above the lanes stand for increasing DNase I concentrations. Marker fragments were generated by double digests of DraI, ClaI, and BamHI, each with ApaI. All samples were electrophoresed in lanes of the same gel. Stippled lines between lanes show where lanes were moved next to each other using Adobe Photoshop CS2.
Pho4- and energy-dependent remodeling of preassembled positioned nucleosomes into a hypersensitive site in vitro.
The PHO5 promoter harbors two Pho4 binding sites: a low-affinity site in the short hypersensitive site between nucleosomes −2 and −3 (UASp1, Fig. 1A) and a high-affinity site within nucleosome −2 (UASp2). In the experiments described above, we added Pho4, histones, and yeast extract to the DNA template at the same time so that Pho4 was present during chromatin assembly. Therefore, the observed hypersensitive site might, in theory, reflect the prevention of forming nucleosome −2 due to UASp2-bound Pho4, rather than Pho4-induced remodeling of preassembled nucleosomes. In order to test if Pho4 could remodel preassembled nucleosomes, we turned to a modified version of the chromatin assembly protocol. Here, chromatin is first assembled by salt gradient dialysis, which deposits nucleosomes but does not lead to in vivo-like nucleosome positioning (27, 69). Proper positioning is generated in a second step by incubation with yeast extract in the presence of energy (27). This protocol is more suitable for side-by-side comparisons, as a large batch of salt gradient dialysis chromatin can be prepared and stored, so that all further manipulations are done with the same starting material.
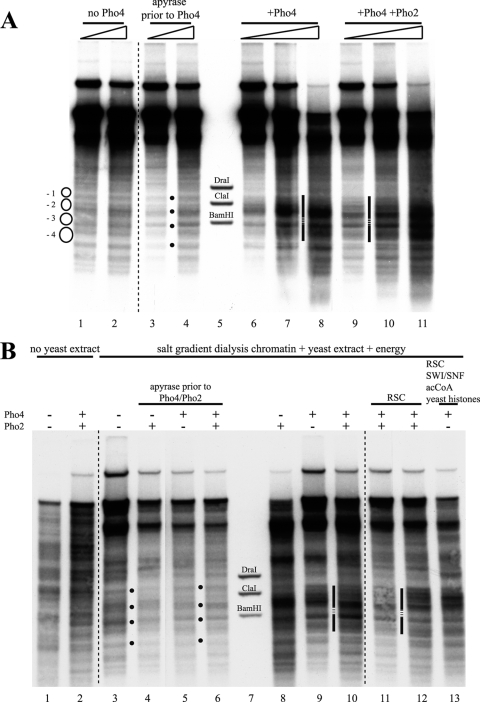
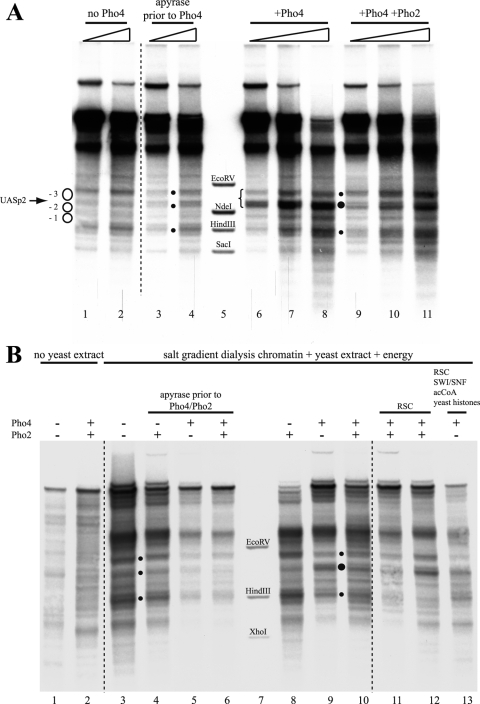
The addition of Pho4 to chromatin with nucleosomes properly prepositioned by the yeast extract generated the same extensive hypersensitive site observed previously in the de novo assembly assay (Fig. 3 A, lanes 6 to 8). Thus, Pho4 does not just interfere with nucleosome assembly but can also induce the remodeling of already positioned nucleosomes. Importantly, the hypersensitive site was not generated if ATP was removed from the reaction mixture by incubation with apyrase prior to the addition of Pho4 (Fig. 3A, lanes 3 and 4). This indicated that Pho4-induced hypersensitivity is not merely the result of Pho4 binding and argued that ATP-dependent remodelers in the extract were involved. The addition of Pho2 in roughly stoichiometric amounts relative to Pho4 (Pho2-to-Pho4 molar ratio of about 0.75) did not change much the appearance of the DNase I-hypersensitive site (Fig. 3A, lanes 9 to 11). In vivo, Pho4 is regulated by phosphorylation at multiple sites, which promotes nuclear export and inhibits nuclear import but also inhibits its interaction with Pho2 (31). Pho2 binds to the PHO5 promoter in close proximity to both UASp1 and UASp2 (6), and the interaction between Pho4 and Pho2 helps Pho4 binding to the promoter and increases the transactivation potential (5). As we did not know whether the interaction of Pho4 with Pho2 is necessary for efficient binding of Pho4 in our in vitro system and as we could not exclude phosphorylation of Pho4 by a kinase in the yeast extract, we used a Pho4 mutant, Pho4-SPA6. This mutant cannot be phosphorylated at the site at which phosphorylation impairs Pho4-Pho2 interaction (57). However, this precaution proved unnecessary as experiments with purified wild-type Pho4 and Pho4-SPA6 produced identical results (Ertel, Hertel, and Korber, unpublished). For simplicity, we therefore use the term “Pho4” for both throughout this paper.
FIG. 3.
Pho4- and energy-dependent remodeling of prepositioned nucleosomes at the PHO5 promoter in vitro. (A) DNase I indirect end-labeling analysis of the PHO5 promoter region in chromatin treated as indicated after preassembly by salt gradient dialysis and incubation with yeast extract (from strain YS27 cpf1) in the presence of energy. The schematic, black dots, vertical bars, stippled lines, ramps, and markers are as in Fig. 2. (B) Same as panel A but with treatment of salt gradient dialysis chromatin as indicated above the lanes. Only for the sample in lane 12 were Pho4 and Pho2 added at the same time as the yeast extract. For all of the samples in lanes 3 to 6, 8 to 11, and 13, salt gradient dialysis chromatin was first treated with yeast extract and energy to yield the pattern shown in lane 3 and then further treated for 1 h at 30°C as indicated. In all DNase I mapping experiments, a range of DNase I concentrations was used, but due to space limitations, only one representative concentration is shown for each condition. acCoA, acetyl-CoA.
