Abstract
The nuclear and oncogenic BCL-3 protein activates or represses gene transcription when bound to NF-κB proteins p50 and p52, yet the molecules that specifically interact with BCL-3 and drive BCL-3-mediated effects on gene expression remain largely uncharacterized. Moreover, GSK3-mediated phosphorylation of BCL-3 triggers its degradation through the proteasome, but the proteins involved in this degradative pathway are poorly characterized. Biochemical purification of interacting partners of BCL-3 led to the identification of CtBP as a molecule required for the ability of BCL-3 to repress gene transcription. CtBP is also required for the oncogenic potential of BCL-3 and for its ability to inhibit UV-mediated cell apoptosis in keratinocytes. We also defined the E3 ligase TBLR1 as a protein involved in BCL-3 degradation through a GSK3-independent pathway. Thus, our data demonstrate that the LSD1/CtBP complex is required for the repressing abilities of an oncogenic IκB protein, and they establish a functional link between the E3 ligase TBLR1 and NF-κB.
Numerous oncogenic proteins are aberrantly expressed because the molecules involved in their degradation are not functioning properly (22, 24, 44). BCL-3 is an IκB protein whose degradation through the proteasome requires GSK3-mediated phosphorylation, yet the E3 ligase involved in this pathway is unknown (42). BCL-3 was originally identified through molecular cloning of the breakpoint of the t(14;19) chromosomal translocation found in a subset of human B-cell chronic lymphotic leukemias (27). This translocation triggers BCL-3 overexpression and, consequently, the deregulation of many genes involved in survival and cell proliferation (30). Aberrant BCL-3 expression has also been reported in multiple myelomas and subtypes of lymphomas, even in the absence of any t(14;19) chromosomal translocation (6, 7, 26, 29). Deregulated BCL-3 expression has also been seen in breast and nasopharyngeal cancers, hepatocarcinomas, and familial cylindromatosis. Familial cylindromatosis is a genetic disease characterized by benign tumors of hair-follicle keratinocytes due to some loss-of-function mutations of CYLD, a deubiquitinating enzyme that limits the nuclear import of BCL-3 (4, 12, 25, 31, 41).
Insight into the role of BCL-3 in the immune system has been provided through the analysis of bcl-3-deficient mice. These mice have defects in the organogenesis of Peyer's patches, in germinal-center formation, and in the microarchitecture of the spleen and lymph nodes, and they exhibit select defects upon immunological challenge (16, 32, 36). A role for BCL-3 in the central tolerance of the thymus was also revealed in NF-κB2-deficient mice (51). Mice deficient in both bcl-3 and NF-κB2 show a complete block in secondary lymphoid organogenesis as well as defects in thymic stromal cells, and they develop lymphocytic infiltrates in multiple organs (51). This severe phenotype, combined with the overlapping phenotypes of mice deficient in bcl-3 or p52, support the notion that BCL-3 and p52 have redundant biological effects in vivo (15, 16, 51).
BCL-3 activates gene transcription by removing the inhibitory NF-κB p50 and p52 homodimers from DNA and/or by acting as a coactivator for a subset of NF-κB target genes (13, 17, 23). BCL-3 can also negatively regulate lipopolysaccharide (LPS)-induced tumor necrosis factor alpha (TNF-α) synthesis in macrophages when bound to histone deacetylase 1 (HDAC1) and HDAC3 and/or because BCL-3 prevents the degradative polyubiquitination of p50 inhibitory homodimers (8, 21, 46).
To learn more about the regulation of BCL-3, we have purified BCL-3-associated proteins and have identified CtBP, LSD1, and TBLR1 as proteins that all bind to the N-terminal domain of this oncoprotein. CtBP is crucial in the biology of BCL-3: this corepressor is required for the stabilization of BCL-3 and for the ability of BCL-3 to repress gene transcription, to transform cells, and to inhibit UV-mediated cell apoptosis in transformed keratinocytes. Moreover, the half-life of BCL-3 is extended in TBLR1-depleted cells due to altered polyubiquitination, thus defining TBLR1 as a key molecule for BCL-3 degradation. Therefore, our data identified the N-terminal domain of BCL-3 as an essential region for the degradation, transcriptional activity, and oncogenic potential of this protein and defined CtBP and TBLR1 as key regulators of different properties of the BCL-3 oncoprotein.
MATERIALS AND METHODS
Cell culture, biological reagents, and treatments.
293, Phoenix, NIH 3T3, HeLa, and Karpas cells were cultured as described previously (42). HaCat cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% glutamine, and 1% penicillin-streptomycin.
MG132 and LiCl were purchased from A&E Scientific (Marcq, Belgium) and Sigma (St. Louis, MO), respectively. Polyclonal antibodies against hemagglutinin (HA), HDAC3, Hsp90, Myc, BCL-3, IκBα, IκBβ, and IκBɛ as well as monoclonal antibodies against ubiquitin, CtBP, and Myc were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-FLAG antibodies and beads were purchased from Sigma. Polyclonal antibodies against LSD1, p52/p100, and p105/p50 were from Abcam (Cambridge, United Kingdom) and Millipore (Temecula, CA), respectively. The monoclonal anti-TBLR1 antibody was from Abnova (Taipei, Taiwan). GFP, TBLR1, and CtBP small interfering RNAs (siRNAs) were purchased from Dharmacon (Lafayette, CO), whereas LSD1 siRNAs were from Eurogentec (Liege, Belgium).
Plasmids, retroviral constructs, and lentiviral constructs.
pMT2T p50, pMT2T BCL-3, pMT2T BCL-3 K13-26R, pMT2T BCL-3 MTS, pMT2T BCL-3 ΔN (which lacks the first 112 amino acids), and FLAG-BCL-3, -ΔN, -ΔC, and -ΔNΔC have been described previously, as has FLAG-IκBα (5, 42, 43). BCL-3 ΔN10 and -ΔN30 constructs were generated by PCR, whereas BCL-3 ANK M123, BCL-3 LAV, and BCL-3 5KR mutants were generated by mutagenesis. The HA-BCL-3-expressing construct was generated by subcloning the corresponding cDNA into the HA-pcDNA3 vector. The HA-p52-, HA-p100-, and HA-p105-expressing constructs were gifts from E. Dejardin (University of Liege, Liege, Belgium) (14). The pBabe BCL-3 and RasV12 constructs have been described previously, whereas the pBabe BCL-3 LAV mutant was generated by subcloning the corresponding cDNA coding sequence into the pBabe construct. The BCL-3 lentiviral construct was generated by subcloning the BCL-3 cDNA into the pLL3.7 lentivirus construct (a gift from L. van Parijs). FLAG-p52 was generated by PCR using pMT2T p52 as the template. Myc-TBLR1 was generated by reverse transcription-PCR (RT-PCR) using total RNAs from 293 cells and subsequent cloning into the pCMV-Myc vector (Invitrogen, Carlsbad, CA). The Myc-TBLR1 ΔN mutant, which lacks the first 100 amino acids, was generated by PCR using Myc-TBLR1 as the template. Myc-CtBP, FLAG-CtBP, and FLAG-CoREST were kindly provided by E. Olson (Southwestern Medical Center, University of Texas, Dallas, TX), J. Mymryk (Department of Oncology, London Regional Cancer Program, and Department of Biochemistry, University of Western Ontario, London, Ontario, Canada), and G. Gill (Department of Pathology, Harvard Medical School, Boston, MA), respectively. The HDAC3 expression constructs have been described previously (43).
Immunoprecipitations and immunofluorescence.
Immunoprecipitations involving ectopically expressed and/or endogenous proteins, as well as immunofluorescence, were performed as previously described (35, 42).
Retroviral transduction and formation of foci.
Infection of NIH 3T3 cells was performed according to instructions from G. Nolan's laboratory (http://www.stanford.edu/group/nolan/retroviral_systems/retsys.html). For focus assays, 1.5 × 106 infected NIH 3T3 cells were seeded in a 10-cm-diameter plate, and the medium was changed every 3 days. The cell cultures were followed for 3 weeks, and colonies were visualized with Giemsa staining.
Lentiviral infections, Affymetrix microarray analysis, and real-time PCR.
Infections of HaCat cells using the control or BCL-3 pLL3.7-expressing constructs were carried out as previously described (35). For Affymetrix microarrays, total RNAs were extracted from the resulting HaCat cells cultured in triplicate, using the RNeasy Mini kit from Qiagen (Valencia, CA). The integrity of the RNAs from those six distinct experimental conditions was checked with the Agilent Bioanalyzer using the RNA 6000 Nano kit (Agilent, Santa Clara, CA). Biotin labeling of cRNA, subsequent hybridization on the GeneChip Human Genome U133A 2.0 (Affymetrix, Santa Clara, CA), and scanning were performed according to the manufacturer's instructions.
