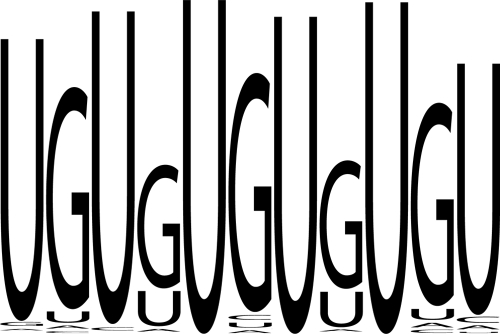Abstract
CUG-repeat binding protein 1 (CUGBP1) mediates selective mRNA decay by binding to GU-rich elements (GREs) containing the sequence UGUUUGUUUGU found in the 3′ untranslated region (UTR) of short-lived transcripts. We used an anti-CUGBP1 antibody to immunoprecipitate CUGBP1 from HeLa cytoplasmic extracts and analyzed the associated transcripts using oligonucleotide microarrays. We identified 613 putative mRNA targets of CUGBP1 and found that the UGUUUGUUUGU GRE sequence and a GU-repeat sequence were both highly enriched in the 3′ UTRs of these targets. We showed that CUGBP1 bound specifically to the GU-repeat sequence and that insertion of this sequence into the 3′ UTR of a beta-globin reporter transcript conferred instability to the transcript. Based on these results, we redefined the GRE to include this GU-repeat sequence. Our results suggest that CUGBP1 coordinately regulates the mRNA decay of a network of transcripts involved in cell growth, cell motility, and apoptosis.
RNA-binding proteins that regulate gene expression at posttranscriptional levels do not usually act on a single target transcript but coordinately regulate multiple transcripts, creating regulatory networks or regulons that are defined by RNA-binding proteins and their target transcripts. Regulons integrate intrinsic and extrinsic signals to coordinately modulate gene expression to regulate distinct cellular processes. The CUG-repeat binding protein 1 (CUGBP1) regulon coordinately regulates the expression of multiple genes at posttranscriptional levels. CUGBP1, a member of the CELF (CUGBP and embryonic lethal abnormal vision-like factor) family of RNA-binding proteins, was first identified as a protein that binds the CUG-repeat sequences of the myotonin protein kinase (25). In subsequent studies, CUGBP1 was shown to be multifunctional, regulating many posttranscriptional processes including alternative splicing, deadenylation, mRNA decay, and translation (reviewed in reference 29). For example, alternative splicing events and translational control in muscle development are steered by the action of CUGBP1 (4, 6, 9, 20, 26). In Xenopus laevis embryonal development, the CUGBP1 homologue embryo deadenylation element-binding protein (EDEN-BP) regulates translational repression in oocytes and deadenylation of maternal RNAs in fertilized eggs (18). Knockout of ETR1, the CUGBP1 homologue in Caenorhabditis elegans, is lethal and impairs muscle development (12). Knockout of CUGBP1 in mice is lethal in most cases, but the few mice that are born display severe fertility defects (8). Thus, the CUGBP1 regulon coordinates gene expression at multiple posttranscriptional levels.
To fulfill all these different functions, CUGBP1 interacts with different RNAs in different contexts, and several different RNA target sequences have been proposed. In translational regulation, CUGBP1 binds to a CUG/CCG sequence in the 5′ untranslated region (UTR) of the CCAAT/enhancer-binding protein β (C/EBPβ) to produce a different isoform of C/EBPβ (27). CUGBP1 also binds to the AU-rich element of tumor necrosis factor alpha (TNF-α) mRNA (13) and the GC-rich sequence in the 5′ UTR of p21 mRNA (5). In X. laevis oocytes EDEN-BP interacts with a U(A/G)-repeat mRNA sequence, leading to rapid deadenylation and translational activation (18).
Recently, the sequence UGUUUGUUUGU, referred to as a GU-rich element (GRE), was identified as a CUGBP1 consensus binding sequence that mediated rapid mRNA decay. This UGUUUGUUUGU consensus sequence was significantly enriched in 3′ UTRs of unstable mRNAs expressed in primary human T cells and functioned as an mRNA decay element when inserted into the 3′ UTR of reporter transcripts through a mechanism that depended on binding by CUGBP1 (30). In X. laevis, similar GU-rich sequence elements are targets of EDEN-BP, (2), suggesting that this RNA-binding interaction was conserved over evolution. A GU-repeat sequence was also established as a CUGBP1 binding motif by yeast three-hybrid screens (22), a SELEX (systematic evolution of ligands by exponential enrichment) approach (10), in vitro binding studies (14), and nuclear magnetic resonance (NMR) structure analysis (28). Although it has been known for several years that the GU-repeat sequence binds to CUGBP1, a biological consequence of this binding has not been demonstrated.
Although the RNA-binding activity and posttranscriptional regulatory functions of CUGBP1 have been characterized, only a limited number of human CUGBP1 target transcripts have been identified. Since CUGBP1 appears to define an evolutionarily conserved posttranscriptional regulatory network that coordinates gene expression in human cells (29), we undertook a systematic approach to identify CUGBP1 target transcripts in human cells. We performed immunoprecipitation (IP) of CUGBP1 from HeLa cell cytoplasmic extracts and analyzed the coimmunoprecipitated transcripts using oligonucleotide microarrays. This technique has been used successfully to identify targets of other RNA-binding proteins, including HuR (23), AUF1 (11), TIAR (7), TTP (21), and Pum1 (15). Using this approach, we identified 613 putative targets of CUGBP1 and found significant enrichment of the consensus GRE sequence UGUUUGUUUGU as well as a GU-repeat sequence in the 3′ UTR of the CUGBP1 target transcripts. We found that the GU-repeat sequence functioned as an mRNA decay element, and knockdown of CUGBP1 stabilized GU-repeat-containing messages. These results led us to redefine the GRE consensus sequence to include GU repeats. Functional analysis of GRE-containing CUGBP1 target transcripts revealed a posttranscriptional regulatory network that coordinates the expression of transcripts involved in cell cycle and cell growth regulation, cell motility, and apoptosis.
MATERIALS AND METHODS
RNA immunoprecipitation and microarray analysis.
