Abstract
Cytoplasm-to-nucleus translocation of Smad is a fundamental step in transforming growth factor β (TGF-β) signal transduction. Here we identify a subset of nucleoporins that, in conjunction with Msk (Drosophila Imp7/8), specifically mediate activation-induced nuclear translocation of MAD (Drosophila Smad1) but not the constitutive import of proteins harboring a classic nuclear localization signal (cNLS) or the spontaneous nuclear import of Medea (Drosophila Smad4). Surprisingly, many of these nucleoporins, including Sec13, Nup75, Nup93, and Nup205, are scaffold nucleoporins considered important for the overall integrity of the nuclear pore complex (NPC) but not known to have cargo-specific functions. We demonstrate that the roles of these nucleoporins in supporting Smad nuclear import are separate from their previously assigned functions in NPC assembly. Furthermore, we uncovered novel pathway-specific functions of Sec13 and Nup93; both Sec13 and Nup93 are able to preferentially interact with the phosphorylated/activated form of MAD, and Nup93 acts to recruit the importin Msk to the nuclear periphery. These findings, together with the observation that Sec13 and Nup93 could interact directly with Msk, suggest their direct involvement in the nuclear import of MAD. Thus, we have delineated the nucleoporin requirement of MAD nuclear import, reflecting a unique trans-NPC mechanism.
Transforming growth factor β (TGF-β) cytokines critically regulate a diverse array of cellular properties in development and homeostasis, through an evolutionarily conserved mechanism that centers around the Smad transcription factors (18, 34). TGF-β induces phosphorylation of the Smad proteins and consequently drives Smads into the nucleus, ensuring that changes in the gene transcription program are strictly signal dependent (24, 26). Two critical elements in nuclear import are the transport receptors (i.e., karyopherins or importins) and the nuclear pore complex (NPC) (28, 30). We recently identified Imp7/8 as the importin for TGF-β-activated Smads, but how the Imp7/8-Smad complex translocates through the NPC has yet to be elucidated (36). The NPC consists of more than 30 evolutionarily conserved nucleoporins, each with a particular localization within the NPC based on biochemical, biophysical, electron microscopy, and computational studies (2, 3, 25). Many nucleoporins contain repeats of phenylalanine-glycine (FG), and the highly hydrophobic and unstructured FG domains occupy the central tunnel of the NPC, constituting a gating mechanism that restricts the movement of macromolecules through the NPC (19). In vivo genetic studies have suggested redundancy among FG domains in maintaining the permeability barrier as well as nuclear import (29, 38). On the other hand, many of the non-FG nucleoporins assemble into subcomplexes within the NPC, most prominently the Nup107-160 (consisting of Nup107, Nup133, Nup75, Sec13, and Seh1, etc.) and Nup53-93 (containing Nup53, Nup93, and Nup205, etc.) complexes, and they are believed to serve mostly as scaffolds for NPC assembly or anchoring to the nuclear envelope (11, 13, 14, 27, 32). How importins and the NPC function in a concerted manner in nuclear import is still highly debatable. Importins are capable of direct interaction with FG-nucleoporins, and this provides the foundation for current models of nuclear import (6, 20, 22, 28, 30). However, whether non-FG nucleoporins participate directly in the nuclear translocation process remained unknown.
Emerging evidence suggests specificity in NPC function. Genetic studies of yeast revealed redundancy among FG-nucleoporins but also showed that a subset of FG-nucleoporins are differentially employed by different importins (29, 31). Mice with a deletion in the non-FG nucleoporin gene nup133 or nup155 developed distinct phenotypes affecting specific cell lineages, arguing for specific functions of these scaffold nucleoporins, although it is unclear whether the phenotypes were due to defects in the nuclear transport of particular cargoes or to something else (16, 39).
Recently, we identified Msk (Imp7 and Imp8 in mammals) as the importin for signal-activated Smads in both Drosophila and mammalian cells (36). In mammalian cells, depletion of Imp7/8 markedly reduced the nuclear import of Smad1, -2, -3, and 4 in response to TGF-β/bone morphogenetic protein (BMP), and overexpression of Imp8 was sufficient to drive the nuclear import of Smad1, -3, and -4 without the need for TGF-β/BMP stimulation. Smad4 and its Drosophila counterpart Medea can enter the nucleus without TGF-β/BMP when cells are treated with the CRM-1 inhibitor leptomycin B (LMB) (21, 33). Interestingly, in Drosophila cells, such spontaneous nuclear import of Medea appears to be independent of either Msk or Impβ (37). Imp7/8 and the classic nuclear localization signal (cNLS)-cargo importin Impβ diverge in primary sequences, while their higher-order structures are believed to be similar (17). However, it remained unclear how Imp7/8 transports Smad through the NPC and whether, as with Impβ, the interplay between Imp7/8 and the NPC occurs mainly through the FG-nucleoporins.
In this study, through RNA interference (RNAi) screening of the Drosophila genome, we uncovered a cohort of nucleoporins that are indispensable for nuclear translocation of signal-activated MAD specifically. For the first time, pathway-specific functions of non-FG scaffold nucleoporins in nuclear import are demonstrated. Such specificity in nucleoporin utilization may reflect different demands of constitutive and signal-induced nuclear import events. Thus, our observations demonstrate novel modes of functional and physical interactions between MAD/Msk and the NPC.
MATERIALS AND METHODS
Tissue culture, transfection, and conditional expression of cDNAs.
Drosophila S2 and S2R+ cells were maintained in Schneider's Drosophila medium (Invitrogen) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 U/ml). Transient transfection was carried out using the Effectene reagent according to the manufacturer's protocol (Qiagen). All expression vectors are under the control of a metallothionein promoter, and expression was induced by CuSO4 (at 0.5 mM for 3 h) (5).
Immunofluorescence staining, confocal microscopy, and image quantification.
Cells were seeded onto coverslips and were induced with CuSO4 (0.5 mM, 3 h) to express various green fluorescent protein (GFP)- or FLAG-tagged proteins for evaluation of their subcellular localizations. Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), followed by permeabilization with 0.2% Triton X-100-PBS, and were stained with appropriate antibodies and 4′,6-diamidino-2-phenylindole (DAPI) according to previously published procedures (36). Prolong Gold (Invitrogen) was used for mounting, and the samples were analyzed by confocal microscopy. All confocal images were collected with a laser-scanning microscope (Leica DMIRE2) at the following wavelengths: UV (DAPI), 488 nm (Alexa Fluor 488 or GFP), 543 nm (Alexa Fluor 546 or red fluorescent protein [RFP]/DsRed2), and 633 nm (Alexa Fluor 633). For low- and high-magnification images, 63×/1.40-numerical-aperture and 100×/1.40-numerical-aperture oil immersion objectives were used. The captured images were processed with Leica confocal microscope software.
Confocal images collected at identical settings were analyzed by NIH ImageJ to measure signal intensities from the nucleus and the cytoplasm. The ratio of the nuclear signal to the cytoplasmic signal was calculated. In each case, at least three separate fields (magnification, ×60) were quantified. Since we mostly used stable cell lines, usually there were more than 50 GFP-positive cells in each field. The standard error (SE) was calculated based on the average of the nuclear/cytoplasmic signal ratios from at least three different fields.
RNAi and rescue experiments.
The whole-genome RNAi screening was carried out at the Drosophila RNAi Screening Center (DRSC) at Harvard Medical School (www.flyrnai.org) by following the format described in our previous publication (36). The images were acquired and analyzed by automated confocal microscopy (Evotech Opera high-content screening system; Perkin-Elmer). The nuclear/cytoplasmic signal ratio was measured using algorithms accompanying the Opera system (Acapella). The design of RNAi constructs was based on recommendations by DRSC, and the sequence information is available through the DRSC web page (www.flyrnai.org). DNA amplicons corresponding to different regions of targeted genes were generated by PCR and were used for in vitro transcription (MEGAscript; Applied Biosystems) to synthesize double-stranded RNA (dsRNA). RNAi in S2R+ cells using the soaking method has been described previously (36). In general, 5 μg dsRNAs was used for each well in a 24-well plate, and the RNAi lasted for 4 days. But for Nup153 and Nup358 RNAi, 2.5 μg dsRNAs was used for 3 days in order to minimize cell lethality.