For a direct comparison of the different in vitro chromatin patterns at the PHO5 promoter, we generated each state from the same preparation of salt gradient dialysis chromatin and examined representative samples in the same gel (Fig. 3B). The DNase I pattern of the salt gradient dialysis chromatin (lane 1) was not changed appreciably by the addition of Pho4 and Pho2 in the absence of yeast extract (lane 2), while the pattern characteristic of closed PHO5 promoter chromatin was generated by incubation of the salt gradient dialysis chromatin with yeast extract and energy (lane 3) (27). If ATP was depleted by the addition of apyrase, this closed pattern was not altered appreciably by the addition of Pho2, Pho4, or both (lanes 4 to 6). However, the addition of Pho4 (lane 9) or Pho4 and Pho2 (lane 10) in the presence of energy generated a hypersensitive site. Pho2 alone (lane 8) usually had less of an effect; i.e., the hypersensitivity generated was more confined to the linker region between nucleosomes −2 and −3. There was some heterogeneity in the band intensities of the hypersensitive sites in different individual experiments, but the same general region was invariably affected, and the pattern was clearly different from the closed promoter state patterns (lanes 3 to 6). The addition of purified RSC remodeling complex (lanes 11 and 12) or of RSC together with the SWI/SNF remodeling complex, acetyl-CoA, and the use of recombinant yeast histones instead of Drosophila embryo histones (lane 13) failed to significantly change the appearance of the hypersensitive site (see also below).
Pho4-induced remodeling at the PHO5 promoter in vitro is very similar to that observed in vivo.
We compared the hypersensitive site generated in vitro with the pattern of the open PHO5 promoter in vivo (Fig. 4 A, compare lanes 6 and 7 with lanes 8 and 9). The overall extent of the hypersensitivity, i.e., the upper and lower borders of the dark smear in the lane, was very similar. However, the in vitro pattern showed a protected region just above the BamHI marker band (stippled stretch in the vertical bar between lanes 6 and 7) that was not visible in vivo, and the in vitro pattern was generally more structured with rather more distinct bands compared to the in vivo hypersensitive site, which consists of a more uniform smear. This may indicate that remodeling in vitro was less extensive across the promoter than in vivo and/or that the pattern reflected a mixture of more and less remodeled chromatin subpopulations.
FIG. 4.
Pho4-induced remodeling at the PHO5 promoter in vitro is similar to but less extensive than in vivo. (A) Same as Fig. 3A but with in vivo samples from wild-type strain CY337 corresponding to the repressed (+Pi) and induced (−Pi) states of the PHO5 promoter electrophoresed alongside in vitro samples. Note that the vertical bar between lanes 6 and 7 highlighting the in vitro hypersensitive region is interrupted by a short stippled stretch at a position of DNA protection that is fully accessible in the induced state in vivo (bar between lanes 8 and 9). (B) ClaI accessibility values for in vitro-assembled chromatin treated as indicated. For columns 4 to 12, Pho4 and/or Pho2 were added after the salt gradient dialysis chromatin was incubated with yeast extract. For columns 13 to 16, they were added together with the yeast extract. Error bars show the variation of two or three independent experiments starting from the same salt gradient dialysis chromatin preparation. In the case of three experiments, the error bars correspond to the standard deviation.
In order to assess the completeness of remodeling in the total population of chromatin templates more quantitatively, we measured the accessibility of the ClaI restriction site within the −2 nucleosome (Fig. 1A). This assay has been used extensively to quantify the degree of PHO5 promoter chromatin remodeling across a cell population in vivo (2, 23). The repressed state in vivo corresponds to 10 to 20% ClaI accessibility and the induced state to 70 to 90% accessibility. The variability in vivo is due to strain background and to the physiology of the induction pathway that entails growth arrest upon phosphate starvation with more or less residual intracellular phosphate (S. Barbarić, A. Schmid, W. Hörz, and P. Korber, unpublished data).
Figure 4B shows the ClaI accessibilities of the various chromatin states, most of them already described in Fig. 3B. The change from the salt gradient dialysis chromatin to the closed PHO5 promoter pattern corresponded to a decrease from about 50% ClaI accessibility in the salt-dialyzed material to about 30% in the extract-treated material (Fig. 4B, columns 1 and 2 versus 3). This indicates that the yeast extract-induced positioning of a nucleosome over the ClaI site in salt gradient dialysis chromatin occurs with an efficiency not dissimilar to that observed in vivo. As suggested by the activator- and energy-dependent generation of the DNase I-hypersensitive site (Fig. 3), there was always more ClaI accessibility upon the addition of Pho4 and/or Pho2 in the presence of energy than after energy depletion (compare columns 4 versus 7, 5 versus 8, and 6 versus 9), although the increase was not dramatic. Addition of Pho2 together with Pho4 also somewhat increased ClaI accessibility (compare columns 6 versus 5 and 9 versus 8), although hardly any change was visible in the DNase I patterns (Fig. 3). It is important to point out that DNase I indirect end labeling involves low digestion regimens and therefore mainly scores the most nuclease-sensitive chromatin species, whereas ClaI digestion monitors the total population of chromatin templates. A ClaI accessibility of 40 to 50% for chromatin samples that showed the DNase I-hypersensitive site (lanes 9 and 10 in Fig. 3B correspond to columns 8 and 9 in Fig. 4B) argues for a subpopulation mixture of well-remodeled chromatin templates that are detected as hypersensitive to DNase I and accessible to ClaI, and less remodeled templates that are not accessible to ClaI and do not contribute to the DNase I pattern.
We wished to separate such subpopulations and therefore fractionated chromatin generated in vitro with or without Pho4 by differential MgCl2 precipitation or by sucrose gradient ultracentrifugation (Ertel and Korber, unpublished). However, none of the obtained fractions of chromatin treated with Pho4 were significantly more remodeled than the others (Ertel and Korber, unpublished). Thus, chromatin templates that were effectively remodeled by Pho4 in vitro could not be readily separated from templates that were refractive to remodeling.
In summary, remodeling induced by Pho4 and Pho2 in the presence of ATP in vitro affected pretty much the same region in terms of DNase I sensitivity as in vivo and led to an activator- and energy-dependent increase in ClaI accessibility, similar to—though less pronounced than—chromatin opening in vivo.
The extent of remodeling in vitro was not significantly affected by supplementing the yeast extract with remodelers, histone acetylase, acetyl-CoA, or competitor DNA or by using recombinant yeast histones.
When Pho4 and/or Pho2 were added to salt gradient dialysis chromatin together with the yeast extract, i.e., before positioning the nucleosomes into the closed chromatin pattern, the accessibility of the ClaI site became significantly higher (up to 65%) and thus approached the in vivo situation of induced cells (Fig. 4B, columns 13 to 16). This increase was not significantly affected by the addition of acetyl-CoA, RSC, or SWI/SNF (columns 14 to 16). Apparently, the in vitro remodeling system was more efficient at keeping the region of the −2 nucleosome open if Pho4/Pho2 was prebound than it was at opening a −2 nucleosome that had already been positioned by the yeast extract.
We wondered if remodeling in vitro was less efficient because remodeling activities were limiting in the yeast extract. Indeed, supplementation of the yeast extract with exogenous RSC did modestly increase the resulting ClaI accessibility, to 55 to 57% (Fig. 4B, columns 11 and 12), though the appearance in DNase I indirect end labeling did not change significantly (Fig. 3B, lanes 11 to 13).
As PHO5 promoter chromatin opening in vivo is known to involve histone acetylation by Gcn5 (7), we also supplemented the yeast extract with acetyl-CoA plus or minus exogenous recombinant Gcn5. Recombinant Gcn5 was confirmed to increase histone acetylation both in octamers and in salt gradient dialysis chromatin with or without extract by incorporation of labeled acetyl groups (Ertel and Korber, unpublished). However, all this did not significantly increase Pho4-induced DNase I hypersensitivity or ClaI accessibility (Fig. 3B, lane 13) (Ertel and Korber, unpublished).