HeLa Tet-Off cells (Clontech) were cultured in minimal essential medium alpha (Gibco) containing 10% tetracycline (Tet)-free fetal bovine serum (FBS; Clontech), 1% [scap]l-glutamine (Gibco), and 100 units/ml penicillin-streptomycin (Gibco). Cytoplasmic extractions and RNA IP were performed as described previously (24, 30) using an antihemagglutinin (anti-HA) antibody (F7; Santa Cruz), anti-CUGBP1 antibody (3B1; Santa Cruz), or anti-poly(A)-binding protein (anti-PABP) antibody (Immuquest). Three independent RNA IP experiments were performed. For each experiment, RNA was purified from the input and immunoprecipitated material from an equivalent number of HeLa cells using an RNeasy kit (Qiagen) following the manufacturer's recommendations. For the input RNA, 5 μg was used to prepare labeled cRNA for microarray hybridizations. For the IP RNA prepared from an equivalent number of cells, SF9 insect cell RNA was added such that the total amount of RNA totaled 5 μg, and this RNA was used to prepare cRNA using a MessageAMP II RNA amplification kit (Ambion). The cRNAs prepared from input RNA and from each IP were hybridized to U133a Plus-2 microarrays. The microarrays were normalized using the gene content robust multiarray average (GCRMA) algorithm and Genespring, version 9.0, software (Agilent Technologies Inc.). Transcripts were considered to be present in the input, anti-CUGBP1 IP, or anti-PABP IP conditions if the log intensity of their average signal was greater than their average log intensity of the signal on the HA chip, with a P value of less than 0.05 by a Student t test.
Identification of conserved sequences of CUGBP1 target transcripts.
The UTRs of transcripts found to be present in the anti-CUGBP1 IP were analyzed by searching for the frequency of the occurrence of all possible 11-mers. The frequency of occurrence of sequences in the 3′ UTR of transcripts found in the anti-CUGBP1 IP were compared to the frequency of occurrence in all of the 24,820 3′ UTRs extracted from the NCBI database. In addition, a BioProspector search was performed on the 3′ UTRs of the transcripts found in the anti-CUGBP1 IP (http://ai.stanford.edu/∼xsliu/BioProspector/) using the following parameters: motif length of 11 nucleotides and searching the forward strand and background definition human genomic 3′ UTR sequences. Consensus motifs were created with Weblogo software, version 2.8.2 (http://weblogo.berkley.edu) (1).
Electrophoretic mobility shift and supershift assays.
Ribo-oligonucleotides were purchased commercially (Sigma). The sequences for each ribo-oligonucleotide are shown as the shaded sequences in Fig. 1A. Ribonucleotide oligomers were end labeled with [γ32P]ATP (6,000 Ci/mmol) using T4 polynucleotide kinase (Invitrogen) to produce a radioactively labeled probe with a specific activity of 4 × 106 cpm/μg. An electrophoretic mobility shift assay (EMSA) was performed as published previously (17). Each reaction mixture contained 10 μg of HeLa cytoplasmic protein and 10 to 15 fmol of radiolabeled RNA probe in a total volume of 20 to 24 μl. For supershift assays, an anti-CUGBP1 monoclonal antibody (3B1; Santa Cruz Biotechnologies) or an anti-His tag antibody (Santa Cruz Biotechnology, Inc.) was added to the reaction mixtures. The mixtures were separated by electrophoresis under nondenaturing conditions on 5% polyacrylamide gels. The gels were dried and analyzed on a Storm 820 phosphorimager (Amersham Biosciences).
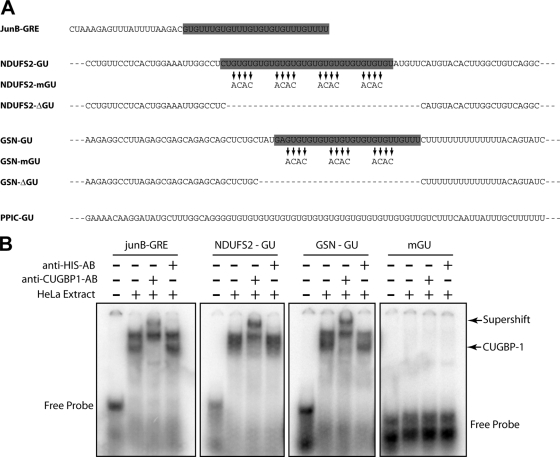
FIG. 1.
CUGBP1 selectively binds to GU-repeat sequences. (A) Sequences from the 3′ UTR of transcripts present in the anti-CUGBP1 IP were cloned into the 3′ UTR of the beta-globin reporter to create the JUNB-GRE (30), NDUFS2-GU, GSN-GU, and PPIC reporter transcripts. The indicated mutations and deletions were introduced to create the NDUFS2-mGU, NDUFS2-ΔGU, GSN-mGU, and GSN-ΔGU transcripts. (B) The GRE sequence of JUNB (JUNB-GRE) or the GU-repeat sequences of NDUFS2 (NDUFS2-GU), GSN (GSN-GU), or a mutated GU-repeat sequence from NDUFS2 (mGU) shown in the shaded boxes in panel A were used as radiolabeled RNA probes in EMSAs. Anti-CUGBP1 or anti-His antibodies were added to the indicated reaction mixtures. RNA-protein complexes were separated by electrophoresis under nondenaturing conditions. Gels were dried and analyzed on a phosphorimager. The positions of migration of the free probe, CUGBP1-containing band (CUGBP1), and the anti-CUGBP1 supershifted band (Supershift) are indicated.
Tet-Off mRNA decay assay.
The decay of beta-globin reporter constructs was performed as described previously (16, 30). The Tet-responsive beta-globin expression construct, pTetBBB (19), was a gift from Ann-Bin Shyu (University of Texas—Houston), and the jun B proto-oncogene (JUNB)-GRE expression construct was described previously (30). Sequences from the 3′ UTR of NADH dehydrogenase Fe-S protein 2 (NDUFS2), gelsolin (GSN), and peptidylprolyl isomerase C (PPIC) were inserted at the unique BglII restriction site in the beta-globin 3′ UTR of pTetBBB to create the NDUFS2-GU, GSN-GU, and PPIC-GU constructs. The NDUFS2-ΔGU (where ΔGU indicates a deletion of the GU-repeat of the NDUFS2-GU reporter), NDUFS2-mGU (where mGU indicates a mutation of every other GUGU to ACAC), GSN-ΔGU, GSN-mGU, and GM1 through GM8 constructs shown in Fig. 1A or 4A were created from the NDUFS2-GU or GSN-GU constructs by site-directed mutagenesis using a QuikChange II XL site-directed mutagenesis kit (Stratagene).