In RNAi rescue experiments, S2R+ cells (0.2 × 106/well in a 24-well plate) were first treated with 5 μg dsRNA targeting the 3′ UTR of either sec13 or nup93 for 2 days and were then transfected with the rescue plasmids (HA-Sec13 and HA-Nup93, driven by the inducible metallothionein promoter) using Effectene. After another 2 days, the cells were induced by CuSO4 (0.5 mM, 3 h) to express GFP-MAD/Punt/Tkv and the rescue constructs and were then processed for immunostaining and imaging.
Nuclear envelope permeability assay.
S2R+ cells were resuspended at 2 × 106 to 4 × 106 cells/ml in ice-cold hypotonic buffer (10 mM HEPES [pH 7.5], 2 mM MgCl2, 25 mM KCl, 2 mM dithiothreitol [DTT], and 200 mM sucrose) supplemented with protease inhibitors. After a 10-min incubation on ice, the cell membrane was disrupted by a Dounce homogenizer (60 strokes, type A pestle). Broken and intact cells were pelleted by centrifugation (1,000 × g, 5 min) and resuspended in transport buffer {20 mM HEPES [pH 7.5], 110 mM potassium acetate [KOAc], 2 mM magnesium acetate [Mg(OAc)2], 5 mM sodium acetate [NaOAc], 0.5 mM EGTA, 250 mM sucrose, 2 mM DTT} containing 0.2 mg/ml 70-kDa Texas red-labeled dextran (Invitrogen), 0.25 mg/ml 500-kDa fluorescein isothiocyanate (FITC)-labeled dextran (Sigma), and 0.2 mg/ml Cascade blue-labeled 3-kDa dextran (Invitrogen). After incubation at room temperature for 5 min, the nuclei were directly imaged without being fixed by confocal microscopy. Only nuclei that were permeable to the 3-kDa dextran (indicating a lysed cell membrane) but not the 500-kDa dextran (suggesting no gross damage to the nuclear envelope during Dounce homogenization) were scored for leakiness toward the 70-kDa dextran.
Protein-protein interactions.
For coimmunoprecipitation, S2 or S2R+ cells were lysed in 50 mM Tris-Cl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, and 2 mM DTT supplemented with protease inhibitors. Cell extracts were incubated with an anti-FLAG or antihemagglutinin (anti-HA) antibody conjugated to agarose beads (Sigma) at 4°C for 16 h, and the beads were washed in the lysis buffer three times before immunoblot analysis.
Recombinant Drosophila Sec13, Nup93, and Msk were produced in Escherichia coli as glutathione S-transferase (GST) fusion proteins and were purified using glutathione-Sepharose 4B beads (GE Healthcare). GST-Msk was digested by thrombin (Novagen) to remove the GST moiety. GST-Sec13 (10 μg) or GST-Nup93 (1 μg) on beads was incubated with recombinant Msk (0.1 μg/μl) at 4°C for 16 h in 20 mM Tris Cl (pH 7.5)-200 mM NaCl-5% glycerol. The beads were washed in the same buffer three times, and the bound proteins were analyzed by immunoblotting. For interaction with MAD, phosphorylated FLAG-MAD was first affinity purified by anti-FLAG agarose beads (Sigma) from stable S2 cells expressing FLAG-MAD and Punt/Tkv. FLAG-MAD was eluted from the anti-FLAG beads by a 3× FLAG peptide in a buffer containing 25 mM Tris Cl (pH 8.0), 250 mM NaCl, 0.05% NP-40, 5% glycerol, and 1 mM DTT. The purified recombinant FLAG-MAD (0.2 μg/μl) in this buffer was incubated at 4°C for 16 h with GST-Sec13 (10 μg) or GST-Nup93 (1 μg) bound to the glutathione-Sepharose beads. The beads were washed 3 times with 20 mM Tris Cl (pH 8.0), 250 mM NaCl, 0.05% NP-40, and 5% glycerol, and the bound proteins were subjected to immunoblot analysis.
For testing of the Nup93-Msk interaction, S2 cells (∼8 × 106) expressing Msk-HA or HA-tagged Msk with a C-terminal deletion (Msk-dC-HA) were purified by an anti-HA affinity matrix (Roche) in 25 mM Tris Cl (pH 8.0), 250 mM NaCl, 0.05% NP-40, 5% glycerol, and 1 mM DTT with protease inhibitors and phosphatase inhibitors. Bound Msk-HA or Msk-dC-HA was then eluted from the beads with HA peptides and was incubated with purified GST-Nup93 (1 μg) on glutathione-Sepharose beads at 4°C for 16 h. The beads were washed three times in the same buffer before immunoblot analysis.
For testing of the effect of nup75 RNAi on the Msk-nucleoporin interaction, S2 cells were transfected with 4 μg dsRNA targeting nup75 using Dharmafect 4 (Thermo Fisher Scientific) and were incubated for 2 days. The same cells were then transfected with the expression vectors using Effectene (Qiagen). Forty-eight hours later, cells were induced with CuSO4 (0.5 mM, 3 h) to express the constructs and were lysed in 20 mM Tris Cl (pH 8.0), 150 mM NaCl, 0.5% NP-40, 5% glycerol, 2 mM DTT, protease inhibitors, and phosphatase inhibitors. The cell extracts were incubated with anti-FLAG-agarose beads (Sigma) at 4°C for 16 h, and the beads were washed in the lysis buffer three times before immunoblot analysis.
Antibodies.
The polyclonal anti-MAD antibody was raised against the N-terminal 160-amino-acid (160-aa) fragment. Rabbit anti-Msk was a gift from L. Perkins (Harvard Medical School), and anti-Sec13 was a gift from B. Fontoura (University of Texas Southwestern Medical School) and W. Hong (IMCB, Singapore). The other antibodies used in the study are anti-phospho-Smad1/5/8 (also recognizing phospho-MAD) (Cell Signaling), MAb414 (Covance), anti-GFP (Zymed), anti-FLAG, and anti-HA (Sigma).
Quantitative real-time RT PCR.
Total RNA was isolated by RNeasy spin columns (Qiagen) and was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was conducted using the iQ SYBR green kit (Bio-Rad), and the threshold values for Rp49 were used as internal standards for quantification.
RESULTS
Identification of nucleoporins required for Msk-mediated nuclear import of MAD.
