Abstract
Epac1 (exchange protein directly activated by cyclic AMP [cAMP]) couples intracellular cAMP to the activation of Rap1, a Ras family GTPase that regulates cell adhesion, proliferation, and differentiation. Using mass spectrometry, we identified the small G protein Ran and Ran binding protein 2 (RanBP2) as potential binding partners of Epac1. Ran is a small G protein best known for its role in nuclear transport and can be found at the nuclear pore through its interaction with RanBP2. Here we demonstrate that Ran-GTP and Epac1 interact with each other in vivo and in vitro. This binding requires a previously uncharacterized Ras association (RA) domain in Epac1. Surprisingly, the interaction of Epac1 with Ran is necessary for the efficient activation of Rap1 by Epac1. We propose that Ran and RanBP2 anchor Epac1 to the nuclear pore, permitting cAMP signals to activate Rap1 at the nuclear envelope.
For many members of the Ras GTPase superfamily, such as Ras and Rap, guanine nucleotide exchange factors (GEFs) outnumber the small GTPases that they activate (13, 34). The abundance of GEFs permits the activation of a small GTPase through different upstream signals.
Among the many GEFs for Rap, Epac1 and Epac2 can directly couple cyclic AMP (cAMP) to Rap activation. Epacs are bona fide targets for the physiological levels of cAMP achieved following hormonal stimulation of the cell (17), binding with an affinity in the micromolar range (1 to 3 μM) (8, 10, 42). By use of genetically encoded cAMP sensors, it has been observed that cAMP is compartmentalized in both time and space within a cell (1, 20). Discrete subpopulations of cAMP can also be regulated following hormonal stimulation at a distance from the adenylate cyclases by the action of phosphodiesterases (1, 20, 47). Therefore, the dynamics of cAMP in the cell may provide subcellular regulation of Epac-Rap signaling.
The localization of Epac proteins themselves can also provide important control of Rap activation. This is particularly relevant for Rap1, one of the Rap family members that is present on both the plasma membrane (PM) and the nuclear envelope (NE) (4). Through specific protein interactions, properly localized Epac proteins may not only function more efficiently but also limit the activation of Rap1 to specific regions within the cell. Direct support for this model comes from detailed structural and functional analysis of Epac proteins in the context of a cell. Both Epac proteins contain homologous regulatory and catalytic regions. Direct binding of cAMP to each regulatory region triggers a conformational change that allows its respective catalytic region to activate Rap1. The catalytic regions of both Epac1 and Epac2 comprise a Ras exchanger motif (REM) domain and a CDC25 homology domain that confers GEF activity toward Rap1. For Epac2, the REM and CDC25 domains are interrupted by a Ras association (RA) domain that interacts with activated, GTP-loaded Ras at an affinity similar to those of the classical Ras effectors, such as Raf-1 and B-Raf (28). Because the locations of Ras and Rap1 can overlap at the plasma membrane, the recruitment of Epac2 to Ras brings it into proximity with a pool of Rap1 at this locale (27). This compartmentalization of Epac2 allows it to activate Rap1 at the PM efficiently in the presence of cAMP.
In contrast to the findings for Epac2, the region between the REM and CDC25 domains in Epac1 contains a putative RA domain for which no binding partner has been identified. Unlike Epac2, Epac1 is predominantly localized to the perinuclear region instead of the PM in multiple cell lines (12, 32, 41, 49). Recent evidence suggests that Epac1 may function in nuclear processes, such as the nuclear transport of DNA-dependent protein kinase (DNA-PK) (21). However, the mechanism for the spatial regulation of Epac1-Rap1 signaling in this subcellular location is unknown. In this study, we identify a novel mechanism underlying the anchored signaling of Epac1 at the nuclear pore via its putative RA domain, and we report for the first time the coupling of the nuclear small GTPase Ran and Rap1 at the NE.
MATERIALS AND METHODS
Plasmids.
Human Epac1 was a gift from Johannes Bos, Utrecht University. Hemagglutinin (HA)-tagged wild-type Ran and RanV19 were gifts from Ian Macara, University of Virginia. Histone 2B-mCherry was obtained from Addgene (Cambridge, MA). GFP-Epac1, Epac2, Flag-Rap1b, and mCherry-RasV12 have been described previously (28, 49). HRas and Rap2b cDNAs were purchased from the Missouri S&T cDNA Resource Center (Rolla, MO) and were subcloned into the pGFP-C1 and/or pmCherry-C1 vector (Clontech, Mountain View, CA). Epac1RA2 was constructed by inserting a PCR product for amino acids (aa) 559 to 720 of Epac2 between the sequences for aa 423 and 586 of Epac1 and was subcloned into the pcDNA3 and pGFP-C1 vectors. GFP-Epac1Δ295 was constructed by subcloning the PCR product for aa 295 to 881 of Epac1 into pGFP-C1. GFP-Epac1Δ673 was constructed by digestion using existing BglII and PstI sites in GFP-Epac1Δ295 and ligation of the blunted ends. GFP-Epac1Δ295RA2 was constructed by subcloning the PCR product for aa 295 to 881 of Epac1RA2 into pGFP-C1. GFP-RanV19-Epac1RA2 was constructed by in-frame insertion of the PCR product of the RanV19 cDNA between the HindIII and EcoRI sites in the GFP-Epac1RA2 vector. mCherry-RanV19-Epac1Δ295RA2 was constructed by in-frame insertion of the PCR product of the RanV19 cDNA between the EcoRI and SalI sites in the mCherry-Epac1Δ295RA2 vector. mCherry-RanV19-CAAX was constructed by in-frame insertion of the PCR product encoding RanV19 between NotI and XhoI, and of the PCR product encoding the CAAX motif of HRas (the last 27 aa at the C terminus) with a stop codon between XhoI and XbaI, in the pmCherry-C1 vector.
Antibodies.
Anti-Rap1a/b, anti-Ran, and unconjugated and agarose-coupled anti-Flag (M2) were from Sigma-Aldrich (St. Louis, MO). The rabbit antiserum against green fluorescent protein (GFP) was from Abcam (Cambridge, MA). Anti-Epac1 and anti-Ras (RAS10) were from Upstate Biotechnology. The anti-HA antibody and anti-nuclear pore complex proteins (MAb414) were from Covance (Princeton, NJ). Anti-RanBP2 (N20), anti-Epac1 (H70), anti-FRS2 (M20), anti-TrkA (763), and anti-Rap2 (V19) were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-RanBP2 (rabbit) from Novus Biologicals (Littleton, CO) and anti-Epac1 (A5) (Santa Cruz) were also used where indicated in the figure legends. Secondary anti-mouse, anti-rabbit, and anti-goat antibodies used in Western blots were purchased from GE Healthcare and Vector Laboratories Inc. (CA). Texas Red- and fluorescein isothiocyanate (FITC)-conjugated secondary antibodies for immunofluorescent microscopy were purchased from Vector Laboratories and Jackson ImmunoResearch Laboratories Inc. (West Grove, PA).
Chemicals.
8-(4-Chloro-phenylthio)-2′-O-methyladenosine-3′,5′-cyclic monophosphate (2OMe) was purchased from Biolog Life Sciences (Bremen, Germany). The 3× Flag peptide, GDP, GTPγS, glutathione peptide, glutathione-agarose beads, and leptomycin B (LMB) were purchased from Sigma-Aldrich (St. Louis, MO).
Cell culture and stable cell lines.
HEK293 cells were cultured in Dulbecco's modified Eagle medium (DMEM) plus 10% fetal bovine serum (FBS), penicillin-streptomycin, and l-glutamine at 37°C under 5% CO2. MEL-24 cells were cultured in Eagle minimum essential medium from ATCC (Manassas, VA) plus 10% FBS and penicillin-streptomycin at 37°C under 5% CO2. Transient transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. For stable cell lines, HEK293 cells were transfected with Flag-Epac1 or the pcDNA3 vector and were selected with 0.5 mg/ml G418 (Invitrogen) for 4 weeks.
Purification of the Epac1-containing complex and mass spectrometry.
Approximately 1 × 107 HEK293-Flag-Epac1 cells or control cells were lysed in lysis buffer (10% glycerol, 1% NP-40, 50 mM Tris-HCl [pH 7.4], 200 mM NaCl, 2 mM MgCl2, 0.5 mM β-glycerolphosphate) supplemented with 1 μM leupeptin, 10 μg/ml soybean trypsin inhibitor, 0.1 μM aprotinin, and 1 mM sodium orthovanadate. Clear lysates were incubated with conjugated anti-Flag (M2) antibodies at 4°C for 4 h. The beads were washed three times in lysis buffer, and bound proteins were eluted at room temperature in 0.1 ml of TBS (50 mM Tris-HCl [pH 7.4], 150 mM NaCl) containing 100 μg/ml of 3× Flag peptides. Eluted proteins were supplemented with 6× Laemmli buffer and were resolved on a sodium dodecyl sulfate-polyacrylamide gradient gel electrophoresis (SDS-PAGE) (5 to 17% acrylamide) gel (Invitrogen) for Imperial blue staining (Thermo Scientific, Rockford, IL). Unique bands from the lane for the HEK293-Flag-Epac1 cells and the corresponding parts in the control lane were excised and subjected to in-gel trypsin digestion. Extracted peptides were analyzed using a ThermoFinnigan LTQ mass spectrometer. All tandem mass spectrometry (MS-MS) samples were analyzed using Sequest (ThermoFinnigan, San Jose, CA; version 27, revision 12), which was set up to search for tryptic peptides in a human subset of the UniProtKB/Swiss-Prot database, version 56_2 (http://www.uniprot.org). Peptide identifications were accepted if they could be established at a >90.0% probability as specified by the Peptide Prophet algorithm (25). Protein identifications were accepted if they contained at least 2 identified peptides and could be established at a >99.0% probability, as assigned by the Protein Prophet algorithm (37).