HeLa Tet-Off cells (15-cm dish) were transfected with 15 μg of the parental BBB reporter plasmid or BBB plasmids containing 3′ UTR inserts. Transfections were performed with 6.25 μg/ml of Lipofectamine 2000 reagent (Invitrogen). Cells were split into four 10-cm dishes the next day. After 48 h, 300 ng/ml of doxycycline was added to stop transcription, and total RNA was isolated after 0, 2, 4, or 6 h. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was generated using a DECAprime kit (Ambion). RNA isolation and Northern blotting to assess expression of beta-globin and GAPDH transcripts were performed as described previously (16). For each point, the hybridization intensity of the beta-globin transcript was normalized to the hybridization intensity of the GAPDH transcript, and the normalized values were used to calculate half-lives using GraphPad Prism software, version 4.03, based on a one-phase exponential model of decay.
siRNA transfection.
CUGBP1 knockdown experiments were performed using a red fluorescent protein targeting a small interfering RNA (siRNA) duplex as a control and two CUGBP1-specific siRNA duplexes described previously (30).
For the Tet-Off mRNA decay assays, HeLa Tet-Off cells (15-cm dish) were transfected with 1 nmol of CUGBP1-specific siRNAs or a control siRNA. After 24 h cells were again transfected with 1 nmol of siRNA and 15 μg of the GSN-GU. Transfections were performed with 6.25 μg/ml of Lipofectamine 2000 reagent (Invitrogen). Cells were split into four 10-cm dishes. After 48 h, 300 ng/ml of doxycycline was added to stop transcription, and total RNA was isolated after 0, 2, 4, or 6 h. GAPDH probe was generated using a DECAprime kit (Ambion). RNA isolation and Northern blotting to assess expression of beta-globin and GAPDH transcripts were performed as described previously (16). For each point, the hybridization intensity of the beta-globin transcript was normalized to the hybridization intensity of the GAPDH transcript, and the normalized values were used to calculate half-lives using GraphPad Prism software, version 4.03, based on a one-phase exponential model of decay. Knockdown of CUGBP1 was assessed by Western blotting for CUGBP1 with an anti-CUGBP1 monoclonal antibody (3B1; Santa Cruz Biotechnologies) and an anti-GAPDH polyclonal antibody (FL355; Santa Cruz Biotechnologies) as a loading control.
For apoptosis assays HeLa Tet-Off cells (six-well plate) were transfected with 50 pmol of siRNA of the CUGBP1-specific or control siRNA on two consecutive days. At 48 h after the last transfection apoptosis was induced with 50 ng/ml TNF-α (Invitrogen/Biosource) and 50 μg/ml cycloheximide (CHX; Acros Organics) for 3 h. Knockdown of CUGBP1 was assessed by Western blotting for CUGBP1 with an anti-CUGBP1 monoclonal antibody (3B1; Santa Cruz Biotechnologies), apoptosis was determined using a rabbit polyclonal antibody against cleaved poly(ADP-ribose) polymerase (PARP; Abcam), and an anti-GAPDH polyclonal antibody (FL355; Santa Cruz Biotechnologies) was used as a loading control. Fluorescence-activated cell sorter (FACS) analysis was performed on a FACSCanto machine (BD) using a phycoerythrin (PE)-coupled anti-activated caspase 3 antibody (BD Pharmingen). Results were analyzed in FlowJo, version 7.6.
RESULTS AND DISCUSSION
We recently showed that CUGBP1 mediated the rapid decay of GRE-containing transcripts in HeLa cells (30) and hypothesized that CUGBP1 coordinately regulates the posttranscriptional expression of a variety of GRE-containing transcripts at the level of RNA decay (29). We therefore performed experiments to identify transcripts associated with CUGBP1 in human cells. In three independent experiments, cytoplasmic lysates from HeLa cells were immunoprecipitated with immobilized anti-CUGBP1, antihemagglutinin (anti-HA), and anti-poly(A)-binding protein (anti-PABP) antibodies. Input RNA and IP RNA were analyzed using Affymetrix U133a Plus-2 microarrays. Signal intensities from each microarray were normalized using the gene content robust multiarray average (GCRMA) algorithm. RNA from the anti-HA IP was considered to be a negative control. For microarrays performed on RNA from the input, CUGBP1 IP, or PABP IP, transcripts were considered to be present if the log intensity of their average signal yielded a P value of less than 0.05 by a Student t test compared to the log intensity values on RNA from the anti-HA IP. We identified 613 transcripts, based on probe identification numbers (IDs), in the RNA from the anti-CUGBP1 IP that were overrepresented compared to the HA control (see Table S1 in the supplemental material). Of these 613 probe IDs, 75 were present in both the input and anti-PABP IP, 19 were present in the anti-PABP IP but not the input, and 59 were present in the input but not the anti-PABP IP. Of note, 477 transcripts were present in the anti-CUGBP1 IP but not the input or anti-PABP IP, suggesting that these transcripts were enriched during the anti-CUGBP1 immunoprecipitation procedure. Transcripts present in the anti-CUGBP1 IP encode proteins that serve various functions, including cell growth and cell cycle regulation, apoptosis, and motility. Table 1 lists a subset of the transcripts present in the anti-CUGBP1 IP that encode proteins that serve important cellular functions, and the complete list of transcripts is found in Table S1 in the supplemental material. In a recent study in Xenopus, RNA IP experiments identified 158 target mRNAs of EDEN-BP, the CUGBP1 homologue in Xenopus (2). These EDEN-BP target transcripts had only minimal overlap with the transcripts we found associated with CUGBP1 in human HeLa cells, but GREs were enriched in both the Xenopus and human experiments. Interestingly, we found that most of the CUGBP1 target transcripts that we identified did not contain a GRE, as defined previously (30), and we decided to use bioinformatics methods to search for other conserved sequences found in the transcripts from the anti-CUGBP1 IP.
TABLE 1.