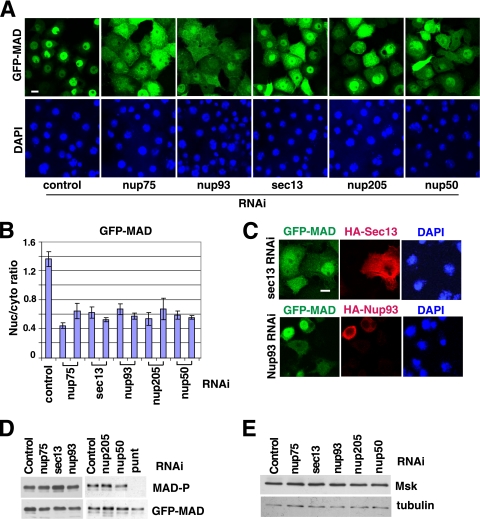
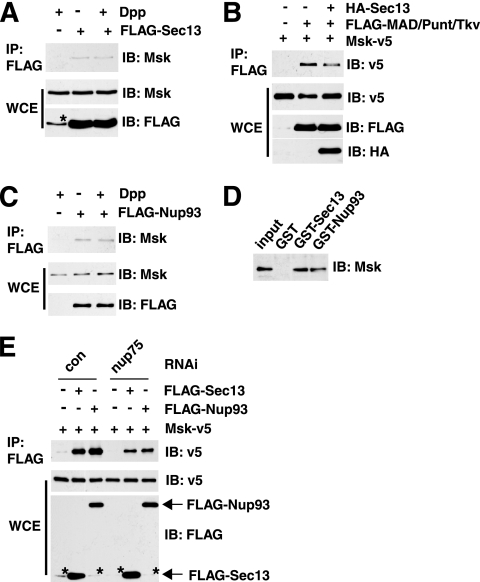
Using a Drosophila S2R+ cell line inducibly coexpressing GFP-MAD and the receptor kinases Punt and Tkv, we established a robust assay to recapitulate phosphorylation/activation-dependent nuclear accumulation of MAD (36). Msk was uncovered as an importin of MAD in a whole-genome RNAi screening using this assay (36). We further evaluated all potential hits by testing two nonoverlapping dsRNAs to eliminate false-positive hits due to off-target RNAi. Among the confirmed hits are nucleoporins Nup75, Nup93, Sec13, Nup205, and Nup50 (Fig. 1 A and B). Moreover, the deficiency in MAD nuclear import caused by sec13 or nup93 RNAi was fully rescued by sec13 or nup93 cDNA, respectively, further confirming the specificity of our RNAi experiments (Fig. 1C). As controls, we determined that RNAi of these nucleoporins did not affect the phosphorylation of MAD, which is a prerequisite for MAD nuclear import, or the expression level of Msk, so the decreased concentration of MAD in the nucleus was directly due to impaired nuclear import (Fig. 1D and E). Sec13 is also a component of the COPII coat, but knockdown of other key players in COPII coat assembly, such as the GTPase Sar-1, did not affect the nuclear import of MAD, arguing that vesicle transport in general is not a requirement for the nuclear targeting of MAD (data not shown) (12).
FIG. 1.
Nucleoporin requirement for nuclear accumulation of activated MAD. (A) S2R+ cells inducibly expressing GFP-MAD and Punt/Tkv (GFP-MAD+R) were treated with the indicated RNAi, and the distribution patterns of GFP-MAD are shown. Nuclei were stained with DAPI. Bar, 10 μM. (B) Two different dsRNA constructs against each nucleoporin hit were tested as described for panel A. The GFP-MAD signals in the nucleus and cytoplasm were quantified, and the ratios were plotted. Data are means ± SEs from ≥3 fields. (C) GFP-MAD+R cells were treated with dsRNAs targeting the 3′ UTR of sec13 or nup93, followed by transfection of HA-Sec13 or HA-Nup93 cDNAs. The distributions of GFP-MAD (green) and the rescue constructs (red) are shown. (D and E) GFP-MAD+R cells were subjected to the indicated RNAi, and the cell extracts were analyzed by immunoblotting with the indicated antibodies.
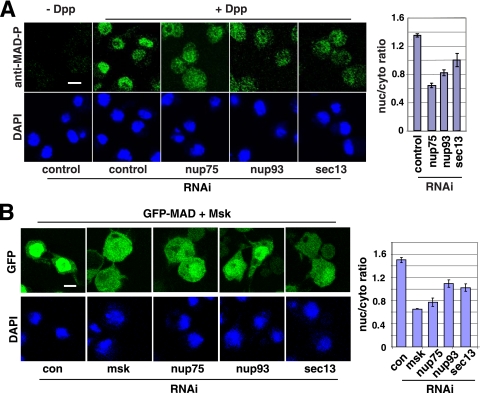
Nup75 and Sec13 are part of the Nup107-160 subcomplex in the NPC, whereas Nup93 and Nup205 are components of the Nup53-93 complex; both subcomplexes are evolutionarily conserved and are believed to serve general structural roles in NPC assembly (27, 32). Thus, the requirement for these non-FG nucleoporins in the nuclear transport of a particular cargo is intriguing. We therefore focused on Sec13 and Nup93 as representatives for further analyses. Available antibodies could barely detect endogenous MAD in S2 or S2R+ cells in immunofluorescence experiments. Fortuitously, in an S2 cell line transfected with MAD driven by an inducible metallothionein promoter (5), without induction, the leaky expression of MAD was sufficient for a phospho-Smad-specific antibody to detect a predominantly nuclear signal only in cells treated with Dpp (a Drosophila TGF-β family cytokine) (Fig. 2 A). In this setting, where there is only limited overexpression of MAD, knockdown of sec13, nup93, and nup75 also significantly reduced nuclear versus cytoplasmic concentrations of phospho-MAD (Fig. 2A, right). These data further support important roles for Sec13, Nup93, and Nup75 in MAD nuclear import in response to Dpp stimulation.
FIG. 2.
Sec13, Nup75, and Nup93 are part of the Msk-dependent nuclear import machinery for activated MAD. (A) (Left) S2 cells were treated with the indicated RNAi, and the phospho-MAD distribution pattern after Dpp treatment (10−9 M, 1 h) was detected by a phospho-MAD-specific antibody. Bar, 10 μM. (Right) Nuclear/cytoplasmic ratios of phospho-MAD signals are plotted; data are means ± SEs for ≥3 fields (P, <0.05 in all cases). (B) (Left) S2R+ cells transfected with GFP-MAD and Msk were subjected to the indicated RNAi, and the distribution patterns of GFP-MAD are shown. Bar, 10 μM. (Right) Nuclear and cytoplasmic GFP signals were quantified, and the ratios are plotted; data are means ± SEs for ≥3 fields (P, <0.005 in all cases).
The dependence on Sec13 and Nup93 for MAD nuclear import raised the question of whether these nucleoporins function in concert with Msk or whether they represent a separate mechanism. We previously found that overexpression of Msk could force the nuclear accumulation of MAD without the need for Dpp activation (37). Since such nuclear import is entirely Msk driven, we could directly test the nucleoporin requirement for Msk. RNAi against sec13, nup93, or Nup75 inhibited the Msk-driven nuclear import of MAD (Fig. 2B; see quantification on the right). Therefore, these non-FG nucleoporins are likely part of the same pathway as Msk, which imports MAD into the nucleus.
Sec13/Nup75/Nup93 are specifically utilized for the nuclear import of MAD.