For real-time PCRs, total RNAs were extracted using the EZNA Total RNA kit (Omega Bio-Tek, Norcross, GA), and cDNAs were synthesized using the RevertAid H Minus First Strand cDNA synthesis kit (Fermentas, Glen Burnie, MD). Subsequent PCRs were carried out using the Power SYBR green PCR master kit (Applied Biosystems, Foster City, CA) on the LightCycler 480 (Roche). Primers, whose sequences are available upon request, were designed using Primer Express software.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were conducted as previously described, with minor modifications (11). Briefly, trypsinized cells were cross-linked with 1% paraformaldehyde, lysed in a sodium dodecyl sulfate (SDS) lysis buffer, and sonicated using the Bioruptor (Diagenode, Liège, Belgium) (11). Extracts were precleared by 1 h of incubation with protein A-herring sperm DNA, and immunoprecipitation was performed by overnight incubation at 4°C with the relevant antibody, using an anti-HA antibody as negative control, followed by 1 h of incubation with protein A-herring sperm DNA. Protein-DNA complexes were washed as per standard ChIP techniques. After elution, proteinase K treatment, and reversal of cross-links, DNA fragments were analyzed by real-time PCR with a SYBR Green PCR master kit (Applied Biosystems, Foster City, CA) on the LightCycler 480 (Roche Applied Sciences, Basel, Switzerland). Input DNA was analyzed simultaneously and used for normalization. For further normalization, the signal obtained from a region within the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene located 2,500 bp downstream of the poly(A) site, where RNA polymerase II (Pol II) is known not to be recruited, was used to compensate for possible fluctuations arising during handling. Primer sequences for the κB site within the PLAUR promoter and for the region within the GAPDH gene are available upon request.
Caspase 3/7 activation assays.
Control, wild-type (WT) BCL-3-, or BCL-3 LAV-expressing HaCat cells were seeded into 24-well plates. The next day, the cells were either left untreated or subjected to UV irradiation (56 J/m2) for 5, 15, or 30 min; then they were cultured in DMEM for 24 h. Activation of caspase 3 and caspase 7 (caspase 3/7) was assessed by using the Caspase Glo 3/7 assay kit (Promega, Madison, WI) according to the protocol provided by the manufacturer.
In-gel tryptic digestion and identification by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS-MS).
In-gel digestion was performed by addition of modified trypsin (Promega, Madison, WI) in 50 mM ammonium bicarbonate at 37°C overnight. The tryptic digests were air dried and placed in formic acid (0.1%) for further MS-MS analysis.
Each in-gel digest of an individual band was analyzed by nano-high-performance liquid chromatography (HPLC) electrospray MS-MS using an XCT ion-trap mass spectrometer (Agilent, Santa Clara, CA). The HPLC separations were performed on an RP C18 Zorbax column (150 mm by 75 μm; particle size, 3.5 μm) from Agilent. The mobile phase was a 90-min gradient mixture formed as follows: mixture A, water-acetonitrile-formic acid (97/3/0.1 [vol/vol/vol]); mixture B, acetonitrile-water-formic acid (90/10/0.1 [vol/vol/vol]). The flow rate was 300 nl/min. The collision energy was set automatically depending on the mass of the parent ion. Each MS full scan was followed by MS-MS scans of the first four most intense peaks detected in the prior MS scan. A list of peptide masses was subsequently introduced into the database for protein identification searches using MASCOT (Matrix Sciences).
RESULTS
CtBP and TBLR1 bind BCL-3 through its N-terminal domain.
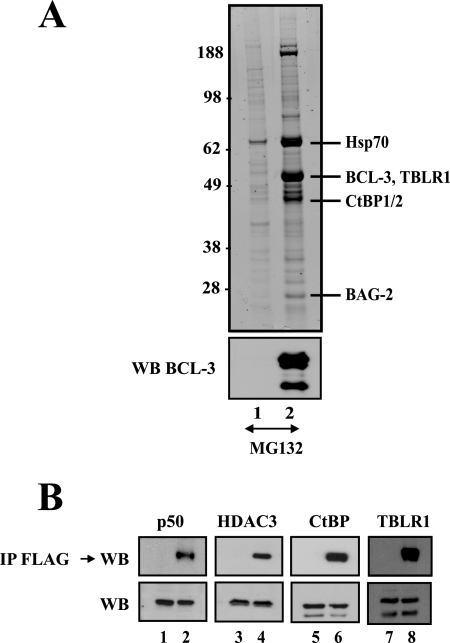
To learn more about BCL-3 function, we initiated a study directed toward identifying its binding partners. Therefore, we created a 293 cell line stably expressing FLAG-BCL-3. This cell line was then treated with MG132, and FLAG-BCL-3 was affinity purified. Copurifying proteins were identified by silver staining and mass spectrometry analysis after separation of BCL-3 complexes by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 1 A). Proteins coprecipitating with BCL-3 included Hsp70, BAG-2, the corepressors CtBP1 and CtBP2, and the TBL1-related protein (TBLR1), initially identified as a component of the nuclear receptor corepressor (N-CoR) complex (Fig. 1A) (48-50). In agreement with previous results, HDAC3 and p50 also bound BCL-3, as evidenced by Western blot analysis performed on the anti-FLAG immunoprecipitates (Fig. 1B). Western blotting with antibodies against CtBP and TBLR1, carried out on those anti-FLAG immunoprecipitates, also confirmed that both proteins are BCL-3-associated proteins (Fig. 1B).
FIG. 1.
Identification of multiple BCL-3-associated proteins through biochemical purification. (A) (Top) Mock-expressing 293 cells or 293 cells stably expressing FLAG-BCL-3, treated with MG132 (20 μM) for 4 h, were subjected to anti-FLAG immunoprecipitation (lanes 1 and 2, respectively). Immunoprecipitated proteins were subsequently released from the beads using the FLAG peptide and were subjected to SDS-PAGE, silver staining, and mass spectrometric analysis for identification. (Bottom) Western blotting (WB) with an antibody against BCL-3 was carried out on the immunoprecipitates. (B) (Top) Western blot analysis of anti-FLAG immunoprecipitates (IP) from MG132-treated mock-expressing or FLAG-BCL-3-expressing cells (odd and even lanes, respectively). Western blotting was carried out with antibodies against p50 and HDAC3 (positive controls) and against CtBP and TBLR1 (identified by mass spectrometry), as indicated, to validate the identifications obtained by mass spectrometry. (Bottom) Western blotting was also carried out with antibodies against p50, HDAC3, CtBP, and TBLR1 by using 5% of the extracts used in immunoprecipitation experiments.
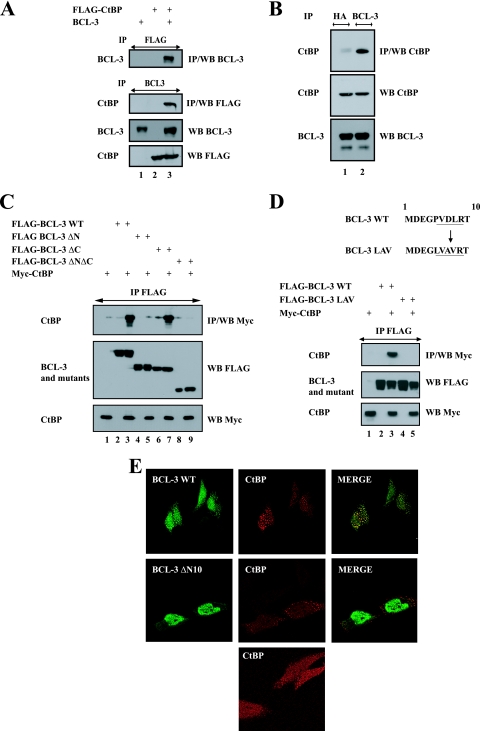
FLAG-CtBP bound overexpressed BCL-3 in 293 cells (Fig. 2 A, top panel, lane 3). Similarly, FLAG-CtBP was also found in the anti-BCL-3 immunoprecipitate (Fig. 2A, second panel from top, lane 3). Importantly, BCL-3 also associated with CtBP at the endogenous level in Karpas cells (Fig. 2B, top panel, lane 2). BCL-3 associates with CtBP independently of its binding to NF-κB proteins, as evidenced by the fact that a BCL-3 mutant that fails to bind to p50 or p52 because of point mutations generated within its ankyrin repeats (BCL-3 ANK M123) still bound CtBP (data not shown). The N-terminal domain of BCL-3 is required for binding to CtBP, as evidenced by the fact that a mutant of BCL-3 that lacks the domain upstream of the ankyrin repeats (BCL-3 ΔN) did not associate with this corepressor, while another mutant, lacking the C-terminal domain downstream of the ankyrin repeats (BCL-3 ΔC), still did (Fig. 2C, top panel, lanes 5 and 7, respectively). Many, if not all, CtBP-interacting proteins harbor a conserved PXDLS motif required for binding to this corepressor (9). Because the first 10 N-terminal amino acids of BCL-3 included a PVDLR sequence from amino acid 5 to 9, we generated a mutant in which this motif was mutated into a LVAVR sequence (BCL-3 LAV) and tested its ability to bind to CtBP. Disruption of this PVDLR motif impaired the binding of BCL-3 to CtBP (Fig. 2D, top panel, lane 5). Thus, BCL-3 binds CtBP through an N-terminal PVDLR sequence. This motif was dispensable for the binding of BCL-3 to HDAC3, even if the entire N-terminal domain was required for this interaction (data not shown). Therefore, BCL-3 binds the corepressors CtBP and HDAC3 through two distinct domains. Of note, strong colocalization of BCL-3 and CtBP was observed upon overexpression of both proteins in HeLa cells (Fig. 2E, top panel on the right). Such colocalization with CtBP was not observed with a mutant lacking the first 10 N-terminal amino acids (BCL-3 ΔN10), which failed to bind to this corepressor (data not shown). BCL-3 actually recruited CtBP to the nucleus in some dots, as evidenced by the fact that CtBP was not as intensively localized in the nucleus when transfected alone (Fig. 2E, bottom panel).