IP and Western blotting.
Cells in 6-cm-diameter plates were lysed at 18 to 24 h after transfection in lysis buffer, and equal amounts of clear cell lysate were subjected to immunoprecipitation (IP) using antibodies as indicated in the figure legends. For IP of the endogenous proteins, lysates from two confluent 15-cm-diameter plates were used for each condition. Lysates were incubated with the indicated antibodies overnight and were then incubated for 2 h with protein A beads (Invitrogen) blocked with 1% bovine serum albumin (BSA). For controls, IP was performed with unrelated purified IgG from the same species (anti-TrkA [rabbit IgG] for Epac1 IP [H70; rabbit] and anti-FRS2 [goat IgG] for RanBP2 IP [goat]). Beads were washed twice in lysis buffer and once in high-salt lysis buffer with NaCl at 300 mM. Bound proteins were eluted with 2× Laemmli buffer and were detected by immunoblotting with antibodies as indicated in the figure legends.
Protein purification.
GST-Epac1 was expressed and purified using the method described previously for GST-Epac2Δ430 (28). pQE-Ran was transformed into Escherichia coli strain M15(pREP4) (Qiagen). A 10-ml volume of overnight culture was used to inoculate 200 ml of LB medium. His-Ran expression was induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 4 h after the cells reached an optical density at 600 nm (OD600) of 0.6. The cell pellet was resuspended in 20 ml lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]) supplemented with phenylmethylsulfonyl fluoride (PMSF) and β-mercaptoethanol and was lysed with a French press. The lysate was cleared by centrifugation, and the supernatant was incubated with 0.5 ml nickel-nitrilotriacetic acid (Ni-NTA) for 1 h at 4°C. The beads were washed twice with 15 ml wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole [pH 8.0]). His-Ran was eluted five times with 0.5 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole [pH 8.0]).
GST pulldown assay.
His-Ran was preloaded with 0.1 mM GTPγS or 1 mM GDP in 200 μl of lysis buffer supplemented with 10 mM EDTA at 30°C for 30 min. The reaction was stopped by MgCl2 at 10 mM. Glutathione S-transferase (GST) alone (0.5 μg) or GST-EPAC1 (0.5 μg) was added to increasing amounts of loaded His-Ran and incubated with 20 μl of GST agarose beads (50% slurry) for 3 h at 4°C. The beads were washed three times with lysis buffer, boiled in 1×SDS loading dye, and subjected to Western blotting.
Rap activation assay and quantification.
GTP-bound Rap1 was assayed with the GST-tagged Ras-binding domain (RBD) of RalGDS as described previously (15). The intensities of the bands from Western blots were quantified with Scion Image (Scion Corp., Frederick, MD). The intensities of Rap1-GTP were normalized to those of total Rap1. All of the experiments were repeated at least three times, and the data from each experiment were expressed as a percentage of the maximum. For Rap2, the GST-RalGDS assay was performed as described above, but Rap2 antibody was used for Western blotting.
Confocal imaging and quantification.
For live-imaging, cells were plated on coverslips coated with poly-d-lysine, transfected on the same day, and used for imaging 12 to 16 h later. The coverslips were clamped into a heated imaging chamber (Warner Instruments, Hamden, CT). All imaging experiments were performed with a Yokogawa CSU-10 Nipkow disk confocal scanning unit (Solamere Technology Group, Salt Lake City, UT) mounted on a Nikon TE2000 PFS microscope with continuous focus compensation. A Spectrum 70C krypton argon laser (Coherent, Santa Clara, CA) was used for excitation, with laser line selection via an acousto-optic tunable filter (Neos Technologies, Melbourne, FL), paired with fast emission filter switching (Applied Scientific Instrumentation Inc., Eugene, OR). The objectives used were a PlanApo 60× (numerical aperture [NA], 1.45] or a 100× [NA, 1.49] objective, heated to physiological temperature with appropriate heater bands (Bioptechs, Butler, PA). Device integration was controlled through MetaMorph (Molecular Devices, Downingtown, PA). Time lapse images (500-ms exposures) were captured on an Orca ER charge-coupled device (CCD) camera (Hamamatsu, Bridgewater, NJ) over the periods of time indicated in the figure legends. Acquired images were quantified using ImageJ, version 1.41o (NIH). For the fluorescence intensity of the perinuclear rim, the NE was traced using a segmented line with a width of 5 pixels, and the mean gray values along the tracing were recorded (R0). This measurement took into consideration both the intensities of individual puncta and the frequency of their presence along the NE. The fluorescence intensities of the cytoplasm (C0) and nucleus (N0) were also recorded as the mean gray values of representative regions within the two compartments. The gray value from the background was designated B0. To quantify the enrichment of Epac1 at the nuclear pore over that at the cytoplasm, more than 50 cells with a wide range of GFP-Epac1 expression levels were quantified, and the absolute rim intensities (R0 − B0) were plotted against the absolute intensities of the cytoplasm (C0 − B0). Linear regression was performed for cells with (C0 − B0) values lower than 40 gray values, and the slope [R = (R0 − B0)/(C0 − B0)] was a constant that reflected the enrichment of GFP-Epac1 at the nuclear pore versus the cytoplasm. This parameter (R) was also used to compare the relative enrichment of wild-type Epac1 and its mutants at the nuclear pore in Fig. 2F, 4E, and 5C. The relative distributions of various constructs within the nucleus were quantified using the ratio N, calculated as (N0 − B0)/(C0 − B0). For each construct, 30 to 40 random cells were measured, and experiments were repeated two to four times using cells of different passages on different days. For the percentage of mCherry-Rap1b-positive cells showing enrichment of GFP-RBDRalGDS at the NE (see Fig. 8C), cells from at least 30 random fields were scored in a blinded fashion, and four experiments were performed and quantified.
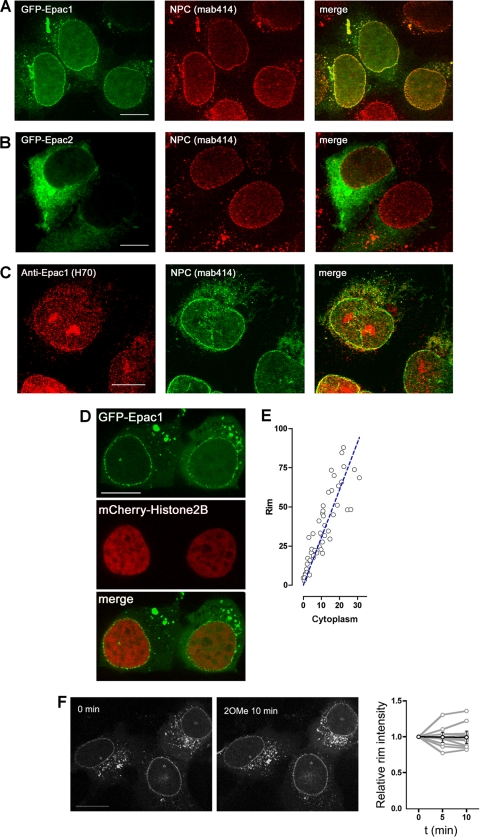
FIG. 2.
Colocalization of Epac1 with the nuclear pore complex (NPC). (A) GFP-Epac1 colocalizes with the NPC. HEK293 cells were transfected with GFP-Epac1 (left) (green) and were stained with MAb414 as a primary antibody and Texas Red as a secondary antibody (center) (red) to identify the NPC. A merged image is also shown (right). (B) GFP-Epac2 does not colocalize with the NPC. HEK293 cells were transfected with GFP-Epac2; they were then treated, and are presented, as for panel A. (C) Colocalization of endogenous Epac1 with the NPC. MEL-24 cells were stained with an anti-Epac1 primary antibody and a Texas Red-coupled secondary antibody (left), and with MAb414 as a primary antibody and an FITC-coupled secondary antibody (center). A merged image is also shown (right). (D) Localization of GFP-Epac1 expressed in HEK293 cells by confocal live imaging. mCherry-Histone2B was cotransfected in order to visualize the nuclear chromatin. Bars, 10 μm. (E) Quantification and correlation of the fluorescence intensities of GFP-Epac1 at the perinuclear rim (y axis) and cytoplasm (x axis) in HEK293 cells over a low range of expression levels (see Materials and Methods for details). (F) Effect of 2OMe on Epac1 localization. HEK293 cells were transfected with GFP-Epac1, serum starved, and treated with 2OMe during confocal live imaging. (Left) Representative localizations of GFP-Epac1 before and after 2OMe treatment. (Right) Quantification of the relative intensities of GFP-Epac1 at the perinuclear rim over time normalized to the intensities at 0 min. Gray lines represent changes in the intensities in individual cells (n = 9); the black line and error bars represent means ± standard errors of the means.