Remodeling at the PHO5 promoter is also known to result in histone eviction in trans (10, 11, 34, 50), which calls for a histone acceptor. We wondered if the presence of more histone acceptor molecules may increase the extent of remodeling. However, supplementing the extract with a 5- to 10-fold mass excess of salmon sperm DNA over chromatinized DNA failed to generate a more extensive hypersensitive site at the PHO5 (and also PHO8; see below) promoter (Ertel and Korber, unpublished).
Finally, the heterologous source of histones might limit the remodeling process in vitro if, for example, they carried inhibiting modifications or impaired the recognition by factors from the yeast extract. Therefore, we also repeated the experiments with recombinant yeast histones. These were much more difficult to handle; i.e., it was more difficult to reproducibly obtain good DNase I patterns of closed PHO5 promoter chromatin in vitro (Ertel and Korber, unpublished). Nonetheless, the Pho4-induced hypersensitive site was very similar to that obtained with histones from Drosophila embryos, and even the combination of using recombinant yeast histones and adding RSC, SWI/SNF, and acetyl-CoA did not make much of a difference (Fig. 3B, lane 13).
PHO8 and PHO84 promoter chromatin was hardly remodeled at all upon addition of Pho4 in vitro.
We previously showed that PHO8 and PHO84 promoter chromatin can also be properly assembled by the yeast extract system (27, 67). We now tested if addition of Pho4 would induce hypersensitive sites, as seen in vivo (3, 67) also at these promoters. In fact, in most of the experiments described so far, plasmids bearing the PHO5 and PHO8 loci, respectively, were present in equimolar amounts in the same tubes, so that these promoters could be simultaneously analyzed on the same blots by differential hybridization, providing excellent internal control.
In striking contrast to remodeling at the PHO5 promoter, addition of Pho4 in the presence of energy to PHO8 promoter chromatin merely led to an increase in the intensity of the band corresponding to the position of UASp2 (Fig. 5 A, lanes 6 to 8). This indicates that Pho4 could access its binding site and maybe caused some widening of the corresponding short hypersensitive region (Fig. 1B). However, there was no generation of a broader hypersensitive site, as seen in vivo (brace in Fig. 5A). None of the various conditions described above for the PHO5 promoter induced a more remodeled pattern at the PHO8 promoter (Fig. 5B) (Ertel, Hertel, and Korber, unpublished).
FIG. 5.
Little or no Pho4-induced chromatin remodeling at the PHO8 promoter in vitro. Panel A shows the same blot as in Fig. 3A, and panel B shows the same blot as in Fig. 3B but with both rehybridized for the PHO8 promoter region. The schematic on the left in panel A corresponds to Fig. 1B. The brace between lanes 5 and 6 shows the region that becomes hypersensitive upon induction in vivo (3). Marker fragments were generated by double digests of EcoRV, NdeI, HindIII, and SacI (panel A) or EcoRV, HindIII, and XhoI (panel B), each with BglII. Dots between lanes mark the bands characteristic for the PHO8 promoter chromatin pattern of the repressed state. Larger dots mark increased sensitivity of the band corresponding to the UASp2 Pho4 binding site.
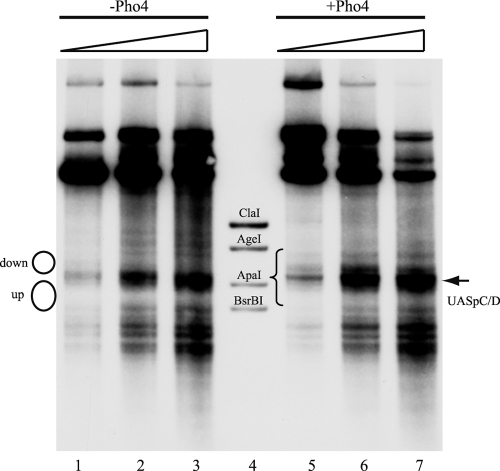
Similarly, we also did not observe much Pho4-induced remodeling of PHO84 promoter chromatin in vitro, besides some broadening of the linker region that contains UASpC/D in the downstream direction, which may indicate some remodeling at the border of the flanking downstream nucleosome (Fig. 6).
FIG. 6.
Little or no Pho4-induced chromatin remodeling at the PHO84 promoter in vitro. DNase I indirect end-labeling analysis of the PHO84 promoter region in chromatin preassembled by salt gradient dialysis, incubated with yeast extract (from strain YS27 cpf1) in the presence of energy and then left untreated or treated with Pho4, as indicated. The schematic on the left corresponds to positioned nucleosomes, as in Fig. 1C. The brace between lanes 4 and 5 shows the region that becomes hypersensitive upon induction in vivo (67). Marker fragments were generated by double digests of ClaI, AgeI, ApaI, and BsrBI, each with SspI. Ramps above the lanes denote increasing DNase I concentrations.
Thus, in contrast to the substantial and in vivo-like remodeling of PHO5 promoter chromatin observed in vitro, the extent of remodeling at the PHO8 and PHO84 promoters was much lower than that observed in vivo.
The intranucleosomal location of a Pho4 binding site in the −2 nucleosome has an auxiliary role in opening PHO5 promoter chromatin in vivo.
We wondered why remodeling of PHO5, but not of PHO8 or PHO84, promoter chromatin was observed in vitro. Previously, we showed that the nucleosomes at the PHO5 promoter are intrinsically less stable than those at the PHO8 promoter (27). However, we also showed that the nucleosome downstream of UASpC/D at the PHO84 promoter is less stable than the upstream nucleosome and similar in stability to the PHO5 promoter nucleosomes (67). In spite of this, the downstream nucleosome in PHO84 was hardly at all remodeled in vitro (Fig. 6). Thus, the difference in nucleosome stability could not in itself explain the differential remodeling at the three PHO promoters in vitro.
Next, we considered the possibility that the intranucleosomal location of a high-affinity Pho4 binding site at the PHO5 promoter might make the difference. We hypothesized that the competition between Pho4 binding to the high-affinity intranucleosomal UASp2 site at the PHO5 promoter on the one hand and formation of the −2 nucleosome on the other hand plays a hitherto unrecognized role in PHO5 promoter remodeling. The PHO8 promoter does not contain an intranucleosomal UASp element, and the PHO84 promoter harbors only two low-affinity intranucleosomal Pho4 sites (UASpB and UASpE) (Fig. 1C), which are largely dispensable for promoter opening in vivo (67). Thus, if the presence of a high-affinity intranucleosomal UASp element were important for effective remodeling under the arguably suboptimal conditions one can obtain in vitro, this could reflect a mechanistically important role for the intranucleosomal location of the UASp2 site in PHO5 promoter chromatin opening also in vivo. We used a variety of PHO5 promoter mutants to explicitly test this hypothesis.