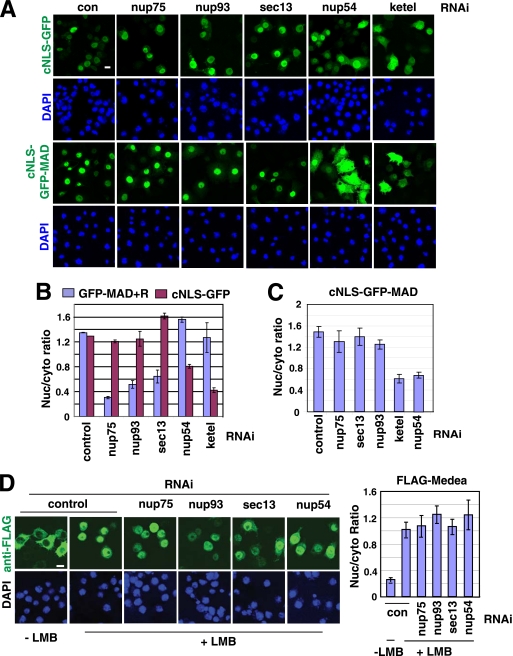
One important question is whether Sec13, Nup93, and Nup75 are uniquely required for MAD or are rather broadly involved in the nuclear import of many cargoes. We tested the nuclear import of cNLS-GFP (two copies of GFP fused to a classic NLS). As expected, the nuclear import of cNLS-GFP was critically dependent on Impβ (Ketel in Drosophila) (Fig. 3 A; see Fig. 3B for quantification). But depletion of Sec13, Nup93, or Nup75 had no effect on the nuclear localization of cNLS-GFP, while, in contrast, it strongly impaired the nuclear import of MAD (Fig. 3A; see Fig. 3B for quantification). The same conclusion was reached when we examined a cell line coexpressing GFP-MAD, Punt/Tkv, and RFP-cNLS so that we could monitor the localizations of GFP-MAD and RFP-cNLS concurrently (data not shown). In contrast, knockdown of nup54 (an FG-nucleoporin) significantly reduced the nuclear import of cNLS-GFP, as previously reported, but had no adverse effect on the nuclear accumulation of MAD (Fig. 3A and B) (23). These observations are not likely due to differences in RNAi efficiency, since quantitative real-time PCR confirmed that knockdowns of sec13, nup75, nup93, and nup54 were equally efficient in the two cell lines expressing GFP-MAD/Punt/Tkv (GFP-MAD+R) or cNLS-GFP (data not shown). More interestingly, when GFP-MAD was fused to a cNLS, it became constitutively nuclear and was completely independent of Sec13, Nup93, or Nup75 for nuclear import but instead relied on Nup54 and Impβ (Fig. 3A;see Fig. 3C for quantification). Again, RNAi efficiencies in cells expressing cNLS-GFP-MAD or GFP-MAD+R were comparable (data not shown). Thus, the cNLS was sufficient to switch MAD to a completely different route across the NPC. cNLS-cargoes are imported by Impα/Impβ independently of Msk/Imp7/8; thus, our results suggest that importins may dictate which nucleoporins to engage, and their modes of translocation through the NPC are likely different (1, 10).
FIG. 3.
Sec13, Nup93, and Nup75 are required specifically for the nuclear import of MAD but not for cNLS-dependent nuclear import. (A) The indicated RNAi experiments were carried out in three S2R+ cell lines inducibly expressing cNLS-GFP (2 copies of GFP fused with a cNLS [PKKKRKVED]), GFP-MAD+R, or cNLS-GFP-MAD. Only images for cNLS-GFP or cNLS-GFP-MAD are shown. Bar, 10 μM. (B and C) The nuclear/cytoplasmic ratio of the GFP signal was quantified and plotted. Data are means ± SEs for ≥3 fields. Compared to activation-induced nuclear translocation of GFP-MAD, constitutive nuclear import of cNLS-GFP-MAD depended on different nucleoporins. (D) (Left) S2R+ cells inducibly expressing FLAG-Medea were subjected to the indicated RNAi, and after treatment with leptomycin B (LMB) (10 ng/ml for 1 h), the distribution pattern of FLAG-Medea was detected by immunofluorescence staining with an anti-FLAG antibody. Bar, 10 μM. (Right) The nuclear/cytoplasmic ratio of the FLAG-Medea signal was quantified. Data are means ± SEs for ≥3 fields.
We further investigated the nucleoporin requirement of Medea, the Drosophila ortholog of Smad4, whose nuclear import is independent of either Msk or Impβ (37). Medea spontaneously undergoes nuclear accumulation upon inhibition of CRM-1 by leptomycin B (LMB), without the need for Dpp stimulation (Fig. 3D). RNAi of sec13, nup75, nup93, or nup54 had little effect on LMB-induced nuclear translocation of Medea (Fig. 3D; see quantification on the right). Real-time PCR showed similar or more efficient knockdown of these nucleoporins in the FLAG-Medea-expressing cell line than in the GFP-MAD+R line (data not shown). Thus, the nucleoporin requirement for the basal-state Medea nuclear import is different from that for either MAD or cNLS-cargoes. Therefore, the results from three types of cargoes strongly suggest that different import pathways utilize different sets of nucleoporins and that non-FG nucleoporins, such as Sec13, Nup93, and Nup75, are specific for Msk-mediated nuclear import.
Physical interactions of non-FG nucleoporins with MAD.
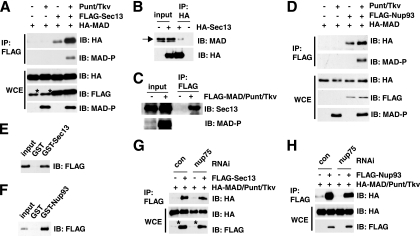
In coimmunoprecipitation experiments, we readily detected an interaction of MAD with Sec13 in S2 cells (Fig. 4 A). More importantly, when the TGF-β receptor kinases Punt and Tkv were cotransfected to induce MAD phosphorylation, the binding of MAD to Sec13 was markedly enhanced (Fig. 4A). Earlier studies of live cells indicated that TGF-β treatment accelerates the rate of Smad nuclear import, yet we previously found that the interaction between the transport factor Msk/Imp7/8 and Smads was rather constitutive (24, 36). Our observations here thus suggest that perhaps association with Sec13 is a rate-limiting step favoring the translocation of activated/phosphorylated MAD into the nucleus. To further test whether a Sec13-MAD interaction could be detected with endogenous proteins, we used anti-MAD and anti-Sec13 antibodies but found that neither could immunoprecipitate (data not shown). However, when only Sec13 was overexpressed, we detected its interaction with endogenous MAD in Dpp-treated cells (Fig. 4B). Likewise, when only MAD was overexpressed with the receptor kinases, we again readily detected the coimmunoprecipitation of MAD with endogenous Sec13 (Fig. 4C).
FIG. 4.
Sec13 and Nup93 interact physically with MAD. (A) Sec13 preferentially interacts with phosphorylated MAD. The indicated expression vectors were transfected into S2 cells, together with Punt/Tkv to phosphorylate MAD. The whole-cell extracts (WCE) were immunoprecipitated (IP) with anti-FLAG, and the bound proteins were analyzed by immunoblotting (IB) with the indicated antibodies. The asterisk marks a protein comigrating with Sec13 that cross-reacts with the anti-FLAG antibody. (B) S2 cells transfected with HA-Sec13 only were treated with Dpp, and the cell lysate was subjected to immunoprecipitation with anti-HA. The bound proteins were examined by IB using anti-MAD. The arrow points to endogenous MAD. The specificity of the anti-MAD antibody has been demonstrated (data not shown). (C) Cell extracts from S2 cells transfected with FLAG-MAD and Punt/Tkv only were immunoprecipitated with anti-FLAG, and the bound proteins were examined by IB with anti-Sec13. (D) Experiments similar to those described for panel A were carried out to examine the Nup93-MAD interaction. (E and F) GST pulldown assays to test direct protein interactions using purified recombinant FLAG-MAD (after phosphorylation by Punt/Tkv) and GST-Sec13 or GST-Nup93, with GST serving as the control. The bound proteins were analyzed by IB with anti-FLAG. (G and H) S2 cells were treated with the indicated RNAi and were transfected with the indicated plasmids. The cell extracts were subjected to IP with anti-FLAG in order to test interactions between MAD and Sec13 or Nup93. Quantitative real-time PCR confirmed that Nup75 knockdown was >70%. The asterisks indicate a band cross-reacting with anti-FLAG.
We also observed an interaction between MAD and Nup93 in coimmunoprecipitation experiments, and phosphorylation of MAD also appeared to enhance its binding to Nup93, although the effect was not as strong as in the case of Sec13 (Fig. 4D; compare to Fig. 4A). Moreover, using purified recombinant proteins (data not shown), we determined that MAD could interact directly with Sec13 or Nup93 (Fig. 4E and F). In such direct binding assays, an excess of Sec13 did not compete away the Nup93 interaction with MAD, suggesting that their association with MAD is not mutually exclusive (data not shown). We also investigated whether Nup75 could contribute to the interaction between MAD and Sec13 or Nup93. After knocking down Nup75 (to less than 30% of its normal level, as measured by real-time reverse transcription-PCR [RT-PCR]), we still detected similar levels of interaction between MAD and Sec13 or Nup93 in coimmunoprecipitation experiments (Fig. 4G and H). Therefore, Nup75 does not appear to regulate the interaction of MAD with Sec13 or Nup93.