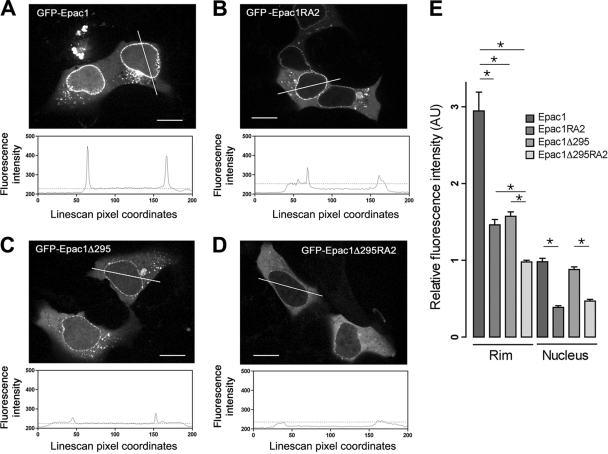
FIG. 4.
Roles of the RA1 domain and regulatory region in Epac1 localization. (A to D) Localization of GFP-tagged Epac1, Epac1RA2, Epac1Δ295, and Epac1Δ295RA2 in HEK293 cells by confocal live imaging. Upper panels show representative distributions of the indicated constructs. Lower panels show the intensity profiles across the lines in the photos. y axis, fluorescence intensity; x axis, line scan pixel coordinates; dotted lines, levels of fluorescence intensity in the cytoplasm. Bars, 10 μm. (E) Quantification of the relative fluorescence intensities of GFP-tagged Epac1 and mutants at the perinuclear rim and within the nucleus (means ± standard errors of the means; *, P < 0.01). AU, artificial units. See Materials and Methods for details.
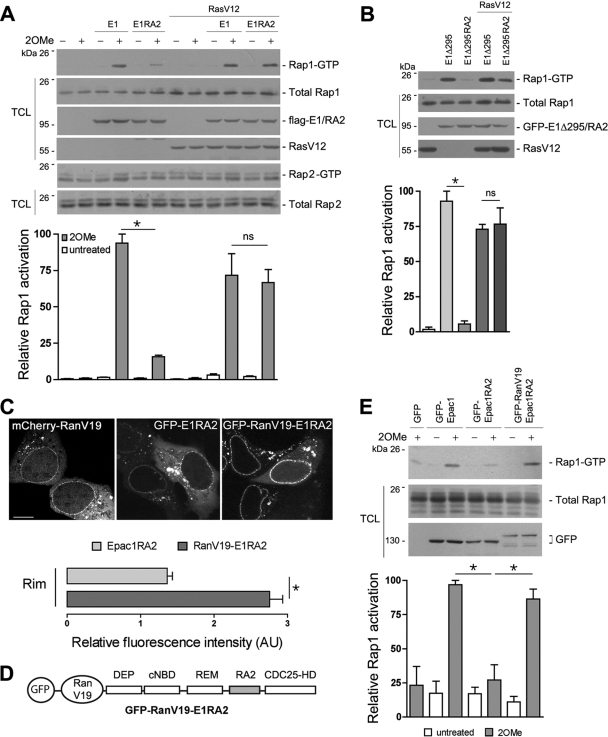
FIG. 5.
Requirement of the RA1 domain for efficient Rap1 activation via Epac1. (A) Activation of endogenous Rap1 and Rap2 by Epac1 (E1) and Epac1RA2 (E1RA2). (Top) Flag-E1 and Flag-E1RA2 were expressed in HEK293 cells in the absence or presence of mCherry-RasV12. Starved cells were treated with 2OMe for 15 min or were left untreated, and the lysates were assayed for Rap1 and Rap2 activation as described in Materials and Methods. Transfected proteins were blotted with anti-Flag and anti-Ras. (Bottom) Quantification of relative Rap1 activation from three independent experiments (means ± standard errors of the means; *, P < 0.05; ns, not significant). (B) GFP-tagged E1Δ295 and E1Δ295RA2 were expressed in HEK293 cells in the absence or presence of mCherry-RasV12. Rap1 activation was assayed, and is presented, as described for panel A. (C) Comparison of the localizations of mCherry-RanV19, GFP-E1RA2, and GFP-RanV19-E1RA2 in HEK293 cells by confocal live imaging. (Top) Representative images. Bar, 10 μm. (Bottom) Quantification of relative fluorescence intensities of GFP-E1RA2 (light shaded bar) and GFP-RanV19-E1RA2 (dark shaded bar) at the perinuclear rim (means ± standard errors of the means; *, P < 0.01). (D) Schematic of the GFP-RanV19-E1RA2 construct. The domains of Epac1 are listed as in Fig. 3A. The RA domain of Epac2 (RA2) is shaded. (E) Rap1 activation by GFP-RanV19-Epac1RA2. GFP-tagged Epac1, Epac1RA2, and RanV19-Epac1RA2 were expressed in HEK293 cells, which were either treated with 2OMe for 15 min or left untreated. Rap1 activation was assayed, and is presented, as described for panel A.
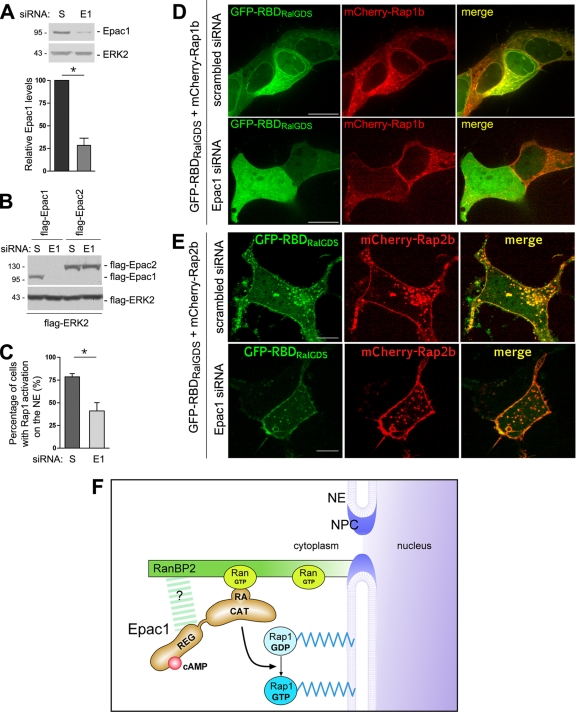
FIG. 8.
Attenuation of Rap1 activation at the NE by depletion of endogenous Epac1. (A) Depletion of Epac1 by siRNA. (Top) Lysates of HEK293 cells transfected with scrambled (S) siRNA or Epac1 siRNA were examined by Western blotting using anti-Epac1 and anti-ERK2 (loading control). (Bottom) Quantification of the results of three experiments (means ± standard errors of the means; *, P < 0.05). (B) Specificity of siRNA for Epac1. HEK293 cells were transfected with Flag-Epac1 or Flag-Epac2 cDNA and either scrambled or Epac1 siRNA, as indicated. The expression of transfected Epac isoforms in the lysates of transfected cells was examined by Western blotting using anti-Flag antibodies. Flag-ERK2 served as a transfection and loading control. (C) Effect of Epac1 depletion on Rap1 activation at the NE. GFP-RBDRalGDS and mCherry-Rap1b were cotransfected with scrambled or Epac1 siRNA in HEK293 cells. The percentages of cells with enrichment of GFP-RBDRalGDS on the NE were quantified from four independent experiments (means ± standard errors of the means; *, P < 0.01). (D) Representative images from the experiment for which results are shown in panel C. The top and bottom panels show cells cotransfected with scrambled and Epac1 siRNA, respectively. (Left) GFP; (center) mCherry; (right) merged images. Bar, 10 μm. (E) Effect of Epac1 depletion on Rap2 activation. GFP-RBDRalGDS and mCherry-Rap2b were cotransfected with scrambled (top) or Epac1 (bottom) siRNA in HEK293 cells. (Left) GFP; (center) mCherry; (right) merged images. Bar, 10 μm. (F) Model for anchored Epac1 signaling at the NE. The NPC is a large multimeric protein complex, shown in cross section, spanning the double bilayer of the NE. Epac1 is localized to the NPC through direct interaction between its RA domain and Ran-GTP, which also associates with RanBP2, the major nucleoporin on the cytoplasmic face of the NPC. Epac1 may also make direct contacts with RanBP2 as shown (question mark). The anchored Epac1 allows cAMP to activate local pools of Rap1 that are tethered on the NE.
Immunofluorescent staining.
MEL-24 cells were grown on poly-d-lysine-coated coverslips, fixed in 4% paraformaldehyde for 15 min, and permeabilized in 0.5% Triton X-100 for 10 min. Cells were incubated first with 1% BSA in phosphate-buffered saline-Tween 20 (PBS-T) for 45 min and then with primary antibodies (as indicated) overnight at 4°C. After extensive washing, cells were incubated with the indicated secondary antibodies overnight at 4°C. Nuclei were stained with 0.1 μg/ml Hoechst stain for 5 min. Coverslips were mounted with Elvanol.
RNA interference.