FIG. 2.
BCL-3 interacts with CtBP through its N-terminal PVDLR motif. (A) (Top two panels) Ectopically expressed CtBP interacts with BCL-3. 293 cells were transfected with the indicated expression plasmids, and cell extracts were subjected to immunoprecipitation (IP) with an anti-FLAG or anti-BCL-3 antibody, followed by Western blot analysis (WB) with an anti-BCL-3 or anti-FLAG antibody. (Bottom two panels) Crude cell extracts were subjected to Western blotting with antibodies against BCL-3 and FLAG, as indicated. (B) (Top) Endogenous CtBP and BCL-3 are associated in vivo. Extracts from Karpas cells were subjected to immunoprecipitation with an anti-HA (negative control) or anti-BCL-3 antibody, followed by Western blotting with an antibody against CtBP. (Center and bottom) Cell extracts were also subjected to Western blot analysis with antibodies against CtBP and BCL-3. (C and D) The PVDLR motif located within the first 10 N-terminal amino acid residues of BCL-3 is required for binding to CtBP. (Top) 293 cells were transfected with the indicated expression plasmids, and crude cell extracts were subjected to anti-FLAG immunoprecipitation, followed by Western blot analysis with an anti-Myc antibody. (Center and bottom) Crude cell extracts were also subjected to Western blotting with antibodies against FLAG and Myc. The PVDLR motif of BCL-3 is shown, as well as the mutations within this motif in the BCL-3 LAV mutant. (E) Wild-type BCL-3, but not the BCL-3 ΔN10 mutant, colocalizes with CtBP. Immunofluorescence analysis with an anti-FLAG or anti-Myc antibody was carried out on HeLa cells cotransfected with a Myc-CtBP expression vector and either wild-type FLAG-BCL-3 or the BCL-3 ΔN10 mutant. The panel at the bottom shows the localization of CtBP when transfected alone into HeLa cells.
The NF-κB protein p52 failed to bind Myc-CtBP (Fig. 3 A, top panel, lane 5). However, p52 bound CtBP in the presence of overexpressed BCL-3 (Fig. 3B, top panel, lane 7), which suggests that the latter IκB protein mediates the binding of p52 to CtBP. The existence of a ternary complex that includes BCL-3, p52, and CtBP was demonstrated in 293 cells (Fig. 3B, top panel, lane 6). We next investigated whether other IκB proteins bind CtBP. Bioinformatic analysis indicated that none of them harbored a conserved PXDLS motif, and this was supported by our data showing that overexpressed IκBα, p100, and p105 failed to bind FLAG-CtBP (Fig. 3C, top panel, lane 5, and Fig. 3D, top panel, lanes 6 and 8, respectively). A similar conclusion also applied with endogenous IκBα, IκBβ, IκBɛ, p100, and p105, which failed to bind CtBP, whereas LSD1, used as a positive control, did (Fig. 3E, lane 2). Taken together, our data suggest that BCL-3 is the only tested IκB family member that binds CtBP.
FIG. 3.
BCL-3 is the only IκB protein that binds CtBP. (A) BCL-3 but not p52 binds CtBP in transfected cells. (Top) 293 cells were transfected with the indicated expression plasmids, and anti-FLAG immunoprecipitation (IP), followed by Western blotting (WB) with an anti-CtBP antibody, was carried out with the cell extracts. (Center and bottom) Western blot analyses with antibodies against FLAG and CtBP were also performed with the crude cell extracts. (B) Evidence for a ternary complex that includes p52, BCL-3, and CtBP. (Top panel) 293 cells were transfected with the indicated expression plasmids, and cell lysates were first subjected to anti-FLAG immunoprecipitation. A FLAG peptide was subsequently added in order to release the immunoprecipitates, which were then immunoprecipitated using the anti-BCL-3 antibody (lanes 1 to 6). The anti-FLAG immunoprecipitate from lane 7 was used directly for Western blotting with an anti-Myc antibody to detect Myc-CtBP, as were the anti-BCL-3 immunoprecipitates from lanes 1 to 6. (Center and bottom panels) Crude cell extracts were also subjected to Western blotting as indicated. (C and D) BCL-3, but not p52, p100, p105, or IκBα, binds CtBP. (Top) 293 cells were transfected with the indicated expression plasmids. Immunoprecipitation with an anti-Myc (C) or anti-FLAG (D) antibody was carried out with the cell extracts, followed by Western blotting with an anti-FLAG (C) or anti-HA (D) antibody (top panels). (Center and bottom) Western blot analyses with antibodies against FLAG (C and D), Myc (C), and HA (D) were also performed with the crude cell extracts. (E) Ectopically expressed CtBP binds LSD1 but not IκBα, IκBβ, IκBɛ, p100, or p105. 293 cells were transfected with FLAG-CtBP or an empty FLAG-expressing construct, as indicated (lanes 4 and 3, respectively), and anti-FLAG immunoprecipitation was carried out on the cell extracts. Western blotting using the indicated antibodies was subsequently performed on the immunoprecipitates (lanes 1 and 2) and on the cell extracts (lanes 3 and 4).
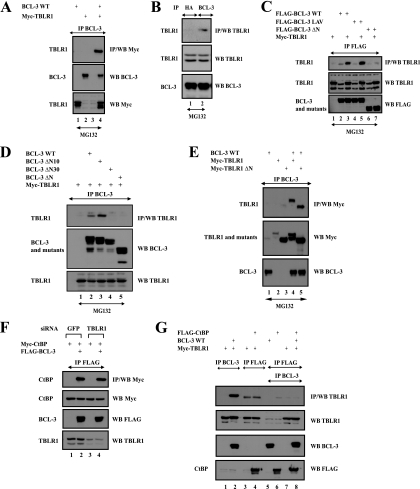
TBLR1 was also identified as a BCL-3-interacting protein (Fig. 1A). BCL-3 bound TBLR1 both in transfected 293 cells and at the endogenous level in Karpas cells (Fig. 4 A, top panel, lane 4, and Fig. 4B, top panel, lane 2, respectively). There again, the N-terminal domain of BCL-3 was required for this association, even if the PVDLR motif was dispensable (Fig. 4C, top panel, lanes 7 and 5, respectively). BCL-3 ΔN10, but not BCL-3 ΔN30 and -ΔN, bound TBLR1 in 293 cells (Fig. 4D, top panel, compare lanes 3 and 5). TBLR1 harbors an N-terminal Lis homology (LisH) motif required for oligomerization, transcriptional repression, and binding to histones H2B and H4 (10, 48). This region was dispensable for binding to BCL-3, as a TBLR1 ΔN mutant lacking the first 100 amino acids, which include this domain, still bound BCL-3 similarly to full-length TBLR1 (Fig. 4E, top panel, lanes 5 and 4, respectively).
FIG. 4.
TBLR1 interacts with BCL-3 through the N-terminal domain but not with CtBP. (A and B) BCL-3 and TBLR1 interact in transfected 293 cells and at the endogenous level. (Top) Cell extracts from 293 (A) or Karpas (B) cells treated with MG132 (20 μM) were subjected to immunoprecipitation (IP) with an anti-HA (B) (negative control) or anti-BCL-3 antibody, followed by Western blot analysis (WB) with an anti-Myc (A) or anti-TBLR1 (B) antibody. (Center and bottom) Crude cell extracts were subjected to Western blotting with antibodies against TBLR1 (B), Myc (A), and BCL-3 (A and B) as well. (C and D) BCL-3 associates with TBLR1 through its N-terminal domain. 293 cells were transfected with the indicated expression plasmids, and cell extracts were subjected to immunoprecipitation and Western blot analysis, as indicated. (E) The N-terminal LisH domain of TBLR1 is dispensable for binding to BCL-3. (Top) 293 cells were transfected with the indicated expression plasmids, and cell extracts were subjected to immunoprecipitation with an anti-BCL-3 antibody, followed by Western blotting with an anti-Myc antibody. (Center and bottom) Cell extracts were also subjected to Western blotting with antibodies against Myc and BCL-3. (F) TBLR1 is dispensable for the association of BCL-3 with CtBP. (Top panel) 293 cells depleted of GFP (negative control) or TBLR1 were transfected with the indicated expression plasmids, and cell extracts were subjected to immunoprecipitation with an anti-FLAG antibody, followed by Western blotting with an anti-Myc antibody. (Center and bottom panels) Cell extracts were subjected to Western blotting with antibodies against Myc, FLAG, and TBLR1 as well. (G) Evidence for distinct CtBP- and TBLR1-containing BCL-3 subcomplexes. (Top panel) 293 cells were transfected with the indicated expression plasmids, and cell lysates were subjected to immunoprecipitation with an anti-BCL-3 (lanes 1 and 2) or anti-FLAG (lanes 3 to 8) antibody. The resulting immunoprecipitates were subsequently subjected to Western blotting with an anti-TBLR1 antibody (lanes 1 to 4) or were released from the beads using a FLAG peptide (lanes 5 to 8). The released materials were subsequently subjected to immunoprecipitation with an anti-BCL-3 antibody followed by Western blotting with an anti-TBLR1 antibody (lanes 5 to 8). (Center and bottom panels) Crude cell extracts were subjected to Western blotting with antibodies against TBLR1, BCL-3, and FLAG as well.