Subset of transcripts present in the anti-CUGBP1 IP
| RefSeq accession no.a | Gene name or description | Gene symbol | Sequence(s)d | P valueb | GO biological function(s)c |
|---|---|---|---|---|---|
| NM_000177 | Gelsolin (amyloidosis, Finnish type) | GSN | TGAGTGTGTGT, TGTGTGTGTGT | 0.003 | Actin filament polymerization, cell cycle |
| NM_002229 | jun B proto-oncogene | JUNB | TGTGTTTGTGT | 0.002 | Cell growth, cell cycle |
| NM_004550 | NADH Dehydrogenase (ubiquinone) Fe-S protein 2 | NDUFS2 | TGTGTGTGTGT | 0.033 | Mitochondrial electron transport |
| NM_000943 | Peptidylprolyl isomerase C (cyclophilin C) | PPIC | TGTGTGTGTGT | 0.049 | Protein folding |
| NM_005171 | Activating transcription factor 1 | ATF1 | TATGTGTGTGT, TGTGTGTGTGT | 0.015 | Cell growth, cell cycle |
| NM_005354 | jun D proto-oncogene | JUND | TGTGTGTGTGT | 0.0005 | Cell growth, cell cycle |
| NM_001769 | CD9 molecule | CD9 | TTTTTGTTTGT, TGTTTGTTTTT | 0.0001 | Motility, cell growth |
| NM_014989 | Regulating synaptic membrane exocytosis 1 | RIMS1 | TGTTTATTTGT, TGTTTGTTGGT | 0.0017 | Protein complex assembly |
| NM_012197 | Rab GTPase activating protein 1 | RABGAP1 | TGTGTGTGTGT | 0.008 | Cell cycle |
| NM_004217 | Aurora kinase B | AURKB | TGTTTGTATGT | 0.045 | Cytokinesis, cell cycle |
| NM_000641 | Interleukin-11 | IL-11 | TGTTTGTTTTT | 0.050 | Cell-cell signaling |
| NM_018685 | Anillin, actin binding protein | ANLN | TGTTTGTTTGT | 0.043 | Cytokinesis |
| NM_001894 | Casein kinase 1, epsilon | CSNK1E | TGTGTGTGTGT | 0.010 | DNA repair |
| NM_001080421 | unc-13 homolog A (C. elegans) | UNC13A | TGTGTGTGTGT, TGTTTGTTTTT | 0.041 | Exocytosis |
| NM_015083 | Cdc2-related kinase, arginine/serine-rich | CRKRS | TGTGTGTGTGT | 0.044 | Protein amino acid phosphorylation |
| NM_002578 | p21 (CDKN1A)-activated kinase 3 | PAK3 | TGTTTGTTTTT | 0.009 | Protein complex assembly |
| NM_001006610 | Seven in absentia homolog 1 (Drosophila) | SIAH1 | TGTGTGCGTGT | 0.007 | Proteolysis |
| NM_002468 | Myeloid differentiation primary response gene 88 | MYD88 | TGGGTGTGTGT, GGTGTGTGTGT | 0.022 | Response to molecule of fungal origin |
| NM_005633 | Son of sevenless homolog 1 (Drosophila) | SOS1 | TGTTTGTGTAT | 0.042 | Signal transduction |
From the NCBI Reference Sequence Project.
P value indicates the statistical significance of the enrichment of the transcript in the anti-CUGBP1 IP compared to the anti-HA IP.
GO, gene ontology.
Some transcripts contain more than one enriched GU-rich sequence.
We extracted the 3′ UTR for each of the 613 CUGBP1 target transcripts and subjected these sequences to a simple overrepresentation algorithm. The overrepresentation algorithm created a list of all possible (411) 11-mers and counted the occurrence of each of these 11-mers in the 3′ UTRs of the genes of interest. Each 11-mer was determined to be present in a given 3′ UTR if it was found in the sequence, allowing for only one mismatched nucleotide. The statistical significance of the overrepresentation of these 11-mers in the genes of interest was determined through a comparison of the rate of appearance of the 11-mers in a background set of 27,472 3′ UTRs from transcripts with known sequences in the NCBI database. The six most frequently encountered 11-mers are shown in Table 2. These consensus sequences included the previously elucidated GRE consensus sequence, UGUUUGUUUGU (30), with a prevalence of 18.69% in the 3′ UTR from transcripts from the CUGBP1 IP compared to only 6.7% in all transcripts (P of 5.35 × 10−26). We also identified a GU-repeat sequence that was highly enriched in transcripts from the anti-CUGP1 IP; this sequence was present in the 3′ UTR in 17.33% of the transcripts from the anti-CUGBP1 IP, compared to 7.6% of all transcripts (P of 2.53 × 10−15). The GU-repeat sequence was previously reported as a sequence capable of binding to CUGBP1 in yeast three-hybrid (22), surface plasmon resonance (14), and SELEX assays (10). In addition to the UGUUUGUUUGU and UGUGUGUGUGU sequences, we also identified a poly(U) sequence that was highly enriched in transcripts from the anti-CUGBP1 IP. A GGA-repeat sequence, the sequence TGGGAATGGTC, and a CA-repeat sequence were also enriched in transcripts from the anti-CUGBP1 IP, with lower statistical significance. As another approach to look for conserved sequences in transcripts from the anti-CUGBP1 IP, a BioProspector search was performed on the 3′ UTRs of these transcripts. The parameters for the BioProspector search included searching only on the forward strand, allowing for multiple occurrences of a motif in a given sequence, and using human 3′ UTR sequences as the background. The results of the BioProspector search included the GU-repeat sequence and the CA-repeat sequence shown in Table 2, which were enriched in transcripts found in the anti-CUGBP1 IP. The BioProspector search also identified several other motifs, but none of the others was enriched in the anti-CUGBP1 IP in a statistically significant manner. These results confirmed our previous report that GRE-containing transcripts are targets of CUGBP1 and suggested that transcripts that contain GU-repeats could also be targets of CUGBP1.
TABLE 2.
Prevalence of motifs in the 3′ UTR of transcripts in the anti-CUGBP1 compared to all transcripts in the genome
| Motif | Prevalence in the anti-CUGBP1 IP (%) | Prevalence in the genome (%) | P value |
|---|---|---|---|
| TGTGTGTGTGT | 17.33 | 7.6 | 2.53 × 10−15 |
| GGAGGAGGAGG | 8.51 | 5.4 | 1.02 × 10−2 |
| TGTTTGTTTGT | 18.69 | 6.7 | 5.35 × 10−26 |
| TTTTTTTTTTT | 38.28 | 21.8 | 8.03 × 10−8 |
| TGGGAATGGTC | 2.58 | 0.8 | 6.28 × 10−6 |
| CACACACACAC | 7.75 | 4.9 | 1.40 × 10−3 |
We identified six distinct sequence motifs enriched in the 3′ UTRs of transcripts found in the anti-CUGBP1 IP (Table 2) and selected two transcripts containing each of these sequences to determine if they could bind to CUGBP1 based on gel shift and supershift assays. For each transcript tested, we designed RNA oligomers that contained the putative sequence elements and surrounding sequences. Using cytoplasmic extracts from HeLa cells, we found that CUGBP1 bound to the GRE sequence from the jun B proto-oncogene (JUNB) as well as the GU-repeat sequences from NADH dehydrogenase Fe-S protein 2 (NDUFS2) and gelsolin (GSN) (Fig. 1). Binding was attributed to CUGBP1 because it was specifically supershifted with an anti-CUGBP1 antibody (Fig. 1B, lanes 3, 7, and 11). This binding was sequence specific because binding did not occur when an RNA probe was used that contained mutations in the GU repeats (Fig. 1B, lanes 14 to 16). Transcripts containing the poly(U) sequence, GGA-repeat sequence, CA-repeat sequence, or the sequence TGGGAATGGTC did not bind to CUGBP1 (data not shown). These experiments confirmed our previous result that CUGBP1 bound to the JUNB GRE (30) and showed that CUGBP1 bound specifically to GU-repeat sequences of NDUFS2 and GSN. These results also confirmed results from other investigators that CUGBP1 recognizes GU repeats (10, 14, 22). From the 613 transcripts identified in the CUGBP1 IP, approximately 36% contain UGUUUGUUUGU or GU-repeat motifs in their 3′ UTRs.