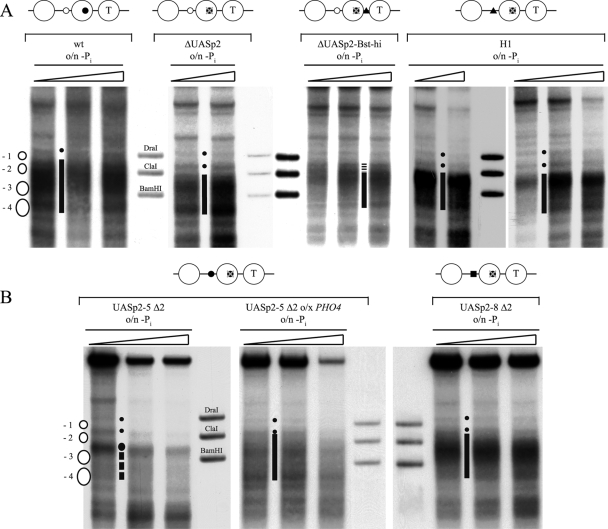
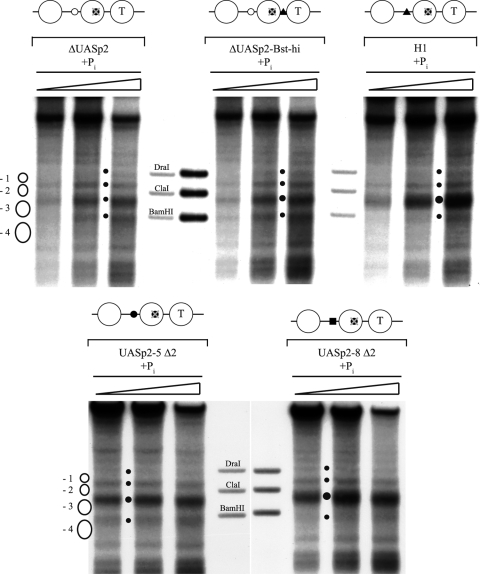
A deletion of the intranucleosomal UASp2 site at the PHO5 promoter by point mutations generating a HindIII site in nucleosome −2 (ΔUASp2) (Fig. 7) severely impaired chromatin opening in vivo (Fig. 8 A; Table 1) and induction of Pho5 acid phosphatase (37 ± 8 U, compared to >400 U for the wild type [4]) after overnight induction in phosphate-free medium. The DNase I indirect end-labeling pattern showed a less extensive hypersensitive site than the wild-type promoter. The hypersensitivity in the region of the −3 and −4 nucleosomes was similar to that in the wild type but did not extend as far into the region of the −2 nucleosome; i.e., the smear of hypersensitivity did not extend upward in the lane beyond the ClaI marker band (Fig. 8A). This incomplete remodeling was confirmed quantitatively by a ClaI accessibility of less than 50% (Table 1). Thus, the PHO5 promoter ΔUASp2 mutant could be remodeled only partially, mostly in the upstream half (nucleosomes −3 and −4, lower part of the gel lane).
FIG. 8.
The intranucleosomal location of a UASp element in the −2 nucleosome is important but not essential for PHO5 promoter chromatin opening in vivo. DNase I indirect end-labeling analysis of the PHO5 promoter region in the (A) wild type (wt) (CY337), ΔUASp2 (CY341), ΔUASp2-Bst-hi (CY337 FE1600), and H1 (CY337 EB1615) (35) configurations and in the (B) UASp2-5 Δ2 (CY339 ura3 pCB-UASp2-5 Δ2), and UASp2-8 Δ2 (CY339 ura3 pCB-UASp2-8 Δ2) configurations after overnight (o/n) incubation in phosphate-free (−Pi) medium. For the UASp2-5 Δ2 mutant, an experiment with incubation in phosphate-free medium for 40 h, i.e., two times overnight, and one experiment with overexpression (o/x) of PHO4 are shown. Small dots between lanes mark the bands corresponding to the linker regions flanking the positioned −1 nucleosome as shown in the schematic on the left, and vertical bars between the lanes highlight the extent of a hypersensitive region, stippled if less extensive. For UASp2-5 Δ2, the stippled vertical line denotes less-pronounced hypersensitivity in the region of the −3 and −4 nucleosomes and the large dot marks the increased hypersensitivity of the linker between the −2 and −3 nucleosomes. Samples from the same chromatin preparation but from two different gels are shown for the H1 mutant promoter. Schematics above the panels are as in Table 1. Markers, ramps, and schematics next to the gels are as in Fig. 2.
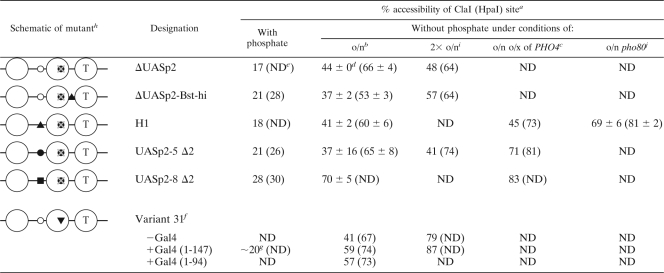
TABLE 1.
ClaI accessibilities of PHO5 promoter UASp mutants under various growth conditions
Accessibility of the ClaI site at the PHO5 promoter and of the HpaI site at the PHO8 promoter (in parentheses), respectively.
Overnight (o/n) induction in phosphate-free medium.
o/x, overexpression.
Errors show the variation of two or three biologically independent experiments.
ND, not determined.
Strain AH2341 grown in 2% raffinose-2% galactose.
Value not determined by PhosphorImager analysis but estimated by eye from band intensities in blot image.
Symbols: ○, low-affinity UASp1; ▪, high-affinity PHO8 UASp2; ▴, high-affinity UASp by point mutation; T, TATA box; •, high-affinity UASp2;  , deleted high-affinity UASp2; ▾, high-affinity Gal4 binding site.
, deleted high-affinity UASp2; ▾, high-affinity Gal4 binding site.
2× o/n, induction twice overnight.
o/n pho80, pho80 deletion and overnight starvation.
However, it could be argued that such a deletion experiment in itself is not sufficient to show an important role for the intranucleosomal location of the UASp element, as it concomitantly removes the only high-affinity UASp element and thus leaves the PHO5 promoter with only the low-affinity UASp1 element. Pho4 binding at only the UASp1 site is apparently insufficient for complete chromatin opening and transactivation. Not even overexpression of PHO4 could recover PHO5 expression in a ΔUASp2 mutant after overnight incubation in phosphate-free medium (49 ± 10 U), although such overexpression did induce chromatin opening in a ΔUASp1 mutant (64). Therefore, we wished to test PHO5 promoter mutants that still contained a high-affinity UASp element, but in a linker region and not intranucleosomally.