TBLR1 was dispensable for the binding of CtBP to BCL-3, as this association was unchanged in TBLR1-depleted BCL-3-expressing 293 cells (Fig. 4F, top panel, compare lanes 2 and 4). Moreover, TBLR1 was found in the anti-BCL-3 but not in the anti-CtBP immunoprecipitate (Fig. 4G, top panel, lanes 2 and 4, respectively), suggesting the existence of two distinct BCL-3 subcomplexes, one with CtBP and the other with TBLR1. This hypothesis is further supported by the fact that we did not detect any ternary complex that would have included CtBP, BCL-3, and TBLR1 (Fig. 4G, top panel, lane 8). Thus, our data suggest that BCL-3 can form distinct subcomplexes.
A role for CtBP in the repressing abilities of BCL-3 and in its stability.
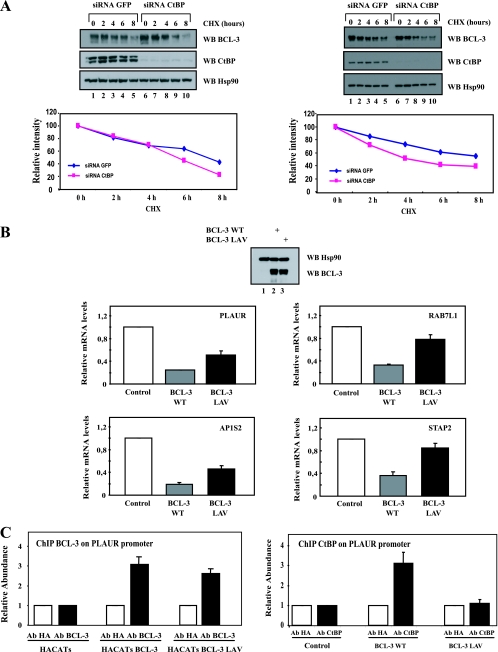
Although CtBP and TBLR1 were defined as proteins associated with BCL-3 through its N-terminal domain, it was unclear how they modulate the transcriptional ability of BCL-3 and/or its stability. To assess the potential role of CtBP in BCL-3 stability, we measured the half-life of BCL-3 in CtBP-depleted cells. CtBP contributes to the stability of BCL-3, as the half-life of BCL-3 was slightly decreased upon CtBP depletion in BCL-3-expressing 293 cells or in Karpas cells (Fig. 5 A, top panels, compare lanes 1 to 5 with lanes 6 to 10). A quantification further supported this observation.
FIG. 5.
CtBP is required for the repressing abilities of BCL-3 and to prevent excessive BCL-3 degradation. (A) CtBP is required for BCL-3 stability. (Top) GFP (negative control)- or CtBP-depleted, BCL-3-expressing 293 (left) or Karpas (right) cells were either left untreated (lanes 1 and 6) or stimulated with cycloheximide (CHX) (50 μg/ml) (lanes 2 to 5 and 7 to 10), and cell extracts were subjected to Western blotting (WB) with antibodies against Hsp90, BCL-3, and CtBP, as indicated. (Bottom) Quantification of BCL-3 levels under control conditions or upon CtBP depletion in BCL-3-expressing 293 cells (left) or in Karpas cells (right). The signal intensity in unstimulated GFP siRNA cells is set to 100%. (B) The N-terminal PVDLR motif is required for the repressing abilities of BCL-3. HaCat cells were infected either with a control lentivirus (negative control) or with a lentivirus expressing WT BCL-3 or BCL-3 LAV. Western blots with antibodies against BCL-3 and Hsp90, performed on extracts from the corresponding experimental conditions in order to ensure comparable levels of WT and mutated BCL-3 products, are shown at the top. Total RNAs from the resulting cells were subjected to real-time PCR in order to assess the mRNA levels of PLAUR, RAB7L1, STAP2, and AP1S2, all genes repressed by WT BCL-3 in transformed keratinocytes (data not shown). The abundance of transcripts in cells infected with the control lentivirus was set to 1, and their levels in cells infected with the other lentivirus were relative to that after normalization with 18S rRNA. Data from three (PLAUR and RAB7L1), four (AP1S2), or five (STAP2) independent experiments (means ± standard deviations) are shown. (C) Chromatin immunoprecipitation assays with an anti-HA (negative control), anti-BCL-3 (left), or anti-CtBP (right) antibody (Ab) were performed using control, WT BCL-3-expressing, or BCL-3 LAV-expressing HaCat cells. Associated DNA was analyzed by real-time PCR using primers derived from the promoter of the PLAUR gene (data not shown). Data from two independent experiments (means ± standard deviations) are shown.
Because CtBP is a corepressor, we next determined whether CtBP modulates the transcriptional potential of BCL-3. We performed microarray analyses in HaCat cells infected with a control lentivirus or a BCL-3-expressing lentivirus in order to identify the genes whose expression is specifically induced or repressed by this IκB protein. We focused our study on the candidates repressed by BCL-3 (data not shown). The levels of several genes, including PLAUR, STAP2, AP1S2, and RAB7L1 (39), were decreased upon BCL-3 overexpression in keratinocytes (data not shown). PLAUR encodes the receptor for urokinase plasminogen activator, whereas RAB7L1 encodes a small GTP binding protein and is a member of the RAS oncogene family-like (40). A link between NF-κB and STAP2, another BCL-3 target gene, was previously established, as this gene encodes a scaffold protein that negatively regulates both canonical and alternative NF-κB-activating pathways by Epstein-Barr virus (EBV)-derived latent membrane protein 1 (LMP1) (19). To address the role of CtBP in the capacity of BCL-3 to repress the expression of these genes, we expressed either a control lentivirus, WT BCL-3, or the BCL-3 LAV mutant in HaCat cells. The two BCL-3 products were expressed at similar levels (Fig. 5B, top panel, compare lanes 2 and 3). We subsequently assessed PLAUR, RAB7L1, STAP2, and AP1S2 mRNA levels. CtBP was required for the repressing abilities of BCL-3, as the BCL-3 LAV mutant did not efficiently repress the expression of all these genes (Fig. 5B). Of note, however, other CtBP-independent mechanisms may account for the repressing potential of BCL-3, as some candidates, such as PLAUR and AP1S2, were still partially repressed in cells expressing BCL-3 LAV, which does not bind to CtBP (Fig. 5B, panels on the left). Therefore, our data suggest that CtBP is a protein required for the repressing abilities of BCL-3. ChIP assay analysis also indicated that BCL-3, BCL-3 LAV, and CtBP were recruited to a κB site (GGGATCCCCT) that we identified within the promoter of PLAUR in vivo (Fig. 5C; also data not shown). The recruitment of CtBP to this κB site was impaired in HaCat cells that overexpressed BCL-3 LAV (Fig. 5C, right). Thus, BCL-3 is required to recruit CtBP on the promoter sequence of PLAUR in vivo, and this mechanism may explain why the BCL-3 LAV mutant, whose recruitment to the PLAUR promoter is very similar to that of WT BCL-3, failed to efficiently repress this gene in keratinocytes.
CoREST and LSD1 are BCL-3-associated proteins.
CtBP is part of a corepressor complex that includes about 50 proteins, such as LSD1, a lysine-specific histone demethylase, and Co-RE1 silencing transcription factor (CoREST) (2, 37, 38). Thus, we next explored whether CoREST and LSD1 also bound BCL-3 by coimmunoprecipitation studies. Ectopically expressed BCL-3 indeed bound CoREST as well as endogenous LSD1 in 293 cells (Fig. 6 A and B, top panels, lanes 2). The PVDLR motif within the first 10 N-terminal amino acids of BCL-3 was required for binding to LSD1 but not to CoREST, as the BCL-3 LAV mutant failed to bind to LSD1 but still interacted with CoREST (Fig. 6B, top panel, lane 3, and Fig. 6A, top panel, lane 4, respectively). Of note, BCL-3 ANK M123 failed to bind LSD1, which indicates that p50 or p52 is required for the association of BCL-3 with LSD1 (Fig. 6B, top panel, lane 9). Importantly, CtBP appears to be required for the binding of BCL-3 to LSD1, as this association was severely compromised in CtBP-depleted 293 cells, while the binding of BCL-3 to p50 remained unchanged upon CtBP depletion (Fig. 6C, top panel and second panel from the top, compare lanes 3 and 4 as well as lanes 5 and 6, respectively).
FIG. 6.