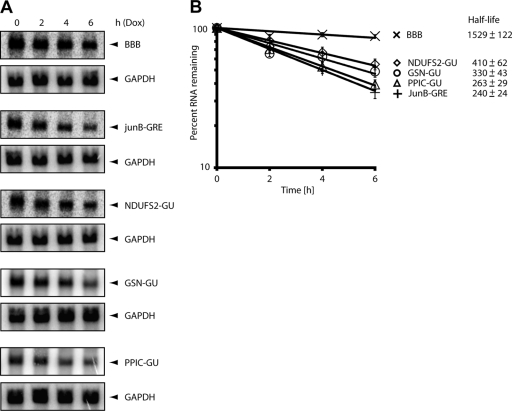
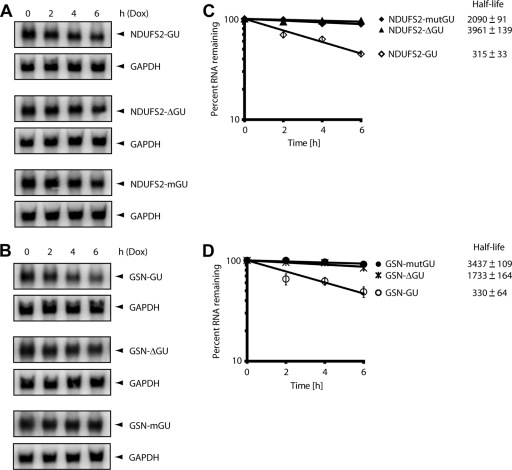
We previously showed that the UGUUUGUUUGU consensus sequence promoted rapid degradation when inserted into the 3′ UTR of an otherwise stable beta-globin reporter in a manner that depended on the presence of CUGBP1 (30). We hypothesized that the GU-repeat sequence that was also enriched in transcripts found in the anti-CUGBP1 IP might also function to regulate CUGBP1-mediated mRNA decay. To test this hypothesis, we inserted GU-repeat-containing sequences from the 3′ UTRs of NDUFS2, GSN, and PPIC into the 3′ UTR of the beta-globin (BBB) reporter to produce the NDUFS2-GU, GSN-GU, and PPIC-GU reporters, respectively, and transfected them into HeLa Tet-Off cells. Doxycycline was added to the medium to stop transcription of the reporter transcripts. Total RNA was extracted after 0, 2, 4, or 6 h and was analyzed by Northern blotting for beta-globin and GAPDH mRNA expression in order to measure the decay of the reporter transcripts (Fig. 2 A). We performed four independent experiments and graphed the mRNA decay results (Fig. 2B). The beta-globin signal was normalized to GAPDH, and the zero time point was set to 100%. The percentage of RNA remaining was plotted against time, and mRNA decay rates were calculated. As expected, the beta-globin transcript was stable, whereas the JUNB-GRE transcripts showed rapid decay, with a half-life of 240 ± 24 min, confirming our previous results (30). All three GU-repeat-containing reporter transcripts, NDUFS2-GU, GSN-GU, and PPIC-GU, displayed rapid decay relative to the stable beta-globin reporter, suggesting that insertion of the GU-repeat sequences was responsible for mediating mRNA decay. To confirm that the GU repeats were responsible for mediating mRNA decay, we deleted or mutated the GU-repeat sequences to determine the effect on mRNA decay. First, we deleted the GU-repeat of the NDUFS2-GU and GSN-GU reporters to generate the NDUFS2-ΔGU and GSN-ΔGU reporters. Next, we mutated every other GUGU to ACAC to generate the NDUFS2-mGU and GSN-mGU reporters (Fig. 1A). We tested the effect of these deletions and mutations on the ability of GU repeats to promote mRNA decay. These reporter plasmids were transfected into HeLa cells, reporter mRNA decay rates were assessed by Northern blotting (Fig. 3 A and B), and results from four independent experiments were plotted (Fig. 3C and D). The NDUFS2-GU and GSN-GU reporters exhibited rapid decay, whereas all of the reporters containing deletions or mutations in the GU repeats were stable. This series of experiments clearly show that the GU-repeat functions to promote decay of the beta-globin reporter transcripts.
FIG. 2.
The GU-repeat sequence mediates rapid decay of the beta-globin reporter transcript. (A) HeLa Tet-Off cells were transfected with the pTetBBB beta-globin reporter construct or with reporter constructs in which GRE-containing sequences from the 3′ UTR of the JUNB transcript (JUNB-GRE) or GU-repeat sequences from the 3′ UTRs of the NDUFS2 (NDUFS2-GU), GSN (GSN-GU), or PPIC (PPIC-GU) were inserted into the beta-globin 3′ UTR. Doxycycline was added to the medium to stop transcription from the Tet-responsive promoter, and total cellular RNA was collected at 0-, 2-, 4-, and 6-h time points. Northern blot analyses were performed to monitor the levels of the GAPDH transcripts and the beta-globin reporter transcripts. (B) The experiment shown in panel A was performed four times, and the Northern blot signals were quantified using a phosphorimager. For each time point, the intensity of the beta-globin band was normalized to the intensity of the GAPDH band, and the band intensity at the zero time point was set to 100%. The percentage of mRNA remaining was plotted over time. The error bars indicate the standard error of the mean from three experiments. Half-lives are indicated in minutes.
FIG. 3.