Human Epac1 (RefSeq number NM_006105) was targeted using a cocktail of three small interfering RNAs (siRNAs) from Ambion Inc. (Austin, TX). The sense sequences were 5′-CCGAGAUGCCCAAUUCUACtt-3′, 5′-GGGAUCUGUCAACGUGGUGtt-3′, and 5′-GGGCACUUCGUGGUACAUUtt-3′. The scrambled or Epac1 siRNAs were transfected alone using Lipofectamine 2000 (Invitrogen) at a final concentration of 100 nM in a course of 3 to 4 days and were then cotransfected with the indicated plasmids overnight before live imaging.
Statistics.
Prism, version 3 (GraphPad Software, La Jolla, CA), was used for data plotting and analysis. Unpaired t tests were performed between groups as indicated, and a P value of <0.05 was regarded as statistically significant.
RESULTS
Isolation of proteins associated with Epac1.
Using HEK293 cell lines stably transfected with Flag-Epac1, or the vector alone as a control, we isolated the proteins that associated with Epac1 by using affinity purification. Analysis of the complex by mass spectrometry identified five proteins as potential Epac1 binding partners (Table 1). Three of these proteins, Ran binding protein 2 (RanBP2, or Nup358), nucleoporin 205 (Nup205), and Nup98, are structural components of the nuclear pore complex (NPC). RanBP2 has four Ran binding sites and forms a filamentous structure on the cytoplasmic side of the NPC (14, 51). The other two proteins, Importin β-1 and Ran, are well known for their roles in nuclear transport and can also bind to multiple nucleoporins (43) to regulate the assembly and function of the NPC and NE (35). These results strongly suggest the presence of Epac1 within a protein complex at the nuclear pore.
TABLE 1.
Mass spectrometry analysis of the proteins associating with Epac1
| Protein identified | Mol mass (kDa)a | No. of assigned spectra (pcDNA3 cells/flag-Epac1 cells) |
|---|---|---|
| RanBP2 | 358 | 0/125 |
| Nup205 | 228 | 0/5 |
| Nup98 | 98 | 0/2 |
| Epac1 | 104 | 0/261 |
| Importin β-1 | 97 | 0/20 |
| Ran | 24 | 0/11 |
See Materials and Methods for details.
Stable localization of Epac1 to the NPC.
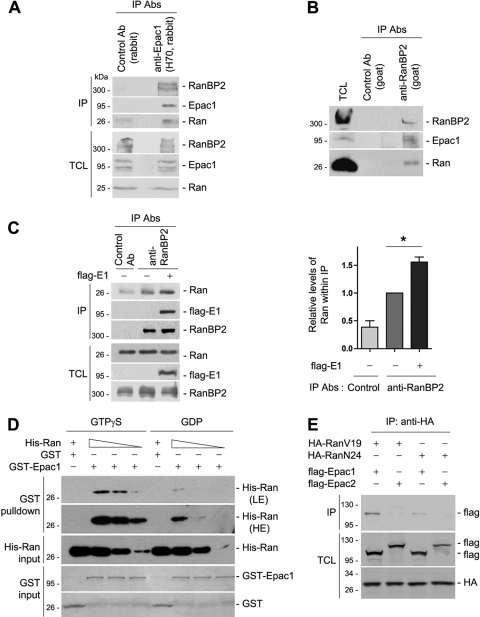
Ran and RanBP2 were the two most represented proteins identified in our analysis after adjustment for their molecular weights. We first examined their abilities to interact with Epac1 in HEK293 cells. RanBP2 and Ran were present within the immunoprecipitate of endogenous Epac1 (Fig. 1 A), and reciprocally, Epac1 and Ran were recovered within the endogenous RanBP2 IP (Fig. 1B). Interestingly, overexpression of Epac1 increased the amount of Ran associated with RanBP2 (Fig. 1C), suggesting that Epac1 may stabilize the interaction between Ran and RanBP2. Because RanBP2 preferentially binds to Ran-GTP (50, 51), Epac1 may actually stabilize Ran in its GTP-loaded form. Therefore, we examined whether Ran and Epac1 interact in a GTP-dependent manner. In a GST pulldown assay in vitro, purified GST-Epac1 bound to GTPγS-loaded His-Ran in a dose-dependent manner, while GST-Epac1 interacted with GDP-loaded His-Ran poorly (Fig. 1D), suggesting a direct role for Ran-GTP in linking Epac1 to RanBP2. To confirm the GTP-dependent binding of Ran to Epac1 in vivo, we used RanV19, a constantly GTP loaded mutant, and RanN24, a constantly GDP loaded mutant. RanV19 bound to Epac1 much better than did RanN24, and neither bound to Epac2 (Fig. 1E).
FIG. 1.
Association of Epac1 with Ran and RanBP2. (A) Coimmunoprecipitation (IP) of endogenous Epac1 with Ran and RanBP2 in HEK293 cells. Cell lysates were subjected to IP using anti-Epac1 (H70) and unrelated rabbit IgG as a control. Western blotting was performed using anti-RanBP2 (Novus), anti-Epac1 (H70), and anti-Ran antibodies (Abs). TCL, total-cell lysates. (B) Coimmunoprecipitation of endogenous RanBP2 with Epac1 and Ran in HEK293 cells. Cell lysates were subjected to IP using an anti-RanBP2 antibody (goat) and unrelated goat IgG as a control. The presence of RanBP2, Ran, and Epac1 within the IP was analyzed by Western blotting using anti-RanBP2 (Novus), anti-Epac1 (A5), and anti-Ran. (C) Effect of Epac1 overexpression on the association of RanBP2 and Ran. IP with anti-RanBP2 was performed as for panel B in the presence (+) or absence (−) of Flag-Epac1 (E1), and the presence of Ran, RanBP2, and Flag-Epac1 within the IP was determined by Western blotting. TCL were also determined by Western blotting. The left panel shows one representative result, and the right panel shows the quantification of the results of five independent experiments normalized to the level of Ran seen in the RanBP2 IP in the absence of transfected Epac1 (means ± standard errors of the means; *, P < 0.05). (D) GTP-dependent interaction between Epac1 and Ran in vitro. Increasing amounts of His-Ran loaded with GTPγS or GDP were incubated with GST or GST-Epac1 and were detected by Western blotting using anti-Ran. LE and HE, low and high exposures. GST and GST-Epac1 levels were shown with Coomassie blue. (E) Association of Ran with Epac1 but not Epac2. HA-RanV19 or HA-RanN24 was coexpressed with Flag-tagged Epac1 or Epac2 in HEK293 cells. IP was performed using anti-HA, and Western blotting was performed using anti-Flag and anti-HA. Data shown are representative of at least three independent experiments.
GFP-Epac1 formed a punctuate rim around the nucleus and colocalized with the staining with MAb414, an antibody recognizing the NPC (Fig. 2 A). In contrast, GFP-Epac2 was distributed diffusely in the cytoplasm and overlapped poorly with the NPC (Fig. 2B). Importantly, staining of the endogenous Epac1 in MEL-24 cells, which express high levels of Epac1 (2), colocalized with MAb414, and was similar to the pattern seen with GFP-Epac1, although additional staining was seen within the nucleus (Fig. 2C). Imaging of GFP-Epac1 in living cells significantly improved the quality of acquired images (Fig. 2D), probably because the NPC and the shape of the nucleus are better preserved under these conditions. We quantified the fluorescence intensity of GFP-Epac1 at the perinuclear rim and found it to be 3-fold the intensity at the cytoplasm (see Materials and Methods) (Fig. 2E).
The perinuclear localization of GFP-Epac1 remained largely unchanged after treatment with the Epac-specific agonist 2OMe (Fig. 2F). Translocation of GFP-Epac1 from the cytoplasm to the PM in response to 2OMe was detected in cells expressing higher levels of GFP-Epac1, consistent with a recent study (40).
Role of the RA domain in Ran-Epac1 association.
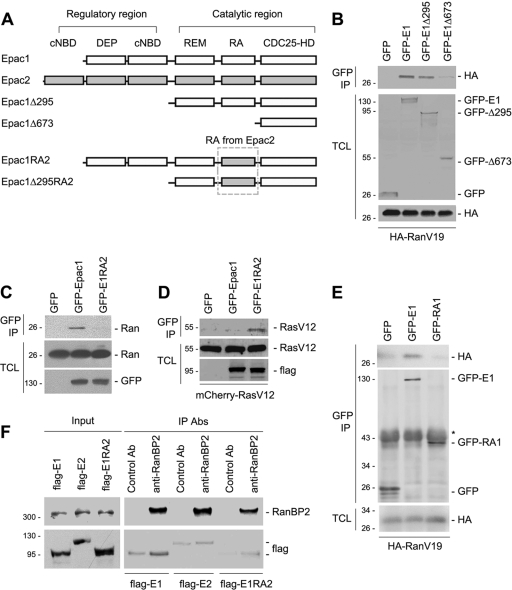
Epac1 and Epac2 are each composed of a regulatory region and a catalytic region, as depicted in Fig. 3A. To determine the domains of Epac1 that bind to Ran, we constructed a series of truncations of Epac1 (Fig. 3A). Deletion of the regulatory region of Epac1 (residues 1 to 295) did not affect the interaction between Epac1Δ295 and RanV19. Further deletion of residues 296 to 673, including the putative RA domain of Epac1 (RA1), disrupted the binding between Epac1Δ673 and RanV19, suggesting a potential Ran binding site within this region (Fig. 3B).
FIG. 3.