CoREST and LSD1 bind BCL-3 through its N-terminal domain. CtBP and LSD1 play roles in the repressing abilities of BCL-3. (A and B) The PVDLR motif of BCL-3 is required for binding to LSD1 but not to CoREST. (Top) Anti-BCL-3 (A) or anti-FLAG (B) immunoprecipitates (IP) with cell extracts derived from 293 cells transfected with the indicated expression plasmids were subjected to Western blot analysis (WB) with an anti-FLAG (A) or anti-LSD1 (B) antibody. (Center and bottom) Crude cell extracts were subjected to Western blotting with antibodies against BCL-3, LSD1, and FLAG, as indicated. (C) The association of BCL-3 with LSD1 is impaired in CtBP-depleted cells. (Top two panels) 293 cells were first transfected with the indicated siRNA targeting either GFP (negative control) or CtBP, as indicated. The resulting cells were transfected with the indicated expression vectors, and cell extracts were subjected to immunoprecipitation with an anti-FLAG antibody, followed by Western blot analysis with an anti-LSD1 or anti-p50 antibody. (Bottom three panels) Crude cell extracts were subjected to Western blotting with an antibody against LSD1, p50, CtBP, or FLAG, as indicated. (D) CtBP and LSD1 contribute to the repressing abilities of BCL-3. (Top) Control or BCL-3-expressing HaCat cells were transfected with a siRNA targeting GFP (control) (lanes 1 and 2) or with a siRNA targeting CtBP (left) or LSD1 (right) (lanes 3 and 4), and cell extracts were subjected to Western blotting with an antibody against Hsp90, BCL-3, CtBP, or LSD1. (Bottom) Total mRNA levels of PLAUR and RAB7L1 were quantified by real-time PCR under all experimental conditions (control or BCL-3-expressing cells, as indicated). The abundance of each transcript in cells infected with the control lentivirus was set to 1, and their levels under the other experimental conditions were relative to that after normalization with 18S rRNA. Data from three independent experiments (means ± standard deviations) are shown.
To further explore the roles of both CtBP and LSD1 in the repressing abilities of BCL-3, we next depleted either CtBP or LSD1 in control or BCL-3-expressing HaCat cells and assessed the expression of several target genes through real-time PCR. Efficient CtBP or LSD1 depletion was achieved through RNA interference (RNAi) (Fig. 6D, bottom panels, compare lanes 3 and 4 with lanes 1 and 2). Those experiments first confirmed that both PLAUR expression and RAB7L1 expression were indeed repressed upon BCL-3 expression in HaCat cells. Moreover, CtBP depletion efficiently restored the expression of both PLAUR and RAB7L1 in BCL-3-expressing cells, thus confirming the requirement of this corepressor for the ability of BCL-3 to repress gene transcription (Fig. 6D). The results obtained upon LSD1 depletion also suggested a role for this lysine-specific histone demethylase in the repressing abilities of BCL-3, although this role may be more promoter specific, compared to a more general role for CtBP. Indeed, whereas LSD1 depletion almost totally restored levels of RAB7L1 in BCL-3-expressing HaCat cells, levels of PLAUR barely changed upon LSD1 depletion in BCL-3 expressing cells (Fig. 6D). Taken together, our data defined a corepressor, CtBP, and a lysine-specific histone demethylase, LSD1, as proteins involved in the repressing abilities of the oncogenic protein BCL-3.
TBLR1 is involved in BCL-3 degradation through a GSK3-independent pathway.
Because TBLR1 acts as an E3 ligase (33), we next explored whether it was involved in BCL-3 degradation and noticed that the half-life of BCL-3 was increased in TBLR1-depleted Karpas cells (Fig. 7 A, top panel, compare lanes 1 to 5 with lanes 6 to 10). A quantification further confirmed this observation (Fig. 7A, panel at the bottom). To strengthen the notion that TBLR1 is involved in BCL-3 polyubiquitination, we depleted TBLR1 in MG132-treated Karpas cells and looked at polyubiquitinated adducts of BCL-3. TBLR1 depletion impaired endogenous BCL-3 polyubiquitination (Fig. 7B, top panel, compare lanes 2 and 4). Of note, some residual polyubiquitinated adducts of BCL-3 were still detected upon TBLR1 depletion in Karpas cells, indicating that some TBLR1-independent pathways may also be required for BCL-3 polyubiquitination.
FIG. 7.
TBLR1 is required for the polyubiquitination of BCL-3. (A) Extended BCL-3 half-life in TBLR1-depleted cells. (Top) TBLR1-depleted Karpas cells were either left untreated (lanes 1 and 6) or stimulated with cycloheximide (CHX) (50 μg/ml) (lanes 2 to 5 and 7 to 10), and cell extracts were subjected to Western blotting (WB) with an antibody against Hsp90, BCL-3, or TBLR1, as indicated. (Bottom) Quantification of BCL-3 levels under control conditions or upon TBLR1 depletion in Karpas cells. The signal intensity in GFP siRNA-treated Karpas cells was set to 100%. (B) Impaired polyubiquitination (poly-Ub) of BCL-3 upon TBLR1 depletion. Karpas cells transfected with a siRNA against GFP or TBLR1 were stimulated with MG132. (Top two panels) Cell extracts were subjected to immunoprecipitation (IP) with an anti-FLAG (negative control) or anti-BCL-3 antibody, followed by Western blotting with an antibody against ubiquitin (Ub) or BCL-3. (Bottom three panels) Cell extracts were subjected to Western blotting with an antibody against BCL-3, Ub, or TBLR1 as well (bottom panels).
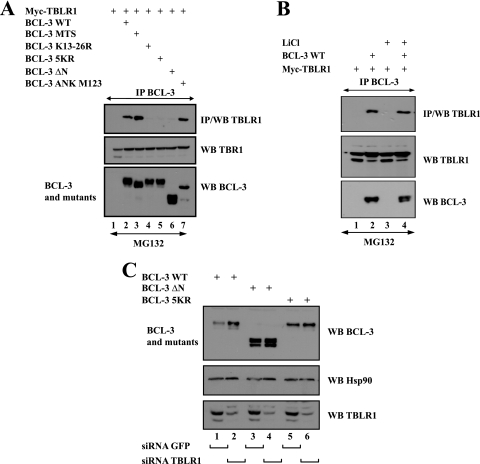
Since lysines 13 and 26 are the residues subjected to the polyubiquitination of BCL-3 (42), we explored whether they were required for the binding of BCL-3 to TBLR1. Whereas WT BCL-3 bound to Myc-TBLR1 in 293 cells, both BCL-3 K13-26R and BCL-3 5KR, where lysine residues were replaced with arginines, did not (Fig. 8 A, top panel, lanes 2, 4, and 5, respectively), suggesting that the N-terminal lysine residues are essential for the binding of BCL-3 to TBLR1. The BCL-3 MTS mutant still bound TBLR1, suggesting that this E3 ligase modulates BCL-3 stability through a GSK3-independent pathway (Fig. 8A, top panel, lane 3). Of note, point mutations within the ankyrin repeats did not prevent the binding of BCL-3 to TBLR1 (Fig. 8A, top panel, lane 7). The notion that TBLR1 targets BCL-3 through a GSK3-independent pathway was further supported by the fact that treatment with LiCl, a GSK3 inhibitor, had no effect on the binding of BCL-3 to TBLR1 in MG132-treated 293 cells transfected with TBLR1 and BCL-3 expression constructs (Fig. 8B, top panel, compare lanes 2 and 4). Importantly, TBLR1 has to bind to BCL-3 in order to destabilize it. Indeed, both the BCL-3 ΔN and 5KR mutants, which do not associate with this E3 ligase (Fig. 8A), were not stabilized in TBLR1 RNAi cells, while the level of WT BCL-3 was increased upon TBLR1 depletion (Fig. 8C, top panel, compare odd with even lanes). Thus, our data suggest that TBLR1 regulates BCL-3 levels by binding to this IκB protein and by promoting its polyubiquitination through a GSK3-independent pathway.
FIG. 8.
BCL-3 requires its N-terminal lysine residues for binding to TBLR1, an E3 ligase acting through a GSK3-independent pathway. (A) The N-terminal domain of BCL-3, which includes lysines 13 and 26, is required for binding to TBLR1. (Top) 293 cells were transfected with the indicated expression plasmids, and immunoprecipitation (IP) with an anti-BCL-3 antibody was carried out on the cell extracts, followed by Western blotting (WB) with an anti-TBLR1 antibody. (Center and bottom) Western blotting with an antibody against TBLR1 or BCL-3 was performed on the crude cell extracts as well. (B) GSK3 inhibition through LiCl treatment does not affect the binding of BCL-3 to TBLR1. 293 cells were transfected with the indicated expression plasmids and were subsequently treated with MG132. (Top) Cell extracts were subjected to immunoprecipitation with an anti-BCL-3 antibody, followed by Western blotting with an anti-TBLR1 antibody. (Center and bottom) Cell extracts were also subjected to Western blotting with an antibody against TBLR1 or BCL-3. (C) WT BCL-3, but not the ΔN or 5KR mutant, is stabilized in TBLR1-depleted cells. 293 cells were transfected with a siRNA targeting TBLR1 or GFP (negative control) as indicated. Cells were subsequently transfected with the indicated expression plasmids, and Western blotting with an antibody against BCL-3, Hsp90, or TBLR1 was performed on the cell extracts.
To learn more about the role of TBLR1 in the regulation of the transcriptional potential of BCL-3, we assessed the consequences of TBLR1 depletion for the levels of several genes repressed by BCL-3 in HaCat cells, namely, PLAUR, RAB7L1, and STAP2. TBLR1 depletion efficiently restored the expression levels of these genes in BCL-3-expressing cells, suggesting that the polyubiquitination of BCL-3 by TBLR1 is required for its repressing abilities (Fig. 9 A). Such a hypothesis was further supported by the fact that the BCL-3 K13-26R mutant, which failed to be polyubiquitinated, did not repress PLAUR or RAB7L1 expression any more (Fig. 9B). Taken together, our data indicate that TBLR1 not only is required for BCL-3 polyubiquitination but also regulates the repressing abilities of this oncogenic protein.