Mutation of the GU-repeat sequence abrogates mRNA decay. (A and B) The decay of the NDUFS2-GU (A) or the GSN-GU (B) beta-globin reporter was compared to the decay of these reporters in which the GU-repeats were deleted (NDUFS2-ΔGU and GSN-ΔGU) or mutated (NDUFS2-mGU and GSN-mGU). HeLa Tet-Off cells were transfected with the indicated beta-globin reporter constructs. Doxycycline (Dox) was added to the medium to stop transcription from the Tet-responsive promoter, and total cellular RNA was collected at 0-, 2-, 4-, and 6-h time points. Northern blot analyses were performed to monitor the levels of the GAPDH transcripts and the beta-globin reporter transcripts. (C and D) The experiments shown in panels A and B were performed four times, and the Northern blot signals were quantified using a phosphorimager. For each time point, the intensity of the beta-globin band was normalized to the intensity of the GAPDH band, and the band intensity at the zero time point was set to 100%. The percentage of mRNA remaining was plotted over time. The error bars indicate the standard error of the mean from three experiments. Half-lives are indicated in minutes.
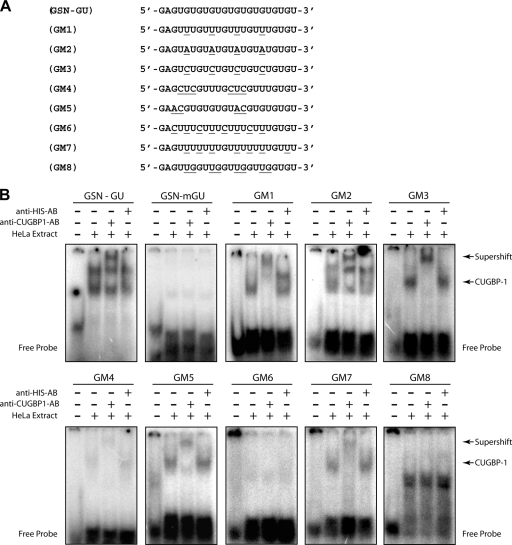
We identified GU repeats as CUGBP1 targets using a very stringent bioinformatics analysis allowing for only one mismatch per 11-mer. To further characterize the sequence requirements of CUGBP1 binding to RNA, we tested RNA oligomers (termed GM1 to GM8) containing several point mutations in the GSN GU-repeat sequence (Fig. 4 A) for their ability to bind to CUGBP1 using gel shift and supershift assays (Fig. 4B). The GM1 oligomer, which contained the previously described consensus GRE (30), and GM2, which contained the EDEN-BP binding motif (18), bound to CUGBP1, as confirmed by supershift using a specific anti-CUGBP1 antibody. GM3, which contained a mutation of every other G to C in the GU-repeat sequence also bound to CUGBP1, whereas GM6 and GM8, which contained mutations that disrupted all UGU triplets, did not bind to CUGBP1. GM5, which contained two AC-dinucleotide disruptions of the GU-repeat sequence also bound to CUGBP1, suggesting that discontinuous GU repeats may be capable of CUGBP1 binding, as suggested earlier (13). Interestingly, GM4, which contained two CUG sequences disrupting the GU repeat sequence, had very little binding activity.
FIG. 4.
CUGBP1 binding activity allows mismatches in the GU-repeat sequence. (A) Mutations and were introduced into the GSN-GU motif to create GM1 through GM8, as indicated. Mutated nucleotides are underlined. (B) The sequences depicted in panel A were used as radiolabeled RNA probes in EMSAs. The GSN-mGU sequence was used as a negative control. Anti-CUGBP1 or anti-His antibodies were added to the indicated reaction mixtures. RNA-protein complexes were separated by electrophoresis under nondenaturing conditions. Gels were dried and analyzed on a phosphorimager. The positions of migration of the free probe, CUGBP1-containing band (CUGBP1), and the anti-CUGBP1 supershifted band (Supershift) are indicated.
To determine if CUGBP1 binding correlated with mRNA decay, the BBB-GSN-GU reporter plasmid and reporter plasmids containing mutations in the GRE were transfected into HeLa Tet-Off cells; decay rates of these reporters were assessed by Northern blotting, and four independent experiments were analyzed. The reporter transcript containing the wild-type gelsolin GU-repeat sequence (BBB-GSN-GU) and the GM3 sequence, which bound well to CUGBP1, decayed rapidly (half-life of 540 ± 10 min). In contrast, the reporter transcript containing the GM4 sequence, which bound poorly to CUGBP1, was quite stable (half life of 1,386 ± 36 min) and had a decay rate that was similar to that of the control reporter that lacked a GU-rich sequence (ΔGU; half-life of 1,733 ± 164 min). These data suggest that CUGBP1 binding by these sequences correlated with decay activity.
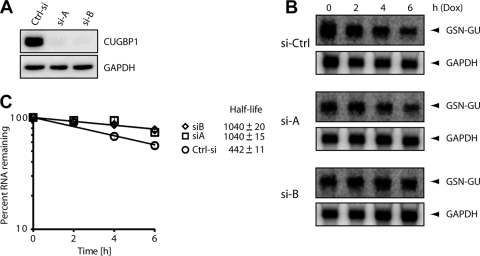
To verify that GU-repeat-mediated decay is mediated by CUGBP1, we used siRNA-mediated knockdown of CUGBP1 and assessed the decay of GU-repeat-containing reporter transcripts. We performed sequential knockdown of CUGBP1 using two independent siRNAs and, as a control, red fluorescent protein siRNA, followed by transfection of the GSN-GU-repeat containing reporter. Both siRNAs led to a 90% to 99% decrease in CUGBP1 levels (Fig. 5 A). In the same cells, reporter decay was measured over a 6-h time course by Northern blotting (Fig. 5B), and the results of four independent experiments were plotted (Fig. 5C). In cells transfected with the control siRNA, the GSN-GU reporter transcript half-life was 442 ± 12 min, and knockdown of CUGBP1 with either CUGBP1-specific siRNA significantly stabilized the transcript (P < 0.01). This experiment clearly shows that the GU-repeat sequence and the GRE are regulated by a CUGBP1-dependent mechanism.
FIG. 5.