We generated either a PHO5 promoter that has both the low-affinity UASp1 site and a high-affinity UASp element in the linker between nucleosomes −1 and −2 (ΔUASp2-Bst-hi mutant, Fig. 7), or we replaced the low-affinity UASp1 site with high-affinity sites. For the latter approach, we used the high-affinity UASp2 element of either the PHO5 or the PHO8 promoter, including surrounding sequences comprising the whole Pho4 footprints (3, 6, 65) (constructs UASp2-5 Δ2 and UASp2-8 Δ2, respectively [Fig. 7]), or we used the H1 mutant generated in the O'Shea group, where the low-affinity E box CACGTTT of UASp1 is converted by point mutations to a high-affinity CACGTGG motif (Fig. 7) (35). The endogenous intranucleosomal UASp2 element is mutated into a HindIII site in all of these mutants. Importantly, the results presented by Lam et al. (35) confirmed the prediction that the point mutations in the H1 mutant indeed conferred higher Pho4 binding affinity than the wild-type UASp1 element. Similarly, the much higher acid phosphatase activity after overnight induction in phosphate-free medium obtained with the ΔUASp2-Bst-hi mutant than with the ΔUASp2 mutant (223 ± 4 versus 37 ± 8 U, respectively)—notably, at a similar degree of chromatin remodeling (Table 1)—confirmed that the artificial high-affinity E box CACGTGG motif that was introduced in the linker between nucleosomes −1 and −2 indeed corresponded to a functional UASp element. Further, proper nucleosome positioning under high-phosphate conditions was confirmed for all of these mutant promoters (Fig. 9; Table 1). The higher affinity of the UASp elements between nucleosomes −2 and −3 in the H1, UASp2-5 Δ2, and UASp2-8 Δ2 mutants resulted in an increased hypersensitivity of the sHS2 region (Fig. 9) and in a mildly increased ClaI accessibility for the UASp2-8 Δ2 mutant (Table 1), probably due to the low constitutive amounts of nuclear Pho4 (42).
FIG. 9.
PHO5 promoter UASp mutants show mostly undisturbed chromatin patterns in the repressed state. DNase I indirect end-labeling analysis of the indicated PHO5 promoter mutant regions under repressive (+Pi) conditions. The strains are the same as in Fig. 8. The schematics above the lanes are the same as in Table 1. The schematics on the left and the marker bands are the same as in Fig. 2. Small dots between the lanes mark the linker regions between positioned nucleosomes. Larger dots mark enhanced hypersensitivity of sHS2 in mutants compared to the wild type. Ramps above the lanes denote increasing DNase I concentrations.
We tested all of these PHO5 promoter mutants for chromatin opening after overnight induction in phosphate-free medium. Intriguingly, we observed significant but only partial promoter chromatin opening for the ΔUASp2-Bst-hi, H1, and UASp2-5 Δ2 mutants. In these three mutants, the ClaI accessibility was similarly low as in the ΔUASp2 mutant (Table 1), while the DNase I patterns showed some variation. For the H1 mutant, it was very similar to that of the ΔUASp2 mutant (Fig. 8A); i.e., the smear of the hypersensitive site hardly extended downstream (upward in the lane) of the ClaI marker band. For the UASp2-5 Δ2 mutant, it seemed even less open, with less pronounced hypersensitivity in the region of the −3 and −4 nucleosomes, so that the increased hypersensitivity of the linker between the −2 and −3 nucleosomes stood out prominently (Fig. 8B). The pattern of the ΔUASp2-Bst-hi mutant appeared to be more remodeled, similar to that of the wild type (Fig. 8A). As mentioned above, these differently extensive DNase I hypersensitivities are not necessarily in contrast to the equally low ClaI accessibilities. DNase I indirect end labeling employs limited degrees of digestion and preferentially scores the most nuclease-sensitive chromatin subpopulation. Therefore, DNase I patterns do not necessarily reflect the total or even average chromatin state, whereas ClaI digestion scores all chromatin templates. Further, a smear in the DNase I pattern may reflect repositioning but not necessarily eviction of promoter nucleosomes. For example, a gcn5 mutant in a pho80 background under +Pi conditions shows increased DNase I hypersensitivity across the entire PHO5 promoter without concomitantly increased restriction enzyme accessibilities (24). This chromatin state corresponds to repositioned but retained nucleosomes, as was confirmed later by the unchanged topology of PHO5 promoter chromatin minicircles (11).
In contrast to the ΔUASp2-Bst-hi, H1, and UASp2-5 Δ2 mutants, the UASp2-8 Δ2 mutant promoter was already fully remodeled, according to both DNase I mapping and ClaI assay, after overnight induction in phosphate-free medium (Fig. 8B; Table 1).
In the light of incomplete remodeling at most PHO5 mutant promoters, we controlled for successful induction of the PHO regulon by monitoring HpaI accessibility within the coregulated but unchanged PHO8 promoter. HpaI accessibility in this promoter increases upon induction to more than 60%, usually 70 to 90%, depending on the strain background and induction conditions (3, 4, 32). All PHO5 mutants were well induced at the PHO8 promoter by this criterion, although the ΔUASp2-Bst-hi mutant, and maybe also the H1 and ΔUASp2 mutants, for unknown reasons, was reproducibly somewhat at the lower limit of full induction. In order to test if stronger induction conditions could eventually open the mutant promoters, we increased the induction potential by prolonged incubation in phosphate-free medium two times overnight, by overexpression of PHO4 (19), or by deletion of the PHO80 gene encoding a negative regulator of PHO5 (30) (Table 1). Prolonged induction led to considerably more—although still not complete—chromatin opening in the ΔUASp2-Bst-hi mutant, in contrast to the ΔUASp2 and UASp2-5 Δ2 mutants (Table 1). Upon overexpression of PHO4, the UASp2-5 Δ2, but not the H1, mutant showed substantially more promoter opening (Fig. 8B; Table 1), while in both cases more complete remodeling occurred at PHO8, as measured by the increase in HpaI accessibility (Table 1). Nonetheless, even the H1 mutant promoter became more completely remodeled upon induction by the combination of the pho80 deletion and overnight starvation in phosphate-free medium.
Collectively, four of five PHO5 promoter UASp mutants without an intranucleosomal Pho4 binding site were severely impaired in chromatin opening after overnight induction in phosphate-free medium, which corresponds to standard “full” induction conditions. This was the case even though three of these mutants harbored a high-affinity linker Pho4 binding site. Evidently, moving the high-affinity binding site out of nucleosome −2 into a linker impaired remodeling of PHO5 promoter chromatin. This argues for an important role for the intranucleosomal location of the high-affinity Pho4 binding site in the −2 nucleosome.
Nonetheless, this role was not essential, as complete remodeling could be achieved in the UASp2-8 Δ2 mutant under standard induction conditions and in the UASp2-5 Δ2 and H1 mutants (and to a lesser degree also in the ΔUASp2-Bst-hi mutant) under forced induction conditions. Apparently, the quality of the UASp elements seemed to be important. The high-affinity linker binding sites inserted in the UASp2-5 Δ2 and H1 mutants were not as potent as that inserted in the UASp2-8 Δ2 mutant.
Mere binding competition at the −2 nucleosome can increase Pho4-dependent remodeling from UASp1.