Physical interactions of non-FG nucleoporins with Msk.
Importins are believed to interact mostly with FG-nucleoporins in mediating nuclear transport. Our observations thus raised an interesting question: whether importins such as Msk could also interact with non-FG structural nucleoporins. Indeed, we found that Sec13 interacted with endogenous Msk in coimmunoprecipitation experiments (Fig. 5 A). But Dpp treatment had no discernible effect on this interaction. Moreover, overexpression of Sec13 had a marginal impact on the interaction between Msk and MAD, so Sec13 is not likely to regulate the loading of phospho-MAD onto Msk (Fig. 5B). We also detected coimmunoprecipitation between endogenous Msk and overexpressed Nup93 in a Dpp-independent manner (Fig. 5C). Msk could bind directly to Sec13 or Nup93, as indicated by GST pulldown experiments (Fig. 5D) using purified recombinant proteins (data not shown). Interestingly, when Nup75 was knocked down to below 30% of its normal level (judging by real-time RT-PCR analysis), the coimmunoprecipitation between Msk and Sec13 or Nup93 was significantly decreased (Fig. 5E). Therefore, even though Msk is capable of direct interaction with Sec13 and Nup93, in the context of the whole NPC, Nup75 may further facilitate such association.
FIG. 5.
Physical interaction between Msk and Sec13 or Nup93. (A) S2 cells were transfected with FLAG-Sec13, and the lysate was used for immunoprecipitation (IP) with anti-FLAG. The endogenous Msk was detected by immunoblotting (IB). (B) Sec13 overexpression did not significantly affect the Msk-MAD interaction. S2 cells were transfected with the indicated expression vectors. The whole-cell extracts (WCE) were immunoprecipitated with anti-FLAG, and the bound proteins were analyzed by IB as indicated. (C) FLAG-Nup93 was expressed in S2 cells, and anti-FLAG immunoprecipitation was used to detect its interaction with endogenous Msk. (D) GST pulldown experiment testing direct interactions between purified recombinant Msk and GST-Sec13 or GST-Nup93, with GST as the control. Proteins bound to GST beads were analyzed by IB with anti-Msk. (E) S2 cells were subject to the indicated RNAi and were then transfected with the indicated expression vectors. IP was carried out to test for interactions between Msk-V5 and FLAG-Sec13 or FLAG-Nup93. RNAi knockdown of Nup75 resulted in decreased Sec13-Msk and Nup93-Msk interactions. Quantitative real-time PCR confirmed that Nup75 knockdown was >70%. The asterisk indicates a band cross-reacting with anti-FLAG.
These biochemical observations reinforced the RNAi data and suggest that Sec13 and Nup93 are directly involved in the nuclear import of MAD. The currently prevailing model is that importins interact mainly with the FG-nucleoporins to transport cargoes through the NPC (20, 28, 30). Our finding here that Msk binds directly to structural non-FG nucleoporins suggests a new mode of importin-NPC interaction that is important for the nuclear import of MAD.
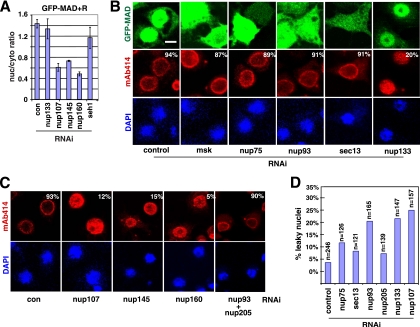
Functional distinction between Sec13/Nup75 and other components of the Nup107-160 subcomplex.
Nup75 and Sec13 are part of the Nup107-160 subcomplex (11, 32). Non-FG nucleoporins of the Nup107-160 complex have been suggested to be critical for NPC assembly and integrity from yeast to mammals; thus, it is surprising that Sec13 and Nup75 are required for the nuclear import of specific cargoes. A comprehensive analysis of the Nup107-160 complex showed that Nup107, Nup145, and Nup160, but not Seh1 or Nup133, were also required for MAD nuclear import (Fig. 6 A). The lack of effect on MAD nuclear import was not likely due to inefficient Seh1 or Nup133 depletion, since we tested two different RNAi constructs with similar results, and at least for nup133 RNAi there was a strong phenotype in NPC assembly, as expected (Fig. 6B).
FIG. 6.
Impact of RNAi against non-FG nucleoporins on MAb414 pattern and nuclear envelope permeability. (A) GFP-MAD+R cells were treated with the indicated RNAi, and the quantification of the nuclear/cytoplasmic concentration of GFP-MAD is plotted. Data are means ± SEs for ≥3 fields. (B) GFP-MAD+R cells were treated with the indicated RNAi and were then induced to express GFP-MAD and Punt/Tkv. The cells were stained with MAb414, and the images for GFP-MAD (green), MAb414 (red), and DAPI (blue) were captured by confocal microscopy. The percentages of cells exhibiting a normal MAb414 staining pattern (>50 cells scored) are shown. Bar, 5 μM. (C) GFP-MAD+R cells were treated with the indicated RNAi and were immunostained with MAb414 (red). The percentages of cells exhibiting a normal MAb414 staining pattern (>50 cells scored) are shown. (D) GFP-MAD+R cells were treated with the indicated RNAi, and the permeability of the nuclear envelope was tested by dextran exclusion assays. The percentages of nuclei permeable to the 70-kDa but not the 500-kDa dextran are plotted.
The integrity of the NPC is commonly analyzed with the monoclonal antibody MAb414, which recognizes a subset of FG-nucleoporins and exhibits a predominantly nuclear rim pattern in yeast and vertebrate cells (7) (Fig. 6B). Somewhat surprisingly, the MAb414 staining pattern was unchanged after knockdown of sec13 or nup75, but in the same cells, the nuclear translocation of MAD was clearly impaired (Fig. 6B). In contrast, knockdown of other Nup107-160 members, including nup107, nup133, nup145, and nup160, resulted in a diffuse MAb414 pattern throughout the nuclei, indicating defects in NPC assembly, as expected (Fig. 6C). It is known that RNAi against nup107 or nup133 results in concomitant loss of several other nucleoporins in the Nup107-160 complex, which might have contributed to the more severe defects in NPC integrity (4, 32). Immunoblot analysis confirmed that sec13 RNAi was highly robust, and knockdown of other Nup107-160 complex components did not cause concomitant loss of Sec13 (data not shown). A normal MAb414 pattern does not necessarily indicate intact NPC assembly, so roles for Sec13 and Nup75 in NPC structure cannot be ruled out. However, our observations did distinguish Sec13/Nup75 from Nup107/Nup145/Nup160 in terms of their requirement in maintaining a normal MAb414 pattern.
Nup133 knockdown clearly affected the MAb414 pattern, as expected for a core component of Nup107-160, but remarkably, in the same cells, nuclear accumulation of GFP-MAD was largely intact (Fig. 6B). This is compelling evidence that components of the Nup107-160 subcomplex individually serve different functions in the process of MAD nuclear transport and NPC assembly. These observations strongly argue that Sec13 and Nup75 are important for the nuclear import of MAD not because they are part of the Nup107-160 complex but because they have unique individual functions.
The Nup53-93 complex is critically involved in the anchoring of the NPC to the nuclear envelope (14). Previous studies of mammalian cells showed aberrant MAb414 staining upon nup93 RNAi (13). However, MAb414 staining seemed normal in S2R+ cells after nup93 RNAi, even though the nuclear import of MAD was clearly inhibited in the same cells (Fig. 6B). Even when we simultaneously knocked down nup93 and nup205, the MAb414 staining pattern still remained normal (Fig. 6C). Interestingly, depletion of Nup93 or Nup205 also failed to affect NPC formation in Caenorhabditis elegans (9). Therefore, we again observed differential dependence on Nup93 in the nuclear import of MAD and the assembly of the NPC.