FIG. 9.
TBLR1 contributes to the repressing abilities of BCL-3. (A) The repressing abilities of BCL-3 on the expression of PLAUR, RAB7L1, and STAP2 are abolished in TBLR1-depleted cells. (Top) Control or BCL-3-expressing HaCat cells were transfected with a siRNA targeting GFP (control) or TBLR1, as indicated, and cell extracts were subjected to Western blotting (WB) with an antibody against BCL-3, TBLR1, or Hsp90, as indicated. (Bottom) Total RNAs from the resulting cells were subjected to real-time PCR to assess the mRNA levels of PLAUR, RAB7L1, and STAP2. The abundance of each transcript in control cells infected with the control lentivirus was set to 1, and their levels under the other experimental conditions were relative to that after normalization with 18S rRNA. Data from three independent experiments (means ± standard deviations) are shown. (B) Lysines 13 and 26 of BCL-3 are required for its repressing potential. (Top) Western blotting with antibodies against Hsp90 and BCL-3 was performed on extracts from HaCat cells expressing either the control lentivirus, WT BCL-3, or the BCL-3 K13-26R mutant, as indicated. (Bottom) PLAUR and RAB7L1 mRNA levels were assessed by real-time PCR. Data from five independent experiments (means ± standard deviations) are shown.
CtBP is required for the oncogenic potential of BCL-3.
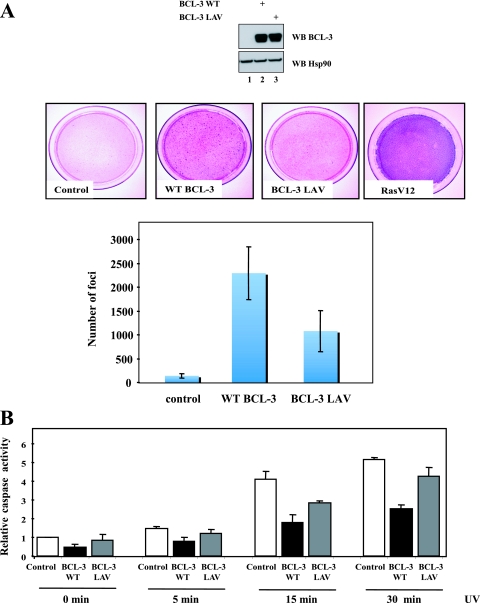
Although our results defined CtBP as a candidate required for the repressing abilities of BCL-3 and for its stability, it was not clear whether this corepressor had any role in the oncogenic potential of BCL-3. We addressed this issue by assessing the ability of WT BCL-3 or the BCL-3 LAV mutant to form colonies when expressed in NIH 3T3 cells through retroviral transductions. The two BCL-3 products were expressed at similar levels in these cells (Fig. 10 A, top panel, lanes 2 and 3). In agreement with previous results (42), BCL-3 formed colonies when expressed in NIH 3T3 cells, but not as strongly as RasV12, used as a positive control (Fig. 10A). Interestingly, colony formation was impaired in NIH 3T3 cells that expressed the LAV mutant, indicating that the binding of BCL-3 to CtBP is required for its oncogenic potential in vitro (Fig. 10A). BCL-3 actually promotes cell proliferation through cyclin D1 induction in keratinocytes and also inhibits cell apoptosis by inducing the expression of HDM2, which acts as an E3 ligase of p53 (20, 25). Therefore, we explored whether the ability of BCL-3 to prevent apoptosis in HaCat cells required CtBP. To address this issue, control, WT BCL-3-expressing, or BCL-3 LAV-expressing cells were either left unstimulated or subjected to UV treatment, and caspase 3/7 activation was subsequently assessed. As expected, BCL-3 expression in HaCat cells inhibited cell apoptosis by preventing caspase 3/7 activation (Fig. 10B). Interestingly, the BCL-3 LAV mutant failed to efficiently inhibit caspase 3/7 upon UV treatment, indicating that the binding of BCL-3 to CtBP is required for its ability to prevent cell apoptosis (Fig. 10B). Taken together, our data defined CtBP as a key molecule that mediates the oncogenic potential of BCL-3.
FIG. 10.
CtBP is required for the transforming potential of BCL-3 and also for its ability to inhibit caspase 3/7 activation in UV-treated transformed keratinocytes. (A) (Top) Extracts from NIH 3T3 cells infected with a control pBabe retrovirus or a with a pBabe construct expressing WT BCL-3 or BCL-3 LAV were subjected to Western blotting (WB) with antibodies against BCL-3 and Hsp90, as indicated. (Center and bottom) Focus formation assays with NIH 3T3 cells infected with the control vector, the BCL-3 vector, the LAV mutant, or RasV12, used as a positive control. Focus formation was visualized after coloration with Giemsa stain. Foci were quantified using ImageJ software, in a binary mode (black and white), from 4 dishes per experimental condition (two distinct infections performed in duplicate for each experimental condition). The histogram shows the number of foci per dish (means ± standard deviations). (B) Inhibition of UV-mediated cell apoptosis by BCL-3 relies on its interaction with CtBP. Control, WT BCL-3-expressing, or BCL-3 LAV-expressing HaCat cells were either left unstimulated or treated with UV radiation for the indicated times, and caspase 3/7 activities were quantified and plotted. Caspase 3/7 activities in unstimulated cells infected with the control lentivirus were set to 1, and their levels under the other experimental conditions were relative to that after normalization of caspase activities with protein concentrations. Data from three independent experiments (means ± standard deviations) are shown.
DISCUSSION
We report the identification of multiple BCL-3-associated proteins that are critical for the degradation of BCL-3, for its ability to repress the expression of several genes in keratinocytes, and for its oncogenic potential. Those interactions require the N-terminal domain of BCL-3, which therefore acts as a key element for the regulation of BCL-3 activity. We identified a PVDLR motif within the first 10 N-terminal amino acids of BCL-3 that is required for binding to the corepressor CtBP. Mutation of this motif has dramatic consequences for BCL-3 functions: it abolishes the ability of BCL-3 to repress the expression of several genes in keratinocytes and also severely impairs the oncogenic potential of BCL-3, at least by interfering with its capacity to prevent UV-mediated cell apoptosis. Because this motif is absent in the other NF-κB/IκB proteins that we tested, it is not surprising to see that those candidates failed to bind to CtBP. Thus, CtBP can be defined as a corepressor that specifically modulates the activity of a single IκB protein rather than as a candidate that universally regulates NF-κB activation through binding to various members of this family. We demonstrated that CtBP is a key factor for the recruitment of additional BCL-3-interacting proteins, such as LSD1. Still, the recruitment of all BCL-3-interacting proteins via the N-terminal domain of BCL-3 does not exclusively rely on CtBP; other transcriptional corepressors, such as HDAC3, bind to BCL-3 through an N-terminal region distinct from the PVDLR motif. This observation may explain why the repressing abilities of BCL-3 do not rely exclusively on CtBP and LSD1. Indeed, the BCL-3 LAV mutant still harbors some residual repressing abilities when recruited to PLAUR or AP1S2 promoters, for example (Fig. 5B). Therefore, the repressing abilities of BCL-3 actually involve the recruitment of histone deacetylases, such as HDAC3, as well as CtBP-associated K9-histone H3 methylase and LSD1 demethylase activities (38), through distinct subregions within the N-terminal domain of BCL-3.
Our data also indicate that CtBP protects BCL-3 from excessive degradation. This may be due to the ability of CtBP to constitutively bind BCL-3, similarly to NF-κB proteins p50 and p52, which also stabilize BCL-3 through constitutive interactions with the ankyrin repeats of this oncogenic protein.
BCL-3 acts as an oncogenic protein through multiple mechanisms that include the induction of cyclin D1 expression in keratinocytes and also the limiting of cell apoptosis through induction of the E3 ligase of p53, HDM2 (20, 25). In support of the latter mechanism, we show here that BCL-3 limits cell apoptosis in UV-stimulated keratinocytes. Interestingly, this pathway relies critically on CtBP, as a BCL-3 mutant lacking the CtBP-interacting motif did not efficiently prevent UV-mediated apoptosis in HaCat cells. Our data are in agreement with previous studies defining CtBP as a protein that promotes cell survival and tumorigenesis by suppressing the expression of proapoptotic candidates, many of which are p53-dependent genes (3, 18). Our results therefore indicate that the oncogenic potential of BCL-3 also results from its ability to repress gene transcription, although the identities of BCL-3 target genes that specifically mediate this function remain to be extensively characterized. Among them is STAP2, which encodes a scaffold protein that limits EBV-mediated NF-κB activation through both the canonical and the alternative pathways (19). Therefore, this finding indicates that the oncogenic protein BCL-3 positively regulates both canonical and alternative NF-κB-activating pathways by repressing the expression of an inhibitory protein, namely, STAP2.