The transactivator Pho4 comprises both a DNA binding domain and an activation domain, the latter being essential for PHO5 promoter chromatin opening (61). Now that we had evidence of an important role for the intranucleosomal location of a UASp element in the −2 nucleosome, we wondered if this role mainly reflected targeting of cofactor recruitment by the Pho4 activation domain to this site or if just binding competition between a DNA binding domain and the nucleosome could also be important by itself. In order to distinguish these possibilities, we turned to a PHO5 promoter variant generated earlier by Gudrun Ertinger in the Hörz group (17). Strain AH2341 harbors the so-called variant 31 PHO5 promoter, where the UASp2 element is replaced with a strong consensus Gal4 binding site (Fig. 7). With regard to Pho4 binding, this mutant is effectively a ΔUASp2 mutant. Indeed, chromatin opening is similarly incomplete after overnight induction in phosphate-free medium also for this version of a ΔUASp2 mutant (Table 1). However, for reasons given below, we used raffinose and galactose instead of glucose as carbon sources, which causes slower growth and therefore delayed depletion of intracellular phosphate pools. Thus, overnight induction under these conditions corresponds to weaker induction than under standard conditions. We tested prolonged induction for two times overnight and observed full chromatin opening at the variant 31 PHO5 promoter (Table 1). Thus, strain AH2341 eventually achieved full chromatin opening, even of a ΔUASp2 PHO5 promoter, whereas strain CY341 did not, even after prolonged induction (Table 1). The discrepancy between chromatin opening of the two different ΔUASp2 constructs in strains CY341 and AH2341, respectively, can be explained by the different strain backgrounds. Notoriously, although for unknown reasons, PHO5 promoter opening in the AH background is much more efficient than in the CY background (almost 100% ClaI accessibility versus about 70%, respectively, after overnight induction in phosphate-free medium).
Nonetheless, as overnight induction in strain AH2341 corresponded to suboptimal induction conditions, we could still test the auxiliary effect of DNA binding competition. We expressed the Gal4 DNA binding domain—either the Gal4(1-147) or the Gal4(1-94) construct (17)—from a plasmid in strain AH2341, which is a gal4 deletion mutant, induced the GAL system by growth in galactose, and monitored if chromatin opening upon phosphate starvation overnight was now more efficient than without a Gal4 construct under otherwise identical conditions. Indeed, ClaI accessibility was markedly increased in the presence of a Gal4 DNA binding domain (Table 1). Importantly, expression of a Gal4 DNA binding domain under +Pi conditions in the presence of galactose was not able to remodel the −2 nucleosome, and the degree of PHO regulon induction was always similar after overnight incubation, as shown by about 70% HpaI accessibility in all cases (Table 1). Thus, remodeling did fully depend on the action of Pho4 but was enhanced by the presence of a Gal4 DNA binding domain.
DISCUSSION
We demonstrate Pho4-induced remodeling of properly positioned nucleosomes at the PHO5 promoter in vitro. It is now clear that such remodeling is ATP dependent, pointing to the requirement of a chromatin remodeler in the process, which was previously mainly inferred from data indicating the involvement of remodeler ATPases in vivo (4, 16, 43, 59). This in vitro system opens the opportunity to elucidate biochemically the mechanism of PHO5 promoter chromatin remodeling.
Nonetheless, at present, this system does not recapitulate all aspects and not the full potential of the in vivo remodeling process. For example, acetyl-CoA dependency of chromatin remodeling could not be observed, even though PHO5 promoter opening strongly depends on the histone acetyltransferase Gcn5 in vivo (7, 24) and transient hyperacetylation is observed during the induction of both the PHO5 and PHO8 promoters (49, 50). However, PHO5 promoter chromatin can become fully remodeled in vivo also without Gcn5, although at a lower rate (7). Thus, it seems that our in vitro system captures the basic mechanistic level of chromatin remodeling leading to chromatin opening but not the intricacies that involve histone acetylation and govern the kinetics of remodeling in vivo. It will be a future challenge to reconstitute the missing features of PHO5 promoter chromatin remodeling.
The suboptimal remodeling potential of our in vitro system was also reflected in the finding that the promoters of PHO8 and PHO84 were hardly remodeled at all by the addition of Pho4 to the extract. This led us to investigate the so-far-unrecognized role of the intranucleosomal location of a UASp element in PHO5 promoter chromatin remodeling in vivo. In the past, the use of suboptimal induction conditions in vivo was very fruitful in recognizing the role of cofactors in the remodeling mechanism at the PHO5 promoter. For example, the involvement of the remodeler ATPases Snf2 and Ino80, of the histone acetyltransferase Gcn5, and of the histone chaperone Asf1 became apparent only under suboptimal induction conditions, such as the use of low-phosphate instead of phosphate-free medium, or during early time points of induction kinetics (1, 4, 7, 32, 43). Now we were guided by suboptimal remodeling conditions in vitro.
Interestingly, the role of the intranucleosomal UASp element touches upon an old discussion in yeast promoter chromatin remodeling research. Early studies suggested that binding of a transactivator like Gal4 to intranucleosomal binding sites can completely disrupt a nucleosome in vitro (68) and in vivo (40). This process was further stimulated in the presence of histone acceptors such as the histone chaperone nucleoplasmin (13) and did not require the Gal4 activation domain. These results suggested that simple binding competition between specific DNA binding factors and histones might help explain nucleosome remodeling in vivo. However, remodeling of PHO8 promoter chromatin does occur without an intranucleosomal UASp site via Pho4 binding adjacent to the positioned nucleosome (3). Further, for the PHO5 promoter, it was explicitly shown that the DNA binding activity of Pho4, i.e., a Pho4 mutant without its activation domain, was not sufficient for either stable binding to the intranucleosomal UASp2 element or PHO5 promoter opening, even if the protein was overexpressed (61). Nonetheless, transient binding of Pho4 to UASp2 in the −2 nucleosome, maybe in a transiently altered nucleosomal state or in a window of opportunity during replication, was assumed to happen during the chromatin remodeling process, as overexpression of PHO4 could eventually open the promoter even in the absence of UASp1 (64). Very recently, Jessica Tyler's group reported conditions where a transient interaction of Pho4 with the −2 nucleosome appeared to be monitored by sequential chromatin immunoprecipitation (48). In any case, the activation domain of Pho4 or other transactivators like Gal4 (58) is required for chromatin opening in vivo. Through their activation domains, these proteins are now thought to result in chromatin remodeling more or less exclusively by recruiting remodelers and histone-modifying cofactors (41, 44, 45). We do not wish to challenge the idea that ATP-dependent remodelers are principal agents in nucleosome remodeling. However, it has remained possible, and was never previously assessed for the PHO5 promoter, that the actual binding of Pho4 to its intranucleosomal UASp element is necessary for, or at least plays a role in, the remodeling process.
Indeed, our new data show that remodeling of the −2 nucleosome is severely impaired if the intranucleosomal UASp2 element is deleted. In three mutants (ΔUASp2-Bst-hi, H1, and UASp2-5 Δ2 mutants), this could not be compensated for under standard induction conditions by a newly introduced high-affinity linker UASp element. Interestingly, with the ΔUASp2 and H1 mutants, we observed for the very first time a clear uncoupling of the remodeling of different nucleosomes at the PHO5 promoter. So far, the four—or even five (29)—PHO5 promoter nucleosomes have always appeared to behave more or less as a concerted unit, also called a “chromatin microdomain” (60, 64), which became affected by remodeling either as a whole or not at all. However, in the ΔUASp2 and H1 mutants, the −3 and −4 nucleosomes were extensively remodeled, but the −1 and −2 nucleosomes were not. This argues that the −3 and −4 nucleosomes can be readily remodeled from a neighboring UASp site but that efficient remodeling of the −2 nucleosome depends significantly on the intranucleosomal location of a UASp element in the −2 nucleosome itself. Importantly, this intranucleosomal site need not be involved in recruiting cofactors but already enhanced remodeling just by binding of a specific DNA binding domain, as in the case of the Gal4 binding site in combination with a Gal4 DNA binding domain [strain AH2341 with plasmid YCpGal4(1-147) or YCpGal4(1-94)]. Further, the intranucleosomal UASp element need not necessarily be as strong as UASp2 since a swap of UASp1 and UASp2 in strain YS70 (Fig. 7) was previously observed to allow complete promoter opening (64). In summary, we conclude that the intranucleosomal location of a UAS element critically contributes to the completeness of remodeling of the −2 nucleosome via binding competition between the activator and the histones.