Impact of RNAi against non-FG nucleoporins on nuclear envelope permeability.
MAb414 is limited in its ability to detect changes in the NPC. We therefore used a dextran exclusion assay to examine the permeability of the nuclear envelope. We found that RNAi against sec13 or nup75 had only a minor effect, whereas knockdown of nup133 or nup107 resulted in a significantly higher number of leaky nuclei (Fig. 6D) (see Materials and Methods for scoring criteria). This again indicated functional differences between Sec13/Nup75 and other members of the Nup107-160 complex. Knockdown of nup93 increased the leakiness of the nuclear envelope, in agreement with previous reports (Fig. 6D) (9). But a leaky nuclear envelope per se does not necessarily lead to impaired MAD nuclear import, since RNAi against nup133 did not affect the nuclear concentration of MAD. Although it is in the same subcomplex with Nup93, Nup205 did not appear to be critical for maintaining the permeability barrier (Fig. 6D). Therefore, the defect in MAD nuclear import after RNAi against Sec13, Nup75, Nup93, or Nup205 was not due to a compromised NPC permeability barrier.
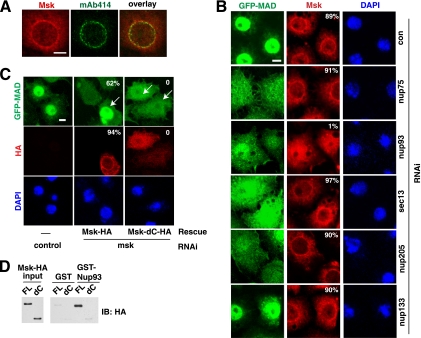
Nup93 regulates the subcellular localization of Msk.
We further investigated how non-FG nucleoporins functionally interact with Msk. Confirming an earlier report, we found endogenous Msk concentrated around the nuclear rim, with some overlap with MAb414 staining (Fig. 7 A) (15). The immunofluorescence staining was highly specific, since msk RNAi abolished the signals (data not shown). Activation of the Dpp pathway by Punt/Tkv did not change the distribution pattern of Msk (data not shown). Interestingly, we detected a completely different Msk pattern after nup93 knockdown: the prominent nuclear rim pattern was absent, and Msk was distributed evenly in both the nucleus and the cytoplasm (Fig. 7B). In contrast, RNAi against nup75 or sec13 did not alter Msk distribution, although nuclear import of MAD was inhibited just as strongly as in the case of nup93 RNAi (Fig. 7B). We found that even though knockdown of Nup133 disrupted the MAb414 pattern (Fig. 6B), the distribution of Nup93 appeared to be unaffected (data not shown), and indeed, Msk was still localized to the nuclear periphery after Nup133 knockdown (Fig. 7B). Interestingly, even though Nup205 is part of the same subcomplex as Nup93, its knockdown did not affect Msk distribution but did cause much-reduced MAD nuclear import (Fig. 7B). Thus, clearly the role of Nup93 in MAD nuclear import is very different from those of Sec13, Nup75, and Nup205. This again illustrates the distinct functional roles that non-FG nucleoporins play in MAD nuclear transport. The fact that knockdown of these non-FG nucleoporins affects different aspects of MAD nuclear translocation is also a strong argument against the possibility that the defect in MAD nuclear transport is due to general disruption of the NPC.
FIG. 7.
The perinuclear localization of Msk depends on Nup93 and is important for Msk function in the nuclear import of MAD. (A) Double immunofluorescence staining of endogenous Msk (red) and NPC (green; by MAb414) in S2R+ cells. (B) GFP-MAD+R cells were treated with the indicated RNAi, and after induction of GFP-MAD/Punt/Tkv expression, the cells were stained with anti-Msk (red). Shown are confocal images of GFP-MAD (green), endogenous Msk (red), and DAPI (blue). The percentages of cells exhibiting a nuclear rim pattern of Msk staining (>50 cells scored) are shown. (C) The perinuclear localization of Msk is dependent on its C-terminal region and correlates with its ability to import MAD into the nucleus. GFP-MAD+R cells were subjected to RNAi targeting the 5′ UTR of Msk and were then transfected with rescue vectors (arrows) expressing HA-tagged full-length Msk or HA-tagged Msk with a C-terminal deletion (Msk-dC; aa 1 to 761). Confocal images of GFP-MAD (green), Msk-HA or Msk-dC-HA (red), and DAPI (blue) are shown. The percentages of cells with normal GFP-MAD nuclear import (green) (>50 cells scored) or exhibiting a nuclear rim pattern of Msk staining (red) (>50 cells scored) are shown. Bars, 5 μM. (D) Recombinant full-length Msk-HA (FL) and Msk-dC-HA (dC) were purified and tested for direct interaction with GST-Nup93.
Mutational analysis of Msk further supported the idea that perinuclear localization of Msk is essential for its function as a Smad nuclear import factor. Removal of the C-terminal 290 amino acids of Msk abolished its function, since it could no longer rescue the msk RNAi phenotype, although the deletion mutant was still capable of interacting with MAD (Fig. 7C; also data not shown). In agreement with the inability to import MAD, this deletion mutant (Msk-dC; aa 1 to 761) was diffusely distributed and was no longer concentrated in the perinuclear space, in contrast to full-length Msk (Fig. 7C). Thus, our data suggest that the localization of Msk to the perinuclear space is important for its ability to transport MAD into the nucleus.
Interestingly, when we examined the direct interaction between purified recombinant Msk and Nup93, we found that Msk-dC failed to interact with Nup93 (Fig. 7D). Therefore, combining the RNAi and protein interaction data, we arrived at the hypothesis that Nup93 may recruit Msk to the nuclear periphery by binding to the C-terminal 290 amino acids of Msk. However, one caveat is that we cannot rule out the possibility that the Msk C terminus is required for interaction with another factor that directly anchors Msk to the nuclear periphery.
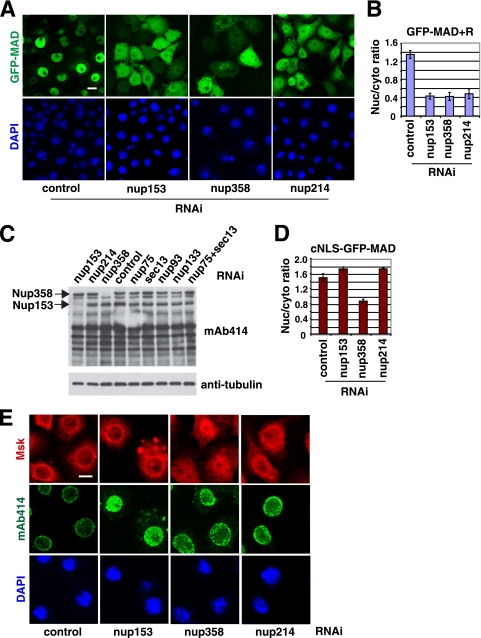
Distinct roles of FG-nucleoporins in the nuclear import of MAD.