Role of the RA domain in the association of Ran and Epac1. (A) Domain structures of Epac1, Epac2, and the mutants used in the study. cNBD, cyclic-nucleotide-binding domain; DEP, Dishevelled, Egl-10, pleckstrin domain; REM, Ras exchange motif; RA, Ras association domain; CDC25-HD, CDC25-homology domain. Epac1 has one and Epac2 has two cNBDs. (B) Loss of Ran association with Epac1Δ673. HA-RanV19 was coexpressed either with GFP alone or with GFP tagged Epac1 (E1), E1Δ295, or E1Δ673 in HEK293 cells. Immunoprecipitation (IP) was performed using anti-GFP, and Western blotting was performed using anti-GFP and anti-HA. TCL, total-cell lysates. (C) Requirement of the RA domain of Epac1 for Ran-Epac1 association. GFP, GFP-tagged Epac1, and GFP-tagged Epac1RA2 (E1RA2) were expressed in HEK293 cells. IP was performed using anti-GFP, and Western blotting was performed using anti-Ran and anti-GFP. (D) Interaction of E1RA2 and RasV12. pcDNA3, Flag-Epac1, or E1RA2 was coexpressed with mCherry-RasV12 in HEK293 cells. IP was performed using an anti-Flag antibody, and proteins were detected by Western blotting using anti-Ras and anti-Flag. (E) The Epac1 RA domain (RA1) requires additional sequences to bind RanV19. GFP, GFP-tagged Epac1 (E1), or the GFP-tagged RA1 domain alone was cotransfected with HA-RanV19 into HEK293 cells, followed by IP with anti-GFP and Western blotting using anti-HA and anti-GFP. (F) The association of Epac1 and RanBP2 requires an intact RA1 domain of Epac1. HEK293 cells were transfected with Flag-tagged Epac1, Epac2, or Epac1RA2, as indicated, and were subjected to IP using a control antibody (Ab) or anti-RanBP2. (Top) Levels of RanBP2 within the input and IP. (Bottom) Levels of Flag-tagged Epac1, Epac2, and Epac1RA2 within the IP.
To examine whether Ran interacted with the RA1 domain, we generated a chimera (Epac1RA2) by swapping the RA1 domain for the analogous RA domain from Epac2 (RA2), as shown in Fig. 3A. Because the overall conformation should be preserved in Epac1RA2, any impaired binding of this mutant could be attributed to the loss of the RA1 domain. GFP-Epac1RA2 lost the ability to interact with endogenous Ran (Fig. 3C) but acquired the ability to bind to RasV12, a constitutively GTP loaded mutant (Fig. 3D). However, when expressed alone, the RA1 domain was unable to bind to Ran (Fig. 3E), suggesting that additional regions elsewhere in Epac1 were needed to stabilize RA1-Ran binding. Precedents for such bipartite interactions include the binding of RACK1 to cAMP phosphodiesterase-4D5 (5). Like the Ran-Epac1 interaction, the RanBP2-Epac1 interaction also required the RA1 domain of Epac1. In Fig. 3F, we show that RanBP2 binding to Epac1 was robust for Epac1, but the binding was greatly reduced for Epac2 and Epac1RA2, both of which show no interaction with Ran-GTP. These results support a model in which the interaction of Epac1 and RanBP2 is, at least partially, mediated by Ran-GTP.
To determine whether the Epac1 RA domain participates in the localization of Epac1 at the NPC, we compared the distributions of GFP-Epac1 and GFP-Epac1RA2. The fluorescence intensity of GFP-Epac1RA2 on the perinuclear rim was significantly lower than that of GFP-Epac1 (Fig. 4A, B, and E). Interestingly, GFP-Epac1Δ295 exhibited decreased fluorescence at the perinuclear rim as well (Fig. 4C and E), suggesting that the regulatory region also contributes to the localization of Epac1, perhaps by interacting with RanBP2 directly or with other proteins within the NPC. This might account for the residual level of GFP-Epac1RA2 seen at the nuclear pore. Indeed, the mutant lacking both the regulatory region and RA1 (GFP-Epac1Δ295RA2) was not detected at the nuclear pore (Fig. 4D and E). Consistent with these findings, the PM-targeted chimera RanV19-CAAX was not sufficient to redistribute GFP-Epac1 to the PM (data not shown), suggesting that, in addition to Ran binding, other protein interactions within the NPC, including RanBP2, are required for localizing Epac1 at that locale. In addition, both GFP-Epac1RA2 and GFP-Epac1Δ295RA2 were excluded from the nucleus (Fig. 4B, D, and E), supporting a role for RA1 in the localization of Epac1 within the nucleus.
Ran-Epac1 interaction is crucial for Epac1-mediated Rap1 activation.
The anchoring of Epac1 to the NPC may activate a specific pool of Rap1. Indeed, the cAMP-dependent activation of Rap1 via Epac1 was significantly stronger than that seen with Epac1RA2 (Fig. 5A). The loss of Rap1 activation by Epac1RA2 was not due to a structural defect caused by swapping RA domains per se, because Epac1RA2 could be fully activated in the presence of RasV12 (Fig. 5A). This is comparable to the enhancement of Epac2-dependent Rap1 activation by RasV12 (28) and may reflect the ability of RA2 to target Epac1RA2 to the PM. This strong dependence of Rap1 activation on the RA domain of Epac1 was not seen with Rap2 (Fig. 5A), in part because of the high basal activity of Rap2, as previously reported (38).
A similar dependence on the RA1 domain for Rap1 activation was seen in experiments using Epac1Δ295. This truncation construct lacks the regulatory region, making it constitutively active. Therefore, although the localization of Epac1Δ295 at the NPC was reduced (Fig. 4C), it could still activate Rap1 robustly (Fig. 5B). Like Epac1RA2, Epac1Δ295RA2 was incapable of activating Rap1, which could be rescued following expression of RasV12 (Fig. 5B). These results suggest that the Ran-Epac1 interaction is necessary for efficient Rap1 activation by Epac1.
To test whether the lack of Rap1 activation by Epac1RA2 was due to loss of localization at the NPC, we artificially tethered Epac1RA2 to the NPC by attaching it to RanV19, which is localized to the nuclear pore (22, 29) (Fig. 5C). This chimera is depicted in Fig. 5D. As expected, the resultant construct, GFP-RanV19-Epac1RA2, was highly enriched at the nuclear pore (Fig. 5C). Importantly, its ability to activate Rap1 was also significantly enhanced over that of Epac1RA2 (Fig. 5E). These data support a model in which the localization of Epac1 to the nuclear pore enhances the activation of Rap1 by Epac1.
Epac1 activates Rap1 on the nuclear envelope.
Activation of endogenous Rap (Rap1 and Rap2) can be directly visualized in cells using a Ras binding domain from RalGDS linked to GFP (referred to below as GFP-RBDRalGDS) (3). In cells expressing mCherry-Epac1 and treated with 2OMe, the level of GFP-RBDRalGDS decreased within the cytoplasm and nucleus, while it increased at the NE and PM (Fig. 6 A).
FIG. 6.
Activation of endogenous Rap at the nuclear envelope (NE). (A) Dynamic activation of Rap by Epac1 at the NE. HEK293 cells were cotransfected with GFP-RBDRalGDS and either mCherry-vector (Vector), mCherry-Epac1 (Epac1), or mCherry-Epac1-CAAX (Epac1-CAAX). The cells were treated with 2OMe after serum starvation. Confocal images acquired at the indicated time points are shown (representative of 11 cells for each condition from three independent experiments). Arrowheads indicate the NE. (B) Line scans were performed for the stack of time lapse images at the indicated region of interest (yellow box in panel A, center row, leftmost panel) and were plotted as a colored surface using MatLab. The color scale and the z axis indicate the relative intensities. The x and y axes indicate time (in minutes) and pixel coordinates, respectively. (C) Subcellular localization of Rap activation by Epac1 mutants. mCherry (i), mCherry-tagged Epac1Δ295 (ii), Epac1Δ295RA2 (iii), or RanV19-Epac1Δ295RA2 (iv) was cotransfected with GFP-RBDRalGDS into HEK293 cells. (Top row) GFP; (center row) mCherry; (bottom row) merged images. All images presented were from confocal live imaging and are representative of cells examined in two to three independent experiments (number of cells examined in each condition > 100). Bars, 10 μm.
The kinetics of the redistribution of GFP-RBDRalGDS within the cell is represented in graphic form as well (Fig. 6B), showing that stimulation with 2OMe triggers the translocation of GFP-RBDRalGDS to both the NE and the PM over a similar time course, with maximal levels reached at both sites in less than 9 min. To ask whether Epac1 localization at the NE was required for Rap activation at the NE, we tethered Epac1 to the PM using the chimera Epac1-CAAX (49). Activation of Epac1-CAAX by 2OMe increased GFP-RBDRalGDS levels at the PM, but not at the NE (Fig. 6A, lower panels). These data also demonstrate that the distinct pools of Rap at the NE and the PM can be activated independently of each other. In the absence of transfected Epac1, translocation of GFP-RBDRalGDS was not observed, possibly because the levels of endogenous Epac1 and Rap1 were too low to redirect GFP-RBDRalGDS upon 2OMe stimulation (Fig. 6A).