This view does not exclude the possibility that recruitment of remodelers to this site also plays an important role. It also does not contradict the notion that the degree of remodeling depends on the degree of cofactor recruitment, which in turn depends on the amount of activation domain function present at the promoter, which again depends on the number and affinity of UAS elements. Nonetheless, our data for promoter variant 31 show clearly that the completeness of chromatin remodeling can be enhanced by DNA binding competition within a nucleosome without changing the level of Pho4 occupancy or introducing another activation domain-bearing factor.
The critical role of intranucleosomal DNA binding competition represents a novel and somewhat unexpected facet of the PHO5 promoter chromatin remodeling mechanism. Indeed, in light of the above argument on the (lack of) acetylation dependence of the remodeling mechanism per se, this facet may be even more basic to remodeling than Gcn5-mediated histone acetylation. Nonetheless, we also generated a PHO5 promoter mutant (UASp2-8 Δ2) where insertion of a strong UASp element in the linker between the −2 and −3 nucleosomes was sufficient for remodeling of the −2 nucleosome, similar to the PHO8 promoter, where a single high-affinity UASp element in a linker region is also sufficient for nucleosome remodeling (42). Moreover, the weaker promoter mutants (especially the H1 and UASp2-5 Δ2 mutants) did eventually become remodeled under certain forced induction conditions (pho80 background or overexpression of PHO4) and even the ΔUASp2 mutant promoter could be fully opened in the AH background. Thus, we still agree with previous conclusions that such binding competition is not essential for promoter nucleosome remodeling in all contexts, but we argue that it is an important part of the wild-type remodeling mechanism in vivo.
It was recently suggested that nucleosomes might be removed from the PHO5 promoter by a sliding-mediated nucleosome disassembly mechanism (9). It was proposed that a remodeling complex such as RSC might bind a nucleosome, slide it along the PHO5 promoter region, and thereby displace the promoter nucleosomes in a snowplow-like fashion, leaving only the remodeler-bound nucleosome itself. This would explain why, on average, one nucleosome always remains at the promoter, even in the fully remodeled state (9-11, 34). We note that it should not matter for such a mechanism if the remodeler is recruited through a UASp element in the linker or in a nucleosome. However, our results suggest that the −2 nucleosome, in the absence of its intranucleosomal UASp element, is more refractory to disassembly than the nucleosomes at positions −3 and −4, therefore representing a “road block for the snowplow.” Therefore, we suggest considering, at least for the −2 nucleosome, a remodeling mechanism that involves not just sliding but also direct binding competition with the transcriptional activator Pho4.
Acknowledgments
This work was funded by the German Research Community (Deutsche Forschungsgemeinschaft, Transregio 05, to P.K.), by the European Community through the 6th Framework Programme (NET grant within the Network of Excellence The Epigenome to P.K.), and by an in-house grant from Cancer Research UK to J.Q.S. Support from the Amgen Foundation made Gözde Güclüler's stay at the Korber laboratory possible.
We are grateful to Tom Owen-Hughes for providing us with purified yeast SWI/SNF and RSC and to Erin O'Shea for sending plasmids of PHO5 promoter mutants. We also appreciate the work of Gözde Güclüler on PHO5 promoter UASp mutants.
Footnotes
Published ahead of print on 21 June 2010.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Almer, A., H. Rudolph, A. Hinnen, and W. Hörz. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbarić, S., K. D. Fascher, and W. Hörz. 1992. Activation of the weakly regulated PHO8 promoter in S. cerevisiae: chromatin transition and binding sites for the positive regulator protein Pho4. Nucleic Acids Res. 20:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbarić, S., T. Luckenbach, A. Schmid, D. Blaschke, W. Hörz, and P. Korber. 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 282:27610-27621. [DOI] [PubMed] [Google Scholar]
- 5.Barbarić, S., M. Münsterkötter, C. Goding, and W. Hörz. 1998. Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol. Cell. Biol. 18:2629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbarić, S., M. Münsterkötter, J. Svaren, and W. Hörz. 1996. The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 24:4479-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbarić, S., J. Walker, A. Schmid, J. Q. Svejstrup, and W. Hörz. 2001. Increasing the rate of chromatin remodeling and gene activation—a novel role for the histone acetyltransferase Gcn5. EMBO J. 20:4944-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeger, H., J. Griesenbeck, and R. D. Kornberg. 2008. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell 133:716-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 11.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 12.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 13.Chen, H., B. Y. Li, and J. L. Workman. 1994. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor- induced nucleosome disassembly. EMBO J. 13:380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clapier, C. R., and B. R. Cairns. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273-304. [DOI] [PubMed] [Google Scholar]
- 15.Del Rosario, B. C., and L. F. Pemberton. 2008. Nap1 links transcription elongation, chromatin assembly, and messenger RNP complex biogenesis. Mol. Cell. Biol. 28:2113-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhasarathy, A., and M. P. Kladde. 2005. Promoter occupancy is a major determinant of chromatin remodeling enzyme requirements. Mol. Cell. Biol. 25:2698-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ertinger, G. 1998. Rolle der Transkriptionsfaktoren bei der Öffnung der Chromatinstruktur am PHO5-Promoter in Saccharomyces cerevisiae. Ph.D. thesis. Universität München, Munich, Germany.