The mechanisms underlying BCL-3 degradation remain poorly characterized. We previously identified a GSK3-dependent degradative pathway that involves the phosphorylation of two C-terminal residues. We hypothesized that constitutive Akt activity, seen in many hematological malignancies, may ultimately contribute to the increased stability of BCL-3 through GSK3 inhibition (42). Elevated levels of BCL-3 in the nucleus can also be the result of loss-of-function mutations targeting CYLD (25). Similarly, human papillomavirus (HPV)-positive cancers, such as cervical and head and neck malignancies, also have elevated levels of nuclear BCL-3, because the HPV-encoded E6 protein inactivates CYLD under hypoxic conditions (1). It is also tempting to speculate that loss-of-function mutations in E3 ligases that target BCL-3 would cause accumulation of this protein. These results may thus extend the pathological contexts that lead to elevated expression of BCL-3 in cancer.
Many proteins whose GSK3-dependent phosphorylation triggers their subsequent degradation bind to the E3 ligase FBW7 for their K48-linked polyubiquitination. FBW7 substrates, such as c-Myc, c-Jun, Notch1, cyclin D1, and cyclin E, share a so-called Cdc4 phospho-degron (CPD) (28, 45) that we actually found on BCL-3 (A. Keutgens et al., unpublished results). Yet FBW7 does not strongly bind phosphorylated BCL-3, and the half-life of BCL-3 is not enhanced in FBW7-depleted cells (Keutgens et al., unpublished). Thus, the E3 ligase that polyubiquitinates BCL-3 upon phosphorylation by GSK3 remains unknown. Nevertheless, we show here that the E3 ligase TBLR1 is a BCL-3-associated protein and regulates BCL-3 stability by promoting its polyubiquitination in a GSK3-independent pathway. Interestingly, TBLR1 was previously described as an exchange factor that mediates “derepression” by promoting the polyubiquitination and degradation of the N-CoR/SMRT corepressor complex (33, 34). As a result, and even if this protein was initially described as a component of the N-CoR corepressor complex (48-50), TBLR1 promotes transcriptional activation by mediating the recruitment of the ubiquitin-proteasome complex to specific target genes (33). In support of this conclusion, the activation of all NF-κB target genes upon TNF-α stimulation is abolished in TBLR1-deficient cells (33). Therefore, TBLR1 positively regulates the activation of the TNF-α- and NF-κB-dependent genes.
Our data establish an intimate connection between the polyubiquitination of BCL-3 and the regulation of its transcription potential. Indeed, we show that the ability of BCL-3 to repress PLAUR, RAB7L1, and STAP2 expression in HaCat cells is abolished upon TBLR1 depletion. We also show that the BCL-3 mutant that failed to be polyubiquitinated and to interact with TBLR1 also failed to repress gene transcription. Therefore, our data suggest that polyubiquitinated BCL-3 may actually repress gene transcription just before being degraded. This concept is in agreement with a previous study which demonstrated that the polyubiquitination of the coactivator SRC-3 confers a transcription activation function prior to promoting its degradation (47).
We show here that TBLR1 is required for BCL-3 degradation through a GSK3-independent pathway, which means that more than one pathway triggers BCL-3 degradation, as reported for other oncogenic proteins. Interestingly, the contribution of the two degradative pathways to the oncogenic potential of BCL-3 appears to be distinct. Indeed, while a BCL-3 mutant that fails to be degraded because of point mutations of its GSK3 phosphorylated sites turned out to be more oncogenic in vitro and in xenograft studies, another BCL-3 mutant that does not bind TBLR1 (i.e., the K13-26R mutant) formed as many colonies as WT BCL-3 in soft agar, even if these colonies were slightly bigger (42). Why and how two distinct degradative pathways can differentially regulate the oncogenic potential of BCL-3 remains an open issue that certainly deserves further investigation. Knowing how critical the level of BCL-3 is for the regulation of its oncogenic potential, resolution of these issues would be essential for better understanding of the biology of BCL-3. Moreover, beside TBLR1, other E3 ligases may physically interact with BCL-3 in order to trigger its degradative polyubiquitination. Future studies will be dedicated to the extensive characterization of the E3 ligases that promote BCL-3 polyubiquitination.
Acknowledgments
This work was supported by grants from the FNRS, TELEVIE, the Belgian Federation against Cancer, the King Baudouin Foundation, the University of Liege (Concerted Research Action Program [04/09-323] and Fonds Spéciaux), the Inter-University Attraction Pole 6/12 (Federal Ministry of Science), the Centre Anti-Cancéreux, and the Leon Fredericq Foundation (ULg).
We are grateful to L. van Parijs, J. Mymryk, E. Olson, G. Gill, and E. Dejardin for the gifts of the pLL3.7 lentivirus, CtBP, CoREST, p52, p100, and p105 expression constructs. A.K. is a research assistant, P.C. and P.V. are senior research assistants, A.C. is a senior research associate, and K.S. is a temporary postdoctoral researcher at the Belgian National Funds for Scientific Research (FNRS).
Footnotes
Published ahead of print on 14 June 2010.
REFERENCES
- 1.An, J., D. Mo, H. Liu, M. S. Veena, E. S. Srivatsan, R. Massoumi, and M. B. Rettig. 2008. Inactivation of the CYLD deubiquitinase by HPV E6 mediates hypoxia-induced NF-κB activation. Cancer Cell 14:394-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andres, M. E., C. Burger, M. J. Peral-Rubio, E. Battaglioli, M. E. Anderson, J. Grimes, J. Dallman, N. Ballas, and G. Mandel. 1999. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. U. S. A. 96:9873-9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergman, L. M., and J. P. Blaydes. 2006. C-terminal binding proteins: emerging roles in cell survival and tumorigenesis. Apoptosis 11:879-888. [DOI] [PubMed] [Google Scholar]
- 4.Bignell, G. R., W. Warren, S. Seal, M. Takahashi, E. Rapley, R. Barfoot, H. Green, C. Brown, P. J. Biggs, S. R. Lakhani, C. Jones, J. Hansen, E. Blair, B. Hofmann, R. Siebert, G. Turner, D. G. Evans, C. Schrander-Stumpel, F. A. Beemer, A. van Den Ouweland, D. Halley, B. Delpech, M. G. Cleveland, I. Leigh, J. Leisti, and S. Rasmussen. 2000. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25:160-165. [DOI] [PubMed] [Google Scholar]
- 5.Bours, V., G. Franzoso, V. Azarenko, S. Park, T. Kanno, K. Brown, and U. Siebenlist. 1993. The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA-binding p50B homodimers. Cell 72:729-739. [DOI] [PubMed] [Google Scholar]
- 6.Brenne, A. T., U. M. Fagerli, J. D. Shaughnessy, Jr., T. K. Vatsveen, T. B. Ro, H. Hella, F. Zhan, B. Barlogie, A. Sundan, M. Borset, and A. Waage. 2009. High expression of BCL3 in human myeloma cells is associated with increased proliferation and inferior prognosis. Eur. J. Haematol. 82:354-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canoz, O., G. Z. Rassidakis, J. H. Admirand, and L. J. Medeiros. 2004. Immunohistochemical detection of BCL-3 in lymphoid neoplasms: a survey of 353 cases. Mod. Pathol. 17:911-917. [DOI] [PubMed] [Google Scholar]
- 8.Carmody, R. J., Q. Ruan, S. Palmer, B. Hilliard, and Y. H. Chen. 2007. Negative regulation of toll-like receptor signaling by NF-κB p50 ubiquitination blockade. Science 317:675-678. [DOI] [PubMed] [Google Scholar]
- 9.Chinnadurai, G. 2007. Transcriptional regulation by C-terminal binding proteins. Int. J. Biochem. Cell Biol. 39:1593-1607. [DOI] [PubMed] [Google Scholar]
- 10.Choi, H. K., K. C. Choi, H. B. Kang, H. C. Kim, Y. H. Lee, S. Haam, H. G. Park, and H. G. Yoon. 2008. Function of multiple Lis-homology domain/WD-40 repeat-containing proteins in feed-forward transcriptional repression by silencing mediator for retinoic and thyroid receptor/nuclear receptor corepressor complexes. Mol. Endocrinol. 22:1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Close, P., N. Hawkes, I. Cornez, C. Creppe, C. A. Lambert, B. Rogister, U. Siebenlist, M. P. Merville, S. A. Slaugenhaupt, V. Bours, J. Q. Svejstrup, and A. Chariot. 2006. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell 22:521-531. [DOI] [PubMed] [Google Scholar]
- 12.Cogswell, P. C., D. C. Guttridge, W. K. Funkhouser, and A. S. Baldwin, Jr. 2000. Selective activation of NF-κB subunits in human breast cancer: potential roles for NF-κB2/p52 and for Bcl-3. Oncogene 19:1123-1131. [DOI] [PubMed] [Google Scholar]
- 13.Dechend, R., F. Hirano, K. Lehmann, V. Heissmeyer, S. Ansieau, F. G. Wulczyn, C. Scheidereit, and A. Leutz. 1999. The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene 18:3316-3323. [DOI] [PubMed] [Google Scholar]