Enrichment of GFP-RBDRalGDS at the NE was also seen upon transfection of mCherry-Epac1Δ295 but not mCherry-Epac1Δ295RA2 (Fig. 6Cii and iii, top panels). This correlated with the presence of Epac1Δ295 and the absence of Epac1Δ295RA2 at the NPC, respectively (Fig. 6Cii and iii, center panels). Together, these results strengthen the case for a role for RA1 in promoting Rap activation at the NE. Epac1Δ295RA2 could be redirected to the NPC by the addition of RanV19 at the N terminus (mCherry-RanV19-Epac1Δ295RA2) (Fig. 6Civ, center panel). Importantly, mCherry-RanV19-Epac1Δ295RA2 restored the pattern of GFP-RBDRalGDS on the NE (Fig. 6Civ, top panel). We noted that although Epac1Δ295 is not present at the PM (40), it enriched GFP-RBDRalGDS at the PM. This might reflect the trafficking of Rap to the PM after its activation (4).
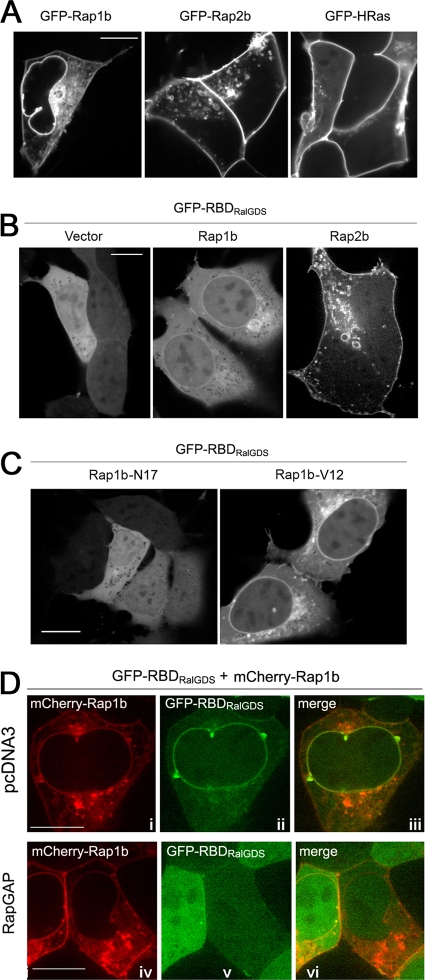
We detected Rap1 on the NE and PM using a GFP-tagged isoform, Rap1b (Fig. 7 A). This pattern is consistent with its localization in COS-1, MDCK, and Jurkat T cells (4). GFP-Rap2b, a Rap2 isoform, was present on the PM but was not seen on the NE (Fig. 7A). The C terminus of Rap2b is similar to that of HRas, which, like Rap2b, was targeted to the PM but not the NE (Fig. 7A).
FIG. 7.
Localization and activation of Rap1b at the NE. (A) Localization of GFP-tagged Rap1b, Rap2b, and H-Ras expressed in HEK293 cells, as indicated. All photos presented were from confocal live imaging. Bars, 10 μm. (B) Distribution of GFP-RBDRalGDS in HEK293 cells cotransfected with pcDNA3, Flag-Rap1b, or Flag-Rap2b, as indicated. (C) GTP dependency of the enrichment of GFP-RBDRalGDS on the NE in HEK293 cells cotransfected with GFP-RBDRalGDS and either Rap1b-N17 or Rap1b-V12, as indicated. (D) Inactivation of Rap1b on the NE by RapGAP. GFP-RBDRalGDS and mCherry-Rap1b were cotransfected into HEK293 cells with pcDNA3 (i to iii) or RapGAP (iv to vi), as indicated. (i and iv) mCherry; (ii and v) GFP; (iii and vi) merged images. All the images are representative of cells examined in two to three independent experiments (number of cells examined in each condition > 100).
Because the GFP-RBDRalGDS reporter does not discriminate between Rap isoforms, isoform-specific activation can be evaluated only following transfection of individual isoforms (3). Ectopic expression of Rap isoforms may also favor the detection of low levels of GEF activity under basal conditions. Indeed, following transfection, Rap1b and Rap2b appeared to be principally activated at the NE and PM, respectively, in the absence of 2OMe (Fig. 7B). For Rap1b, the enrichment of GFP-RBDRalGDS at the NE was GTP dependent, as this pattern was reproduced with Rap1b-V12, which was constitutively GTP loaded, but not with Rap1b-N17, which was constitutively GDP loaded (Fig. 7C). Importantly, the signal of GFP-RBDRalGDS on the NE in the presence of wild-type Rap1b was completely abolished by RapGAP (Fig. 7D), confirming that the distribution of GFP-RBDRalGDS on the NE reflected Rap1b activation.
To ask whether endogenous Epac1 could be responsible for the activation of Rap1b at the NE, we utilized siRNA for Epac1, which reduced the level of endogenous Epac1 by 75% (Fig. 8A), and was selective for Epac1 (Fig. 8B). Among cells cotransfected with scrambled siRNA, 79% had enrichment of GFP-RBDRalGDS on the NE in the presence of mCherry-Rap1b, whereas among cells cotransfected with Epac1 siRNA, 41% showed enrichment of GFP-RBDRalGDS on the NE in the presence of mCherry-Rap1b (Fig. 8C and D). The relatively modest reduction might reflect either incomplete depletion of endogenous Epac1 by siRNA or the presence of other Rap exchangers at the NE. Epac1 siRNA had no effect on the pattern of Rap2b activation (Fig. 8E). Taken together, the data confirm that endogenous Epac1 contributes to the activation of Rap1b at the NE.
DISCUSSION
The perinuclear localization of Epac1 has been reported previously (12, 32, 41, 49), but a mechanism for the anchoring of Epac1 to this location has not been established. Binding of Epac1 to the A-kinase anchoring protein mAKAP has been proposed to localize Epac1 to the perinuclear region in cardiomyocytes (12). However, this localization of Epac1 is also seen in cells that do not express mAKAP, so another mechanism must exist in noncardiac cells. Here, using a proteomic approach, we have identified five potential binding partners that are localized to the nuclear pore complex and are expressed in all cells: RanBP2, Nup205, Nup98, Importin β-1, and Ran. Among these proteins, Ran and RanBP2 were the most represented in this screen. We confirmed that endogenous Epac1 could be coimmunoprecipitated with endogenous RanBP2 and Ran. The binding of transfected Epac1 to Ran is GTP dependent and requires the Ras association (RA1) domain. This is the first characterization of a binding partner for this putative RA1 domain.
RanBP2 has four binding domains for GTP-bound Ran. It is possible that Epac1 binds to Ran-GTP directly and associates with RanBP2 as a consequence of binding to Ran. Three lines of evidence support this model that Ran-GTP binds both Epac1 and RanBP2 in a single complex. One, the expression of Epac1 can increase the level of Ran bound to RanBP2. Two, the maximal localization of Epac1 to the nuclear pore requires the RA1 domain, and this is decreased when this domain is replaced with the RA domain of Epac2 (Epac1RA2). Because a low level of binding of GFP-Epac1RA2 can be detected at the nuclear pore, it is possible that direct interactions between Epac1 and RanBP2 or other components of the NPC may also participate in this targeting of Epac1 (Fig. 8F). Three, Epac1 associates with RanBP2 in vivo, and at a higher level than Epac1RA2, as judged by coprecipitation within RanBP2 pulldowns (Fig. 3F). Importin-β1 was also identified as a binding partner for Epac1. Importin-β1 can bind to multiple nucleoporins, including RanBP2, as well as Ran (9, 11, 19, 48). Therefore, its association with Epac1 could be indirect. Whether Epac1 is a cargo for Importin-β is not known.
The association of Epac1 with Ran and RanBP2 represents a new mechanism of targeting proteins to the nuclear pore. This anchoring of Epac1 is not regulated by cAMP and is seen within the unstimulated cell. Although the binding is constitutive, it does require Ran-GTP. Ran is efficiently GTP loaded within the nucleus through the action of the Ran exchanger RCC1, which is exclusively nuclear (36). This asymmetry sets up the Ran gradient that drives nuclear trafficking (30). Ran-GTP binds RanBP2 at four potential Ran binding sites (50, 51). RanGAP, which promotes the conversion of Ran-GTP to Ran-GDP, also binds to RanBP2 (33). However, increasing the level of RanGAP bound to RanBP2 by transfection did not decrease the level of Ran associating with RanBP2, nor did it disrupt the localization of Epac1 at the NPC (data not shown). We propose that a pool of Ran-GTP that is stably present at the NPC contributes to the constitutive anchoring of Epac1 at this site. Epac1 binding to Ran-GTP/RanBP2 may also protect additional Ran-GTP from inactivation by RanGAP. It is not known whether this ability of Epac1 to regulate the association of Ran-GTP with RanBP2 affects Ran-mediated nuclear transport.
In addition, Epac1 was seen within the nucleus, and the RA domain of Epac1 was critical for this localization. It has been reported recently that blocking nuclear export using LMB increased the nuclear accumulation of Epac1 (21). We observed a modest LMB-dependent increase in the nuclear accumulation of Epac1, but not in that of of Epac1RA2 (data not shown), supporting the idea that the RA1 domain of Epac1 also plays a role in the nuclear localization of Epac1. Ran and RanBP2 have important functions at the chromosome and the mitotic spindle (6, 7, 18, 22, 44), and it is possible that they exist in a nuclear complex with Epac1.