False-negative results due to cell lethality, RNAi efficiency, or functional redundancy are potential pitfalls in RNAi screening. Our previous studies have suggested that the FG-nucleoporins Nup153 and Nup214 play roles in the nuclear import of Smads, but they were not scored as positive hits in our screening (35). However, upon close examination, we noticed that both nup153 and nup358 RNAi resulted in cell lethality. Therefore, we decided to revisit the issue of whether FG-nucleoporins Nup153, Nup214, and Nup358 are important for the nuclear import of Smads. By moderating the RNAi efficiency (i.e., reducing the dsRNA concentration by half and shortening the duration of RNAi from 4 days to 3 days), we considerably improved the viability of the cells treated with nup153 or nup358 RNAi and found that, indeed, Nup153, Nup358, and also Nup214 were required for the nuclear import of MAD (Fig. 8 A; quantification in Fig. 8B). Importantly, by immunoblotting, we detected little change in the Nup358 level upon RNAi against sec13, nup75, or nup93, ruling out the possibility that the impaired MAD nuclear import observed earlier after RNAi against these non-FG nucleoporins was caused by codepletion of Nup358 (Fig. 8C). Although there was a slight decrease in the level of Nup153 upon RNAi against sec13, nup75, or nup93, it was not likely significant enough to affect the nuclear import of MAD, since nup133 RNAi also caused a similar change in the Nup153 level (Fig. 8C), with no impact on MAD nuclear transport (Fig. 7B). Therefore, the requirements for Nup153 and Nup358 in MAD nuclear import are separate from those for the non-FG nucleoporins Sec13, Nup75, and Nup93.
FIG. 8.
FG-nucleoporins Nup153, Nup214, and Nup358 serve different functions for the transport of MAD into the nucleus. (A) GFP-MAD+R cells were treated with the indicated RNAi and were induced to express GFP-MAD and Punt/Tkv. The distribution patterns of GFP-MAD (green) and DAPI (blue) are shown. Bar, 10 μM. (B) Quantification of the nuclear/cytoplasmic ratio of GFP-MAD signals in panel A. Data are means ± SEs for ≥3 fields. (C) GFP-MAD+R cells were treated with the indicated RNAi, and the cell extracts were analyzed by immunoblotting with MAb414 and antitubulin. (D) Quantification of the nuclear/cytoplasmic ratio of cNLS-GFP-MAD in cells treated with the indicated RNAi. Data are means ± SEs for ≥3 fields. (E) S2R+ cells were treated with the indicated RNAi, and the cells were double stained with MAb414 (green) and anti-Msk (red). Bar, 5 μM.
When MAD was fused to a cNLS, its nuclear import became independent of Nup214 and Nup153 but, interestingly, still required Nup358 (Fig. 8D; also data not shown). These observations agree with the findings of a previous study on the requirement of Nup358 for cNLS-dependent nuclear import (23). The same study also found that Nup153 was important for cNLS-mediated nuclear import, but this was not confirmed here, probably because we had to knock down Nup153 only partially (23). Thus, the notion of pathway-specific utilization of nucleoporins applies to FG-nucleoporins as well; Nup358 is involved broadly in the nuclear import of both MAD and the cNLS, while Nup214 may serve more specifically in the nuclear import of MAD. Again, we confirmed that the efficiencies of RNAi were similar in cells expressing cNLS-GFP-MAD and those expressing GFP-MAD+R (data not shown). Furthermore, by double immunofluorescence staining, we found that nup358 knockdown affected the perinuclear localization of Msk but RNAi against nup153 or nup214 had little effect (Fig. 8E). In terms of disrupting the MAb414 pattern, nup153 knockdown had a very severe effect, whereas RNAi against nup358 or nup214 had a weaker impact, consistent with a previous report (Fig. 8E) (23). These data suggest that different FG-nucleoporins play different roles in the nuclear import of MAD, in agreement with our observations with non-FG nucleoporins.
DISCUSSION
In this study we identified a distinct nucleoporin cohort, including both non-FG nucleoporins and FG-nucleoporins, that represents a unique trans-NPC mechanism for signal-activated MAD. Such specificity in nucleoporin utilization may reflect different demands of constitutive and signal-induced nuclear import events. Most unexpectedly, several non-FG nucleoporins, including Sec13, Nup93, Nup75, and Nup205, appear to act in concert with Msk to selectively transport MAD, but not the cNLS-cargo or basal-state Medea, into the nucleus. This is the first indication that beyond their involvement in the general assembly of the NPC, non-FG nucleoporins could play discrete roles in specific nuclear transport pathways. We further identified the distinct functions served by two non-FG scaffold nucleoporins, Sec13 and Nup93, that are critical and specific for the nuclear import of MAD. Our findings suggest a novel functional interplay between the MAD nuclear import machinery and the NPC.
Sec13 is part of the Nup107-160 complex, and Nup93 is part of the Nup53-93 complex; both are scaffolds of the NPC. We emphasize that our findings are not in conflict with the established roles of Sec13 and Nup93 in general NPC assembly but broaden the functions of these non-FG nucleoporins to specific nuclear import pathways. It was somewhat surprising that depletion of Nup75 and Sec13 had little impact on MAb414 staining and nuclear envelope permeability, in contrast to the more severe phenotypes exhibited by the knockdown of other components in the Nup107-160 complex (i.e., nup145, nup107, and nup160). We could hardly detect Sec13 after RNAi (data not shown), so the lack of impact on MAb414 staining could not be attributed to incomplete depletion of Sec13. Therefore, our observations suggest that knocking down individual components of the Nup107-160 complex could lead to different phenotypes regarding MAD nuclear import, the MAb414 staining pattern, and the permeability of the NPC, arguing that each nucleoporin in the Nup107-160 complex serves distinct functions.
The challenging question ahead is how these non-FG nucleoporins mediate the nuclear import of MAD. Interestingly, Sec13 has been shown to dynamically transit between the cytoplasm and the nucleus, and endogenous Sec13 is partitioned among NPC, the intranuclear space, and the endoplasmic reticulum (ER) (8). With our observation that Sec13 preferentially interacts with phosphorylated/activated MAD, it is possible that Sec13 could act as an active trafficker rather than as a stationary component of the NPC to mediate the nuclear import of MAD. Whether phosphorylated MAD reaches the NPC via random diffusion or is guided by particular factors remains an open question, and it will be interesting to investigate whether Sec13 might be involved.
Msk has a characteristic nuclear rim localization pattern that we show here is important for its ability to transport MAD into the nucleus. Two of the nucleoporins that are required for MAD nuclear import, Nup93 and Nup358, appear to be responsible for targeting Msk to the nuclear periphery. Deletion of the C-terminal region of Msk disrupted its nuclear rim distribution and also significantly weakened the Msk-Nup93 interaction. It is unclear whether the same C-terminal deletion of Msk would affect the Msk-Nup358 interaction as well. Thus, the question remains, between Nup93 and Nup358, which one is more directly responsible for recruiting Msk to the nuclear periphery. Interestingly, Impβ is also concentrated to the nuclear periphery, like Msk, but such localization has been shown to depend on Nup153 instead of Nup93 and Nup358 (23). Therefore, different importins are apparently recruited to the NPC through distinct nucleoporins, another direct indication that various nuclear import pathways operate through different modes of interaction with the NPC.
As exemplified by Sec13, Nup93, and Nup358, the nucleoporins implicated in MAD nuclear import serve distinct functions at different stages of the import process. One appealing model is that Msk is positioned by Nup93 and Nup358 to the vicinity of NPC, and perhaps Sec13 engages phosphorylated MAD and, through its own trafficking ability, delivers MAD to Msk, which completes the translocation across the NPC. While our data clearly suggest that Sec13 and Nup93 play roles distinct from those of the other components of the Nup107-160 or Nup53-93 complex, we do not suggest that they function in isolation from the other nucleoporins. Nor can we at this point rule out a possible requirement for other nucleoporins in the nuclear import of MAD. Nevertheless, the direct physical interaction between Sec13/Nup93 and MAD or Msk, as well as the very selective impact of Sec13 and Nup93 RNAi on the nuclear import of MAD, but not other cargoes, is consistent with our interpretation that Sec13 and Nup93 are directly involved in the nuclear import of MAD.