- 18.Fascher, K. D., J. Schmitz, and W. Hörz. 1990. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 9:2523-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fascher, K. D., J. Schmitz, and W. Hörz. 1993. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J. Mol. Biol. 231:658-667. [DOI] [PubMed] [Google Scholar]
- 20.Flaus, A., D. M. Martin, G. J. Barton, and T. Owen-Hughes. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34:2887-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudreau, L., A. Schmid, D. Blaschke, M. Ptashne, and W. Hörz. 1997. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell 89:55-62. [DOI] [PubMed] [Google Scholar]
- 22.Gregory, P. D., S. Barbarić, and W. Hörz. 1998. Analyzing chromatin structure and transcription factor binding in yeast. Methods 15:295-302. [DOI] [PubMed] [Google Scholar]
- 23.Gregory, P. D., S. Barbarić, and W. Hörz. 1999. Restriction nucleases as probes for chromatin structure. Methods Mol. Biol. 119:417-425. [DOI] [PubMed] [Google Scholar]
- 24.Gregory, P. D., A. Schmid, M. Zavari, L. Lui, S. L. Berger, and W. Hörz. 1998. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell 1:495-505. [DOI] [PubMed] [Google Scholar]
- 25.Gregory, P. D., A. Schmid, M. Zavari, M. Münsterkötter, and W. Hörz. 1999. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 18:6407-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haswell, E. S., and E. K. O'Shea. 1999. An in vitro system recapitulates chromatin remodeling at the PHO5 promoter. Mol. Cell. Biol. 19:2817-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hertel, C. B., G. Längst, W. Hörz, and P. Korber. 2005. Nucleosome stability at the yeast PHO5 and PHO8 promoters correlates with differential cofactor requirements for chromatin opening. Mol. Cell. Biol. 25:10755-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imhof, A., and A. P. Wolffe. 1999. Purification and properties of the Xenopus Hat1 acetyltransferase: association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry 38:13085-13093. [DOI] [PubMed] [Google Scholar]
- 29.Jessen, W. J., S. A. Hoose, J. A. Kilgore, and M. P. Kladde. 2006. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat. Struct. Mol. Biol. 13:256-263. [DOI] [PubMed] [Google Scholar]
- 30.Kaffman, A., I. Herskowitz, R. Tjian, and E. K. O'Shea. 1994. Phosphorylation of the transcription factor Pho4 by a cyclin-CDK complex, Pho80-Pho85. Science 263:1153-1156. [DOI] [PubMed] [Google Scholar]
- 31.Komeili, A., and E. K. O'Shea. 1999. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284:977-980. [DOI] [PubMed] [Google Scholar]
- 32.Korber, P., S. Barbarić, T. Luckenbach, A. Schmid, U. J. Schermer, D. Blaschke, and W. Hörz. 2006. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 281:5539-5545. [DOI] [PubMed] [Google Scholar]
- 33.Korber, P., and W. Hörz. 2004. In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication-independent extract system. J. Biol. Chem. 279:35113-35120. [DOI] [PubMed] [Google Scholar]
- 34.Korber, P., T. Luckenbach, D. Blaschke, and W. Hörz. 2004. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 24:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam, F. H., D. J. Steger, and E. K. O'Shea. 2008. Chromatin decouples promoter threshold from dynamic range. Nature 453:246-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 37.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119:1-16. [DOI] [PubMed] [Google Scholar]
- 38.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304:3-19. [DOI] [PubMed] [Google Scholar]
- 39.Mellor, J., W. Jiang, M. Funk, J. Rathjen, C. A. Barnes, T. Hinz, J. H. Hegemann, and P. Philippsen. 1990. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 9:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morse, R. H. 1993. Nucleosome disruption by transcription factor binding in yeast. Science 262:1563-1566. [DOI] [PubMed] [Google Scholar]
- 41.Morse, R. H. 2007. Transcription factor access to promoter elements. J. Cell. Biochem. 102:560-570. [DOI] [PubMed] [Google Scholar]
- 42.Münsterkötter, M., S. Barbarić, and W. Hörz. 2000. Transcriptional regulation of the yeast PHO8 promoter in comparison to the coregulated PHO5 promoter. J. Biol. Chem. 275:22678-22685. [DOI] [PubMed] [Google Scholar]
- 43.Neef, D. W., and M. P. Kladde. 2003. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 23:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4:649-655. [DOI] [PubMed] [Google Scholar]
- 46.Nourani, A., R. T. Utley, S. Allard, and J. Cote. 2004. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 23:2597-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pondugula, S., D. W. Neef, W. P. Voth, R. P. Darst, A. Dhasarathy, M. M. Reynolds, S. Takahata, D. J. Stillman, and M. P. Kladde. 2009. Coupling phosphate homeostasis to cell cycle-specific transcription: mitotic activation of Saccharomyces cerevisiae PHO5 by Mcm1 and Forkhead proteins. Mol. Cell. Biol. 29:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ransom, M., S. K. Williams, M. L. Dechassa, C. Das, J. Linger, M. Adkins, C. Liu, B. Bartholomew, and J. K. Tyler. 2009. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J. Biol. Chem. 284:23461-23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinke, H., P. D. Gregory, and W. Hörz. 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter. Mol. Cell 7:529-538. [DOI] [PubMed] [Google Scholar]
- 50.Reinke, H., and W. Hörz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 51.Reinke, H., and W. Hörz. 2004. Anatomy of a hypersensitive site. Biochim. Biophys. Acta 1677:24-29. [DOI] [PubMed] [Google Scholar]
- 52.Richmond, E., and C. L. Peterson. 1996. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 24:3685-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmid, A., K. D. Fascher, and W. Hörz. 1992. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell 71:853-864. [DOI] [PubMed] [Google Scholar]
- 54.Sengstag, C., and A. Hinnen. 1987. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 15:233-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sengstag, C., and A. Hinnen. 1988. A 28-bp segment of the Saccharomyces cerevisiae PHO5 upstream activator sequence confers phosphate control to the CYC1-lacZ gene fusion. Gene 67:223-228. [DOI] [PubMed] [Google Scholar]
- 56.Simon, R. H., and G. Felsenfeld. 1979. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 6:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Springer, M., D. D. Wykoff, N. Miller, and E. K. O'Shea. 2003. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 1:E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stafford, G. A., and R. H. Morse. 1997. Chromatin remodeling by transcriptional activation domains in a yeast episome. J. Biol. Chem. 272:11526-11534. [DOI] [PubMed] [Google Scholar]
- 59.Steger, D. J., E. S. Haswell, A. L. Miller, S. R. Wente, and E. K. O'Shea. 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299:114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svaren, J., and W. Hörz. 1997. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22:93-97. [DOI] [PubMed] [Google Scholar]
- 61.Svaren, J., J. Schmitz, and W. Hörz. 1994. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 13:4856-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svaren, J., U. Venter, and W. Hörz. 1995. In vivo analysis of nucleosome structure and transcription factor binding in Saccharomyces cerevisiae. Methods Mol. Genet. 6:153-167. [Google Scholar]
- 63.Terrell, A. R., S. Wongwisansri, J. L. Pilon, and P. J. Laybourn. 2002. Reconstitution of nucleosome positioning, remodeling, histone acetylation, and transcriptional activation on the PHO5 promoter. J. Biol. Chem. 277:31038-31047. [DOI] [PubMed] [Google Scholar]
- 64.Venter, U., J. Svaren, J. Schmitz, A. Schmid, and W. Hörz. 1994. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 13:4848-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vogel, K., W. Hörz, and A. Hinnen. 1989. The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol. Cell. Biol. 9:2050-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams, S. K., D. Truong, and J. K. Tyler. 2008. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. U. S. A. 105:9000-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wippo, C. J., B. S. Krstulovic, F. Ertel, S. Musladin, D. Blaschke, S. Sturzl, G. C. Yuan, W. Hörz, P. Korber, and S. Barbarić. 2009. Differential cofactor requirements for histone eviction from two nucleosomes at the yeast PHO84 promoter are determined by intrinsic nucleosome stability. Mol. Cell. Biol. 29:2960-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Workman, J. L., and R. E. Kingston. 1992. Nucleosome core displacement in vitro via a metastable transcription factor nucleosome complex. Science 258:1780-1784. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Y., Z. Moqtaderi, B. P. Rattner, G. Euskirchen, M. Snyder, J. T. Kadonaga, X. S. Liu, and K. Struhl. 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 16:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 25:8985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]