The use of GFP-RBDRalGDS as a reporter for Rap activation is well suited for identifying large pools of Rap-GTP activated by overexpressed GEFs or constitutively active mutants of Rap itself (3). This reporter does not discriminate between Rap isoforms, and cotransfection of specific Rap isoforms is required for the examination of isoform-specific activation. Following transfection of Rap1b (but not of Rap2b), we showed that GFP-RBDRalGDS identified a pool of Rap1 at the nuclear envelope that was activated by endogenous Epac1.
It is possible that a portion of the Rap1-GTP that is activated at the nuclear envelope may translocate to other compartments, including the PM. Although a recent study showed that Epac1 itself could translocate to the PM upon cAMP stimulation to activate Rap1 and affect cell adhesion, this action requires an intact DEP (Dishevelled, Egl-10, pleckstrin) domain (40). The constitutively active truncation construct of Epac1 used in our study lacks the DEP domain. It is located at the nuclear pore and is incapable of translocating to the PM. Although this mutant predominantly increased the levels of Rap1-GTP detected on the nuclear envelope, it also increased the levels of GFP-RBDRalGDS that translocated to the PM. This is consistent with the possibility that Rap1-GTP can translocate to the PM following its activation on the nuclear envelope. Whether this pool of translocated Rap1-GTP behaves similarly to Rap1 directly activated at the PM is not known.
Rap1 and Rap2 have overlapping but distinct functions. In neuronal cells, Rap2 appears to be selectively involved in the retraction of axons and dendrites (16). In nonneuronal cells, only Rap1, not Rap2, can antagonize Ras-dependent functions (23). Preferential regulators and effectors of Rap2b have also been proposed (31, 45). The function of the pool of Rap1 at the nuclear envelope needs further study. Only one previous report that specifically linked Rap signaling to the trafficking of nuclear proteins examined the trafficking of DNA-dependent protein kinase (DNA-PK), and identified a requirement for Rap2, but not Rap1 (21). It is possible that Rap1 affects the nuclear/cytoplasmic trafficking of other proteins. Alternatively, Rap1 could activate signaling cascades to affect nuclear transport in general. One established effector of Rap1 is B-Raf, a kinase upstream of the extracellular signal-regulated kinase (ERK) signaling cascade (24, 39, 46). The possibility that Epac1 could activate a localized B-Raf/ERK signaling cascade is particularly attractive, since ERK phosphorylation of nucleoporins has recently been shown to regulate Ran-dependent transport (26). B-Raf is also required for the formation of the mitotic spindle (6) and, like Ran, RanBP2, and Epac1, is also detected at the mitotic spindle and kinetochore in mitotic cells (6, 41, 44). Therefore, it is possible that localization of Epac1 to Ran/RanBP2 may activate a pool of B-Raf at these sites during mitosis.
Our study identifies fundamental differences between the RA domains of Epac1 and Epac2. We propose that these RA domains have distinct effects on the abilities of Epac1 and Epac2 to activate different pools of Rap1. Epac2 is a bona fide Ras effector, acting as a coincidence detector for Ras-dependent signaling and cAMP (27, 28). This is because Epac2 requires proper targeting to Ras-GTP via its Ras association domain (RA2) for efficient Rap1 activation. In contrast, Epac1 is not able to bind Ras-GTP and does not require recruitment to Ras in order to activate Rap1. Instead, Epac1 requires targeting to Ran and Ran-BP2 for efficient Rap1 activation. We have no evidence that Epac1 activity per se is regulated by Ran-GTP. Rather, anchoring brings Epac1 into the proximity of a pool of Rap1 that can now access Epac1 upon cAMP binding.
In our previous study, we demonstrated the requirement of Ras in the Epac2-dependent activation of Rap1 by introducing a point mutation into its RA domain, which abolished the Ras binding without affecting the catalytic function of Epac2 (28). This mutation caused a charge reversal within the RA domain by changing a lysine to glutamate. Epac1 contains a glutamate at this position (27), explaining the inability of Epac1 to bind to Ras-GTP. Although we did not identify a specific point mutation in Epac1 that is able to block Ran binding, we selectively interfered with Ran binding by swapping the RA domain of Epac1 for that of Epac2. This chimera (Epac1RA2) showed a dramatically reduced level of Rap1 activation that was not due to a loss of catalytic function per se, as the activity of this chimera could be restored by targeting it to Ras-GTP via the chimeric RA2 domain. The decreased Rap1 activation induced by Epac1RA2 correlated with the decreased localization of Epac1RA2 to the nuclear pore, and it could be rescued by redirecting the chimera to the nuclear pore, by linking Epac1RA2 to RanV19. These results and our previous studies of Epac2 suggest that the differential localizations of Epac1 and Epac2 are dictated by their unique RA domains and are necessary for their abilities to activate different pools of Rap1 efficiently in cells.
The ability of one class of small G proteins to signal to other classes of small G proteins promotes cross talk and integration among signaling pathways (34). The presence of both an RA domain and a CDC25 homology domain within a single GEF protein provides one such mechanism. For example, the GEFs Tiam1, Ral-GDS, and PLC-ɛ each contain an RA domain that recruits that GEF to a specific activated small G protein and a CDC25 homology domain that promotes the GTP loading of another small G protein (34). The coupling of Ras to Rap1 by Epac2 (27, 28) and the coupling of Ran to Rap1 by Epac1 also fit into this general mode of signaling. These targeting mechanisms highlight the importance of RA domains of diverse GEFs in coupling small G proteins to each other. While many of the previous examples of RA domains have identified the coupling of small G proteins within the Ras family, this study showing the connection between Ran and Rap1 via Epac1 represents the first example of the regulation of a member of the Ras family by the small G protein Ran.
In conclusion, our study has identified a novel role of Ran in anchoring Epac1 at the nuclear pore and has characterized NE as a novel intracellular site of Rap1 activation (Fig. 8F). This anchoring of Epac1 is mediated partly by its RA domain and is required for the efficient activation of Rap1 by Epac1. Due to this unique localization of Epac1, we propose that Epac1 functions as a cAMP sensor at the nuclear pore, which converts local cAMP elevations into Rap1 activation on the NE. The discovery of Epac1-dependent activation of Rap1 at the NE is likely to reveal a potential role of cAMP and Rap1 in nuclear transport through the nuclear pore and in the assembly and functioning of the NE.
Acknowledgments
We thank Zhiping Wang, Mike Forte, Mihail Iordanov, and Larry David for discussions; John Klimek, Debra McMillen, and Srilatha Tavisala for technical assistance; and Ian Macara and Johannes Bos for sharing of reagents.
This work was funded by NIH NCI grant CA72971 to P.J.S.S., a Vertex Scholarship to C.L., and training grant NIH-5-T32-CA106195-06A1 to Y.L.
Footnotes
Published ahead of print on 14 June 2010.