CUGBP1 is responsible for GU-repeat-mediated mRNA decay. (A) The decay of the GSN-GU beta-globin reporter was measured in cells that expressed a control siRNA (Ctrl-si) or two different siRNAs (siA and siB) directed against CUGBP1. HeLa Tet-Off cells were transfected with the GSN-GU-repeat-containing reporter and the indicated siRNAs. Knockdown efficiency was monitored for each experiment by Western blotting with a specific anti-CUGBP1 antibody. A GAPDH antibody was used as the loading control. (B) Doxycycline (Dox) was added to the medium to stop transcription from the Tet-responsive promoter, and total cellular RNA was collected at the 0-, 2-, 4-, and 6-h time points. Northern blot analyses were performed to monitor the levels of the GAPDH transcripts and the beta-globin reporter transcripts. (C) The experiment shown in panel B was performed four times, and the Northern blot signals were quantified using a phosphorimager. For each time point, the intensity of the beta-globin band was normalized to the intensity of the GAPDH band, and the band intensity at the zero time point was set to 100%. The percentage of mRNA remaining was plotted over time. The error bars indicate the standard error of the mean from three experiments. Half-lives are given in minutes.
We previously defined the GRE as the consensus sequence UGUUUGUUUGU, which was highly enriched in short-lived transcripts expressed in primary human T cells (30), and showed that CUGBP1 mediated mRNA decay by binding to this sequence (30). Our results here suggest that CUGBP1 binds to a larger subset of transcripts that includes transcripts that contain the previously defined GRE as well as transcripts that contain GU repeats. Our finding that GU repeats are able to function as mediators of mRNA decay in a manner that depends on CUGBP1 suggests that we should broaden our definition of the GRE to include GU repeats. Of the 613 transcripts that were identified in the anti-CUGBP1 IP, if we allow one mismatch, 123 transcripts contain the previously defined GRE 11-mer, and 114 contain the GU repeat sequence. We combined these 237 transcripts into a set of transcripts that contain CUGBP1 binding sites and used Weblogo software, version 2.8.2 (1), to create a new consensus GRE sequence based on all UTRs that contained the UGUUUGUUUGU element or the GU-repeat element (Fig. 6).
FIG. 6.
Generation of the GRE consensus sequence. Weblogo was generated by compiling a list of all occurrences of the GU repeat and the GRE found in the 3′ UTRs of those transcripts identified as present in the CUGBP1 IP. This list was uploaded into the program Weblogo, version 2.8.2, to generate the Weblogo (1).
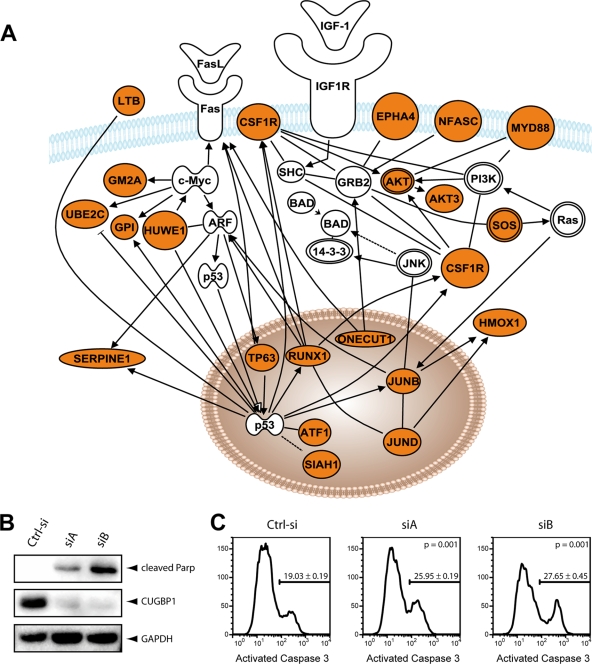
The coordinate regulation of the expression of multiple genes involved in specific functional pathways by specific RNA-binding proteins and cis sequences forms regulatory networks that are known as posttranscriptional regulons. The 237 CUGBP1 target transcripts that contain the GRE consensus (shown in Fig. 6) in their 3′ UTRs define a posttranscriptional regulon. These GRE-containing transcripts encode proteins that participate in specific functional pathways involved in cell growth, apoptosis, and cell motility. For example, transcripts encoding multiple regulators of cell cycle and cell growth were identified as CUGBP1 targets. Also, CUGBP1 targets included transcripts encoding regulators of the extrinsic apoptotic pathway, as suggested previously (8), and included regulators such as Akt and TP63 (Fig. 7 A). To determine if CUGBP1 is a regulator of apoptosis, we transfected HeLa cells sequentially in two rounds within 24 h with either a red fluorescent protein siRNA as a control or two CUGBP1-specific siRNAs, as described previously (30). At 48 h after the second transfection, cells were split, and apoptosis was induced with TNF-α and CHX for 3 h. The knockdown of CUGBP1 resulted in a 90 to 99% reduction in CUGBP1, as demonstrated by Western blotting (Fig. 7B). Probing the same Western blot for the apoptosis marker, cleaved PARP, showed that knockdown of CUGBP1 promoted apoptosis. Aliquots of cells were also stained with a PE-coupled antibody specific for activated caspase 3 and analyzed by flow cytometry. Cells transfected with the control siRNA displayed significantly less apoptosis (19.03% ± 0.19%) than the cells that were transfected the CUGBP1-specific siRNAs (25.95% ± 0.19% and 27.65% ± 0.45%) in four independent experiments (Fig. 7B). These results suggest that CUGBP1 is a regulator of apoptosis.
FIG. 7.
Posttranscriptional regulation of the CUGBP1 target network. (A) The network diagram depicts the coordinate regulation of CUGBP1 target transcripts involved in apoptosis. Transcripts depicted in orange represent GRE-containing transcripts that were enriched in the CUGBP1 targets. These pathways were identified by Ingenuity Pathway Assistant software (Ingenuity Systems, CA). (B) HeLa Tet-Off cells were transfected twice within 24 h with either a control siRNA or two different siRNAs specific for CUGBP1 (siA and siB). Knockdown efficiency was monitored by Western blotting with a specific anti-CUGBP1 antibody. A GAPDH antibody was used as the loading control. Apoptosis was assessed with a specific anti-PARP antibody. (C) Cells were stained for FACS analysis with a PE-labeled anti-active caspase 3 antibody. A representative experiment is shown. Four independent experiments were analyzed. Average percentages of apoptosis, standard error, and P values are indicated.
Other CUGBP1 target transcripts encode regulators of cell motility, such as the small G protein rho or gelsolin. Interestingly, cell growth and cell cycle, apoptosis, and cell motility pathways play important roles in cancer pathogenesis, including cell transformation, abnormal growth, cancer formation, progression, and metastasis (3). Thus, the CUGBP1-GRE regulatory network may play an important role in regulating gene expression during normal cell growth and development by mediating mRNA degradation. Aberrations in the regulation of this network might lead to cancer formation and progression through the abnormal stabilization of critical transcripts.