- 14.Dejardin, E., V. Deregowski, M. Chapelier, N. Jacobs, J. Gielen, M. P. Merville, and V. Bours. 1999. Regulation of NF-κB activity by IκB-related proteins in adenocarcinoma cells. Oncogene 18:2567-2577. [DOI] [PubMed] [Google Scholar]
- 15.Franzoso, G., L. Carlson, L. Poljak, E. W. Shores, S. Epstein, A. Leonardi, A. Grinberg, T. Tran, T. Scharton-Kersten, M. Anver, P. Love, K. Brown, and U. Siebenlist. 1998. Mice deficient in nuclear factor (NF)-κB/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J. Exp. Med. 187:147-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franzoso, G., L. Carlson, T. Scharton-Kersten, E. W. Shores, S. Epstein, A. Grinberg, T. Tran, E. Shacter, A. Leonardi, M. Anver, P. Love, A. Sher, and U. Siebenlist. 1997. Critical roles for the Bcl-3 oncoprotein in T cell-mediated immunity, splenic microarchitecture, and germinal center reactions. Immunity 6:479-490. [DOI] [PubMed] [Google Scholar]
- 17.Fujita, T., G. P. Nolan, H.-C. Liou, M. L. Scott, and D. Baltimore. 1993. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-κB p50 homodimers. Genes Dev. 7:1354-1363. [DOI] [PubMed] [Google Scholar]
- 18.Grooteclaes, M., Q. Deveraux, J. Hildebrand, Q. Zhang, R. H. Goodman, and S. M. Frisch. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. U. S. A. 100:4568-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda, O., Y. Sekine, T. Yasui, K. Oritani, K. Sugiyma, R. Muromoto, N. Ohbayashi, A. Yoshimura, and T. Matsuda. 2008. STAP-2 negatively regulates both canonical and noncanonical NF-κB activation induced by Epstein-Barr virus-derived latent membrane protein 1. Mol. Cell. Biol. 28:5027-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashatus, D., P. Cogswell, and A. S. Baldwin. 2006. Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev. 20:225-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwata, H., Y. Watanabe, H. Miyoshi, M. Yamamoto, T. Kaisho, K. Takeda, and S. Akira. 2003. IL-10-inducible Bcl-3 negatively regulates LPS-induced TNF-α production in macrophages. Blood 102:4123-4129. [DOI] [PubMed] [Google Scholar]
- 22.Lee, D. F., H. P. Kuo, M. Liu, C. K. Chou, W. Xia, Y. Du, J. Shen, C. T. Chen, L. Huo, M. C. Hsu, C. W. Li, Q. Ding, T. L. Liao, C. C. Lai, A. C. Lin, Y. H. Chang, S. F. Tsai, L. Y. Li, and M. C. Hung. 2009. KEAP1 E3 ligase-mediated downregulation of NF-κB signaling by targeting IKKβ. Mol. Cell 36:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung, T. H., A. Hoffmann, and D. Baltimore. 2004. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell 118:453-464. [DOI] [PubMed] [Google Scholar]
- 24.Lin, D. I., O. Barbash, K. G. Kumar, J. D. Weber, J. W. Harper, A. J. Klein-Szanto, A. Rustgi, S. Y. Fuchs, and J. A. Diehl. 2006. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-αB crystallin) complex. Mol. Cell 24:355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massoumi, R., K. Chmielarska, K. Hennecke, A. Pfeifer, and R. Fassler. 2006. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell 125:665-677. [DOI] [PubMed] [Google Scholar]
- 26.Mathas, S., K. Johrens, S. Joos, A. Lietz, F. Hummel, M. Janz, F. Jundt, I. Anagnostopoulos, K. Bommert, P. Lichter, H. Stein, C. Scheidereit, and B. Dorken. 2005. Elevated NF-κB p50 complex formation and Bcl-3 expression in classical Hodgkin, anaplastic large-cell, and other peripheral T-cell lymphomas. Blood 106:4287-4293. [DOI] [PubMed] [Google Scholar]
- 27.McKeithan, T. W., J. D. Rowley, T. B. Shows, and M. O. Diaz. 1987. Cloning of the chromosome translocation breakpoint junction of the t(14;19) in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 84:9257-9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minella, A. C., M. Welcker, and B. E. Clurman. 2005. Ras activity regulates cyclin E degradation by the Fbw7 pathway. Proc. Natl. Acad. Sci. U. S. A. 102:9649-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishikori, M., Y. Maesako, C. Ueda, M. Kurata, T. Uchiyama, and H. Ohno. 2003. High-level expression of BCL3 differentiates t(2;5)(p23;q35)-positive anaplastic large cell lymphoma from Hodgkin disease. Blood 101:2789-2796. [DOI] [PubMed] [Google Scholar]
- 30.Ohno, H., G. Takimoto, and T. W. McKeithan. 1990. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell 60:991-997. [DOI] [PubMed] [Google Scholar]
- 31.O'Neil, B. H., P. Buzkova, H. Farrah, D. Kashatus, H. Sanoff, R. M. Goldberg, A. S. Baldwin, and W. K. Funkhouser. 2007. Expression of nuclear factor-κB family proteins in hepatocellular carcinomas. Oncology 72:97-104. [DOI] [PubMed] [Google Scholar]
- 32.Paxian, S., H. Merkle, M. Riemann, M. Wilda, G. Adler, H. Hameister, S. Liptay, K. Pfeffer, and R. M. Schmid. 2002. Abnormal organogenesis of Peyer's patches in mice deficient for NF-κB1, NF-κB2, and Bcl-3. Gastroenterology 122:1853-1868. [DOI] [PubMed] [Google Scholar]
- 33.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511-526. [DOI] [PubMed] [Google Scholar]
- 34.Perissi, V., C. Scafoglio, J. Zhang, K. A. Ohgi, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 2008. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol. Cell 29:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert, I., M. Aussems, A. Keutgens, X. Zhang, B. Hennuy, P. Viatour, G. Vanstraelen, M. P. Merville, J. P. Chapelle, L. de Leval, F. Lambert, E. Dejardin, A. Gothot, and A. Chariot. 2009. Matrix metalloproteinase-9 gene induction by a truncated oncogenic NF-κB2 protein involves the recruitment of MLL1 and MLL2 H3K4 histone methyltransferase complexes. Oncogene 28:1626-1638. [DOI] [PubMed] [Google Scholar]
- 36.Schwarz, E. M., P. Krimpenfort, A. Berns, and I. M. Verma. 1997. Immunological defects in mice with a targeted disruption in Bcl-3. Genes Dev. 11:187-197. [DOI] [PubMed] [Google Scholar]
- 37.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 38.Shi, Y., J. Sawada, G. Sui, B. Affar el, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, Y. Nakatani, and Y. Shi. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu, F., T. Katagiri, M. Suzuki, T. K. Watanabe, S. Okuno, Y. Kuga, M. Nagata, T. Fujiwara, Y. Nakamura, and E. Takahashi. 1997. Cloning and chromosome assignment to 1q32 of a human cDNA (RAB7L1) encoding a small GTP-binding protein, a member of the RAS superfamily. Cytogenet. Cell Genet. 77:261-263. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu, F., T. K. Watanabe, S. Okuno, Y. Omori, T. Fujiwara, E. Takahashi, and Y. Nakamura. 1997. Isolation of a novel human cDNA (rhoHP1) homologous to rho genes. Biochim. Biophys. Acta 1351:13-16. [DOI] [PubMed] [Google Scholar]
- 41.Thornburg, N. J., R. Pathmanathan, and N. Raab-Traub. 2003. Activation of nuclear factor-κB p50 homodimer/Bcl-3 complexes in nasopharyngeal carcinoma. Cancer Res. 63:8293-8301. [PubMed] [Google Scholar]
- 42.Viatour, P., E. Dejardin, M. Warnier, F. Lair, E. Claudio, F. Bureau, J. C. Marine, M. P. Merville, U. Maurer, D. Green, J. Piette, U. Siebenlist, V. Bours, and A. Chariot. 2004. GSK3-mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol. Cell 16:35-45. [DOI] [PubMed] [Google Scholar]
- 43.Viatour, P., S. Legrand-Poels, C. van Lint, M. Warnier, M. P. Merville, J. Gielen, J. Piette, V. Bours, and A. Chariot. 2003. Cytoplasmic IκBα increases NF-κB-independent transcription through binding to histone deacetylase (HDAC) 1 and HDAC3. J. Biol. Chem. 278:46541-46548. [DOI] [PubMed] [Google Scholar]
- 44.Wei, W., J. Jin, S. Schlisio, J. W. Harper, and W. G. Kaelin, Jr. 2005. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8:25-33. [DOI] [PubMed] [Google Scholar]
- 45.Welcker, M., and B. E. Clurman. 2008. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8:83-93. [DOI] [PubMed] [Google Scholar]
- 46.Wessells, J., M. Baer, H. A. Young, E. Claudio, K. Brown, U. Siebenlist, and P. F. Johnson. 2004. BCL-3 and NF-κB p50 attenuate lipopolysaccharide-induced inflammatory responses in macrophages. J. Biol. Chem. 279:49995-50003. [DOI] [PubMed] [Google Scholar]
- 47.Wu, R. C., Q. Feng, D. M. Lonard, and B. W. O'Malley. 2007. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell 129:1125-1140. [DOI] [PubMed] [Google Scholar]
- 48.Yoon, H. G., D. W. Chan, Z. Q. Huang, J. Li, J. D. Fondell, J. Qin, and J. Wong. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22:1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon, H. G., Y. Choi, P. A. Cole, and J. Wong. 2005. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 25:324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, X., H. Wang, E. Claudio, K. Brown, and U. Siebenlist. 2007. A role for the IκB family member Bcl-3 in the control of central immunologic tolerance. Immunity 27:438-452. [DOI] [PMC free article] [PubMed] [Google Scholar]