REFERENCES
- 1.Baillie, G. S. 2009. Compartmentalized signalling: spatial regulation of cAMP by the action of compartmentalized phosphodiesterases. FEBS J. 276:1790-1799. [DOI] [PubMed] [Google Scholar]
- 2.Baljinnyam, E., K. Iwatsubo, R. Kurotani, X. Wang, C. Ulucan, M. Iwatsubo, D. Lagunoff, and Y. Ishikawa. 2009. Epac increases melanoma cell migration by a heparan sulfate-related mechanism. Am. J. Physiol. Cell Physiol. 297:C802-C813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bivona, T. G., and M. R. Philips. 2005. Analysis of Ras and Rap activation in living cells using fluorescent Ras binding domains. Methods Enzymol. 37:138-145. [DOI] [PubMed] [Google Scholar]
- 4.Bivona, T. G., H. H. Wiener, I. M. Ahearn, J. Silletti, V. K. Chiu, and M. R. Philips. 2004. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 164:461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolger, G. B., G. S. Baillie, X. Li, M. J. Lynch, P. Herzyk, A. Mohamed, L. H. Mitchell, A. McCahill, C. Hundsrucker, E. Klussmann, D. R. Adams, and M. D. Houslay. 2006. Scanning peptide array analyses identify overlapping binding sites for the signalling scaffold proteins, beta-arrestin and RACK1, in cAMP-specific phosphodiesterase PDE4D5. Biochem. J. 398:23-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borysova, M. K., Y. Cui, M. Snyder, and T. M. Guadagno. 2008. Knockdown of B-Raf impairs spindle formation and the mitotic checkpoint in human somatic cells. Cell Cycle 7:2894-2901. [DOI] [PubMed] [Google Scholar]
- 7.Chen, T., T. L. Muratore, C. E. Schaner-Tooley, J. Shabanowitz, D. F. Hunt, and I. G. Macara. 2007. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol. 9:596-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, A. E., F. Selheim, J. de Rooij, S. Dremier, F. Schwede, K. K. Dao, A. Martinez, C. Maenhaut, J. L. Bos, H. G. Genieser, and S. O. Doskeland. 2003. cAMP analog mapping of Epac1 and cAMP kinase. Discriminating analogs demonstrate that Epac and cAMP kinase act synergistically to promote PC-12 cell neurite extension. J. Biol. Chem. 278:35394-35402. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, P. R., and C. Zhang. 2004. Spatial and temporal control of nuclear envelope assembly by Ran GTPase. Symp. Soc. Exp. Biol. 2004:193-204. [PubMed] [Google Scholar]
- 10.Dao, K. K., K. Teigen, R. Kopperud, E. Hodneland, F. Schwede, A. E. Christensen, A. Martinez, and S. O. Doskeland. 2006. Epac1 and cAMP-dependent protein kinase holoenzyme have similar cAMP affinity, but their cAMP domains have distinct structural features and cyclic nucleotide recognition. J. Biol. Chem. 281:21500-21511. [DOI] [PubMed] [Google Scholar]
- 11.Delphin, C., T. Guan, F. Melchior, and L. Gerace. 1997. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol. Biol. Cell 8:2379-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodge-Kafka, K. L., J. Soughayer, G. C. Pare, J. J. Carlisle Michel, L. K. Langeberg, M. S. Kapiloff, and J. D. Scott. 2005. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature 437:574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrhardt, A., G. Ehrhardt, X. Guo, and J. Schrader. 2002. Ras and relatives—job sharing and networking keep an old family together. Exp. Hematol. 30:1089. [DOI] [PubMed] [Google Scholar]
- 14.Fahrenkrog, B., and U. Aebi. 2003. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 4:757-766. [DOI] [PubMed] [Google Scholar]
- 15.Franke, B., J.-W. Akkerman, and J. L. Bos. 1997. Rapid Ca2+-mediated activation of rap1 in human platelets. EMBO J. 16:252-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu, Z., S. H. Lee, A. Simonetta, J. Hansen, M. Sheng, and D. T. Pak. 2007. Differential roles of Rap1 and Rap2 small GTPases in neurite retraction and synapse elimination in hippocampal spiny neurons. J. Neurochem. 100:118-131. [DOI] [PubMed] [Google Scholar]
- 17.Grandoch, M., S. S. Roscioni, and M. Schmidt. 2010. The role of Epac proteins, novel cAMP mediators, in the regulation of immune, lung and neuronal function. Br. J. Pharmacol. 159:265-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao, Y., and I. G. Macara. 2008. Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J. Cell Biol. 182:827-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harel, A., and D. J. Forbes. 2004. Importin beta: conducting a much larger cellular symphony. Mol. Cell 16:319-330. [DOI] [PubMed] [Google Scholar]
- 20.Houslay, M. D. 2010. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem. Sci. 35:91-100. [DOI] [PubMed] [Google Scholar]
- 21.Huston, E., M. J. Lynch, A. Mohamed, D. M. Collins, E. V. Hill, R. MacLeod, E. Krause, G. S. Baillie, and M. D. Houslay. 2008. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc. Nat. Acad. Sci. U. S. A. 105:12791-12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchins, J. R., W. J. Moore, and P. R. Clarke. 2009. Dynamic localisation of Ran GTPase during the cell cycle. BMC Cell Biol. 10:66-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimenez, B., V. Pizon, I. Lerosey, F. Béranger, A. Tavitian, and J. DeGunzburg. 1991. Effects of the ras-related rap2 protein on cellular proliferation. Int. J. Cancer 49:471-479. [DOI] [PubMed] [Google Scholar]
- 24.Kao, S., R. K. Jaiswal, W. Kolch, and G. E. Landreth. 2001. Identification of the mechanisms regulating the differential activation of the MAPK cascade by epidermal growth factor and nerve growth factor in PC12 cells. J. Biol. Chem. 276:18169-18177. [DOI] [PubMed] [Google Scholar]
- 25.Keller, A., A. I. Nesvizhskii, E. Kolker, and R. Aebersold. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383-5392. [DOI] [PubMed] [Google Scholar]
- 26.Kosako, H., N. Yamaguchi, C. Aranami, M. Ushiyama, S. Kose, N. Imamoto, H. Taniguchi, E. Nishida, and S. Hattori. 2009. Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat. Struct. Mol. Biol. 16:1026-1035. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., S. Asuri, J. F. Rebhun, A. F. Castro, N. C. Paranavitana, and L. A. Quilliam. 2006. The RAP1 guanine nucleotide exchange factor Epac2 couples cyclic AMP and Ras signals at the plasma membrane. J. Biol. Chem. 281:2506-2514. [DOI] [PubMed] [Google Scholar]
- 28.Liu, C., M. Takahashi, Y. Li, S. Song, T. J. Dillon, U. Shinde, and P. J. S. Stork. 2008. Ras is required for the cyclic AMP-dependent activation of Rap1 via Epac2. Mol. Cell. Biol. 28:7109-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lounsbury, K. M., S. A. Richards, K. L. Carey, and I. G. Macara. 1996. Mutations within the Ran/TC4 GTPase. Effects on regulatory factor interactions and subcellular localization. J. Biol. Chem. 271:32834-32841. [DOI] [PubMed] [Google Scholar]
- 30.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machida, N., M. Umikawa, K. Takei, N. Sakima, B. E. Myagmar, K. Taira, H. Uezato, Y. Ogawa, and K. Kariya. 2004. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J. Biol. Chem. 279:15711-15714. [DOI] [PubMed] [Google Scholar]
- 32.Magiera, M. M., M. Gupta, C. J. Rundell, N. Satish, I. Ernens, and S. J. Yarwood. 2004. Exchange protein directly activated by cAMP (EPAC) interacts with the light chain (LC) 2 of MAP1A. Biochem. J. 382:803-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 34.Mitin, N., K. L. Rossman, and C. J. Der. 2005. Signaling interplay in Ras superfamily function. Curr. Biol. 15:R563-R574. [DOI] [PubMed] [Google Scholar]
- 35.Moore, M. S. 1998. Ran and nuclear transport. J. Biol. Chem. 273:22857-22860. [DOI] [PubMed] [Google Scholar]
- 36.Nemergut, M. E., and I. G. Macara. 2000. Nuclear import of the Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol. 149:835-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nesvizhskii, A. I., A. Keller, E. Kolker, and R. Aebersold. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646-4658. [DOI] [PubMed] [Google Scholar]
- 38.Ohba, Y., N. Mochizuki, K. Matsuo, S. Yamashita, M. Nakaya, Y. Hashimoto, M. Hamaguchi, T. Kurata, K. Nagashima, and M. Matsuda. 2000. Rap2 as a slowly responding molecular switch in the Rap1 signaling cascade. Mol. Cell. Biol. 20:6074-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohtsuka, T., K. Shimizu, B. Yamamori, S. Kuroda, and Y. Takai. 1996. Activation of brain B-Raf protein kinase by Rap1B small GTP-binding protein. J. Biol. Chem. 271:1258-1261. [DOI] [PubMed] [Google Scholar]
- 40.Ponsioen, B., M. Gloerich, L. Ritsma, H. Rehmann, J. L. Bos, and K. Jalink. 2009. Direct spatial control of Epac1 by cyclic AMP. Mol. Cell. Biol. 29:2521-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiao, J., F. C. Mei, V. L. Popov, L. A. Vergara, and X. Cheng. 2002. Cell cycle-dependent subcellular localization of exchange factor directly activated by cAMP. J. Biol. Chem. 277:26581-26586. [DOI] [PubMed] [Google Scholar]
- 42.Rehmann, H., F. Schwede, S. O. Doskeland, A. Wittinghofer, and J. L. Bos. 2003. Ligand-mediated activation of the cAMP-responsive guanine nucleotide exchange factor Epac. J. Biol. Chem. 278:38548-38556. [DOI] [PubMed] [Google Scholar]
- 43.Rotem, A., R. Gruber, H. Shorer, L. Shaulov, E. Klein, and A. Harel. 2009. Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol. Biol. Cell 20:4031-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salina, D., P. Enarson, J. B. Rattner, and B. Burke. 2003. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162:991-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt, M., S. Evellin, P. A. Weernink, F. von Dorp, H. Rehmann, J. W. Lomasney, and K. H. Jakobs. 2001. A new phospholipase-C-calcium signalling pathway mediated by cyclic AMP and a Rap GTPase. Nat. Cell Biol. 3:1020-1024. [DOI] [PubMed] [Google Scholar]
- 46.Stork, P. J. S. 2003. Does Rap1 deserve a bad Rap? Trends Biochem. Sci. 28:267-275. [DOI] [PubMed] [Google Scholar]
- 47.Terrin, A., G. Di Benedetto, V. Pertegato, Y. F. Cheung, G. Baillie, M. J. Lynch, N. Elvassore, A. Prinz, F. W. Herberg, M. D. Houslay, and M. Zaccolo. 2006. PGE(1) stimulation of HEK293 cells generates multiple contiguous domains with different [cAMP]: role of compartmentalized phosphodiesterases. J. Cell Biol. 175:441-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vetter, I. R., A. Arndt, U. Kutay, D. Gorlich, and A. Wittinghofer. 1999. Structural view of the Ran-Importin beta interaction at 2.3 Å resolution. Cell 97:635-646. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Z., T. J. Dillon, V. Pokala, S. Mishra, K. Labudda, B. Hunter, and P. J. Stork. 2006. Rap1-mediated activation of extracellular signal-regulated kinases by cyclic AMP is dependent on the mode of Rap1 activation. Mol. Cell. Biol. 26:2130-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, J., M. J. Matunis, D. Kraemer, G. Blobel, and E. Coutavas. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270:14209-14213. [DOI] [PubMed] [Google Scholar]
- 51.Yokoyama, N., N. Hayashi, T. Seki, N. Pante, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, U. Aebi, et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376:184-188. [DOI] [PubMed] [Google Scholar]