Our genetic dissection of the Smad nuclear import pathway has important implications for the model of NPC structure and function. The findings in this study depart from the current dogma that puts only FG-nucleoporins at the center of the NPC-importin interplay. The diversity in trans-NPC routes and the pathway-specific involvement of non-FG nucleoporins need to be incorporated into models of NPC function in nuclear transport. It is increasingly clear that there are multiple distinct routes through the NPC that are taken by different importin/cargo complexes. The question, then, is how the NPC can accommodate these different passages. X-ray crystal structure analysis and electron microscopy have suggested that the Nup107-160 complex assumes a Y-shaped topography, raising speculations that such a porous assembly may leave room for additional trans-NPC passages besides the central tunnel, which is densely populated by FG-nucleoporins (4). One could also speculate that maybe the NPC can assume different configurations upon receiving different importin/cargo complexes to enable the translocation process.
Acknowledgments
We thank D. Forbes (University of California at San Diego) and M. Dasso (NICHD, NIH) for antibodies and the Drosophila RNAi Screening Center (Harvard Medical School) for analysis of the whole-genome RNAi screening. We are grateful to Y. T. Ip for insightful discussions and to Q. Xu for critical reading of the manuscript.
This work was funded by NIH (R01CA108509) and the Worcester Foundation for Biomedical Research.
Footnotes
Published ahead of print on 14 June 2010.
REFERENCES
- 1.Adam, S. A., and L. Gerace. 1991. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66:837-847. [DOI] [PubMed] [Google Scholar]
- 2.Alber, F., S. Dokudovskaya, L. M. Veenhoff, W. Zhang, J. Kipper, D. Devos, A. Suprapto, O. Karni-Schmidt, R. Williams, B. T. Chait, M. P. Rout, and A. Sali. 2007. Determining the architectures of macromolecular assemblies. Nature 450:683-694. [DOI] [PubMed] [Google Scholar]
- 3.Alber, F., S. Dokudovskaya, L. M. Veenhoff, W. Zhang, J. Kipper, D. Devos, A. Suprapto, O. Karni-Schmidt, R. Williams, B. T. Chait, A. Sali, and M. P. Rout. 2007. The molecular architecture of the nuclear pore complex. Nature 450:695-701. [DOI] [PubMed] [Google Scholar]
- 4.Boehmer, T., J. Enninga, S. Dales, G. Blobel, and H. Zhong. 2003. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 100:981-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunch, T. A., Y. Grinblat, and L. S. Goldstein. 1988. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 16:1043-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Angelo, M. A., and M. W. Hetzer. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18:456-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, L. I., and G. Blobel. 1986. Identification and characterization of a nuclear pore complex protein. Cell 45:699-709. [DOI] [PubMed] [Google Scholar]
- 8.Enninga, J., A. Levay, and B. M. Fontoura. 2003. Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol. Cell. Biol. 23:7271-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galy, V., I. W. Mattaj, and P. Askjaer. 2003. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol. Biol. Cell 14:5104-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Görlich, D., S. Prehn, R. A. Laskey, and E. Hartmann. 1994. Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79:767-778. [DOI] [PubMed] [Google Scholar]
- 11.Harel, A., A. V. Orjalo, T. Vincent, A. Lachish-Zalait, S. Vasu, S. Shah, E. Zimmerman, M. Elbaum, and D. J. Forbes. 2003. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11:853-864. [DOI] [PubMed] [Google Scholar]
- 12.Haucke, V. 2003. Vesicle budding: a coat for the COPs. Trends Cell Biol. 13:59-60. [DOI] [PubMed] [Google Scholar]
- 13.Hawryluk-Gara, L. A., M. Platani, R. Santarella, R. W. Wozniak, and I. W. Mattaj. 2008. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol. Biol. Cell 19:1753-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawryluk-Gara, L. A., E. K. Shibuya, and R. W. Wozniak. 2005. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol. Biol. Cell 16:2382-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James, B. P., T. A. Bunch, S. Krishnamoorthy, L. A. Perkins, and D. L. Brower. 2007. Nuclear localization of the ERK MAP kinase mediated by Drosophila αPS2βPS integrin and importin-7. Mol. Biol. Cell 18:4190-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupu, F., A. Alves, K. Anderson, V. Doye, and E. Lacy. 2008. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev. Cell 14:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosammaparast, N., and L. F. Pemberton. 2004. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14:547-556. [DOI] [PubMed] [Google Scholar]
- 18.Moustakas, A., and C. H. Heldin. 2009. The regulation of TGFβ signal transduction. Development 136:3699-3714. [DOI] [PubMed] [Google Scholar]
- 19.Patel, S. S., B. J. Belmont, J. M. Sante, and M. F. Rexach. 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129:83-96. [DOI] [PubMed] [Google Scholar]
- 20.Pemberton, L. F., and B. M. Paschal. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187-198. [DOI] [PubMed] [Google Scholar]
- 21.Pierreux, C. E., F. J. Nicolas, and C. S. Hill. 2000. Transforming growth factor beta-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol. 20:9041-9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radu, A., G. Blobel, and M. S. Moore. 1995. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. U. S. A. 92:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabri, N., P. Roth, N. Xylourgidis, F. Sadeghifar, J. Adler, and C. Samakovlis. 2007. Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J. Cell Biol. 178:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmierer, B., A. L. Tournier, P. A. Bates, and C. S. Hill. 2008. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc. Natl. Acad. Sci. U. S. A. 105:6608-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz, T. U. 2005. Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 15:221-226. [DOI] [PubMed] [Google Scholar]
- 26.Shi, Y., and J. Massagué. 2003. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113:685-700. [DOI] [PubMed] [Google Scholar]
- 27.Siniossoglou, S., C. Wimmer, M. Rieger, V. Doye, H. Tekotte, C. Weise, S. Emig, A. Segref, and E. C. Hurt. 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84:265-275. [DOI] [PubMed] [Google Scholar]
- 28.Stewart, M. 2007. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8:195-208. [DOI] [PubMed] [Google Scholar]
- 29.Strawn, L. A., T. Shen, N. Shulga, D. S. Goldfarb, and S. R. Wente. 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6:197-206. [DOI] [PubMed] [Google Scholar]
- 30.Terry, L. J., E. B. Shows, and S. R. Wente. 2007. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318:1412-1416. [DOI] [PubMed] [Google Scholar]
- 31.Terry, L. J., and S. R. Wente. 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178:1121-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walther, T. C., A. Alves, H. Pickersgill, I. Loiodice, M. Hetzer, V. Galy, B. B. Hulsmann, T. Kocher, M. Wilm, T. Allen, I. W. Mattaj, and V. Doye. 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113:195-206. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe, M., N. Masuyama, M. Fukuda, and E. Nishida. 2000. Regulation of intracellular dynamics of Smad4 by its leucine-rich nuclear export signal. EMBO Rep. 1:176-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, M. Y., and C. S. Hill. 2009. Tgf-β superfamily signaling in embryonic development and homeostasis. Dev. Cell 16:329-343. [DOI] [PubMed] [Google Scholar]
- 35.Xu, L., Y. Kang, S. Col, and J. Massagué. 2002. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell 10:271-282. [DOI] [PubMed] [Google Scholar]
- 36.Xu, L., X. Yao, X. Chen, P. Lu, B. Zhang, and Y. T. Ip. 2007. Msk is required for nuclear import of TGF-β/BMP-activated Smads. J. Cell Biol. 178:981-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao, X., X. Chen, C. Cottonham, and L. Xu. 2008. Preferential utilization of Imp7/8 in nuclear import of Smads. J. Biol. Chem. 283:22867-22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeitler, B., and K. Weis. 2004. The FG-repeat asymmetry of the nuclear pore complex is dispensable for bulk nucleocytoplasmic transport in vivo. J. Cell Biol. 167:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X., S. Chen, S. Yoo, S. Chakrabarti, T. Zhang, T. Ke, C. Oberti, S. L. Yong, F. Fang, L. Li, R. de la Fuente, L. Wang, Q. Chen, and Q. K. Wang. 2008. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 135:1017-1027. [DOI] [PubMed] [Google Scholar]