Overall, we have identified 613 new targets for CUGBP1 and have demonstrated that a GU-repeat motif is sufficient to promote rapid decay when present in a transcript's 3′ UTR. This GU repeat and the previously identified UGUUUGUUUGU sequence were used to redefine the GRE, and we found that GREs were present in the 3′ UTRs of 36% or 62% of the transcripts found in the anti-CUGBP1 IP if we allowed one or two mismatches, respectively. For this analysis, we focused on the 3′ UTR because that is where known functional GREs are located. We acknowledge, however, that we could miss functional CUGBP1 target sites in the 5′ UTR or coding regions of the transcripts. When we looked for GREs at any location in the target transcripts, allowing two mismatches, we found that 73% of the transcripts in the anti-CUGBP1 immunoprecipitation contained a GRE. Interestingly, although the GRE was highly enriched in the 3′ UTR of target transcripts, it was not enriched in the 5′ UTR or the coding regions. Since CUGBP1 is reported to be involved in translational regulation, we investigated the role of the GRE in translation, evaluating the migration of reporter transcripts containing GRE sequences or mutated GRE sequences in polysome gradients (see Fig. S1 in the supplemental material). We found similar migration of GRE or mutated GRE-containing transcripts, suggesting that GREs located in the 3′ UTRs of these reporters did not have a major influence on translation in this system.
Recently, CUGBP1 was found to be a posttranscriptional regulator of TNF-α expression via the AU-rich element (31), which contained multiple dispersed UGU sequences that may function as CUGBP1-binding sites. If dispersed UGU sequences do indeed function as CUGBP1 binding sites, these sites would not be included in our consensus definition of the GRE. In addition to the GRE, CUGBP1 is known to bind to CUG repeat sequences (27) although these sequences was not identified in our motif search. Nevertheless, we searched for enrichment of a consensus sequences containing four tandem CUG repeats, allowing two mismatches, in the transcripts found in the anti-CUGBP1 IP. We found this sequences to be enriched in the 5′ UTR (P of 0.02) and 3′ UTR (P of 6 × 10−4) of these transcripts, corresponding to 48 and 100 transcripts, respectively. This CUG repeat sequence was not, however, enriched in the coding region. Thus, of the putative target transcripts contained in the anti-CUGBP1 IP, 83% percent contained GRE or CUG repeat consensus sequences somewhere in the transcript.
Overall, our data suggest that the GREs in the 3′ UTR of transcripts are important targets of CUGBP1 and thereby function to regulate mRNA decay. The CUGBP1 target transcripts that we identified encode important regulators of cell cycle control, apoptosis, and cell motility, as well as proto-oncogenes and other pathways involved in cancer formation and progression. These findings suggest that CUGBP1 and its target transcripts define a posttranscriptional regulatory network that functions to control cellular growth and homeostasis, and disruptions in this network may play a role in the development of cancer.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AIO57484 and AIO72068 to P.R.B. B.R. was funded by a postdoctoral fellowship from the Swiss National Science Foundation. D.B. was supported by MSTP grant 5 T32 GM008244 from the NIH. J.C.J. was supported by an Undergraduate Research Opportunity Program award. I.A.V. was funded through a fellowship from the Lymphoma Research Foundation.
We thank the Microarray Facility of the Academic Health Center of the University of Minnesota and the Minnesota Supercomputing Institute for their services. We thank George Karypis, Cavan S. Reilly, and Wayne Xu for helpful discussions regarding the bioinformatics analysis, Jin-Young Han for providing technical support and reagents, and Ann-Bin Shyu for providing plasmids.
We declare that we have no conflict of interests.
Footnotes
Published ahead of print on 14 June 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graindorge, A., O. Le Tonqueze, R. Thuret, N. Pollet, H. B. Osborne, and Y. Audic. 2008. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 36:1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 4.Huichalaf, C. H., K. Sakai, G. L. Wang, N. A. Timchenko, and L. Timchenko. 2007. Regulation of the promoter of CUG triplet repeat binding protein, Cugbp1, during myogenesis. Gene 396:391-402. [DOI] [PubMed] [Google Scholar]
- 5.Iakova, P., G. L. Wang, L. Timchenko, M. Michalak, O. M. Pereira-Smith, J. R. Smith, and N. A. Timchenko. 2004. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 23:406-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin, J., G. L. Wang, E. Salisbury, L. Timchenko, and N. A. Timchenko. 2009. GSK3β-cyclin D3-CUGBP1-eIF2 pathway in aging and in myotonic dystrophy. Cell Cycle 8:2356-2359. [DOI] [PubMed] [Google Scholar]
- 7.Kim, H. S., Y. Kuwano, M. Zhan, R. Pullmann, Jr., K. Mazan-Mamczarz, H. Li, N. Kedersha, P. Anderson, M. C. Wilce, M. Gorospe, and J. A. Wilce. 2007. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell. Biol. 27:6806-6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kress, C., C. Gautier-Courteille, H. B. Osborne, C. Babinet, and L. Paillard. 2007. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol. Cell. Biol. 27:1146-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ladd, A. N., N. Charlet, and T. A. Cooper. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquis, J., L. Paillard, Y. Audic, B. Cosson, O. Danos, C. Le Bec, and H. B. Osborne. 2006. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 400:291-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazan-Mamczarz, K., Y. Kuwano, M. Zhan, E. J. White, J. L. Martindale, A. Lal, and M. Gorospe. 2009. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 37:204-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milne, C. A., and J. Hodgkin. 1999. ETR-1, a homologue of a protein linked to myotonic dystrophy, is essential for muscle development in Caenorhabditis elegans. Curr. Biol. 9:1243-1246. [DOI] [PubMed] [Google Scholar]
- 13.Moraes, K. C., C. J. Wilusz, and J. Wilusz. 2006. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA 12:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori, D., N. Sasagawa, Y. Kino, and S. Ishiura. 2008. Quantitative analysis of CUG-BP1 binding to RNA repeats. J. Biochem. 143:377-383. [DOI] [PubMed] [Google Scholar]
- 15.Morris, A. R., N. Mukherjee, and J. D. Keene. 2008. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol. Cell. Biol. 28:4093-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogilvie, R. L., M. Abelson, H. H. Hau, I. Vlasova, P. J. Blackshear, and P. R. Bohjanen. 2005. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 174:953-961. [DOI] [PubMed] [Google Scholar]
- 17.Ogilvie, R. L., J. R. Sternjohn, B. Rattenbacher, I. A. Vlasova, D. A. Williams, H. H. Hau, P. J. Blackshear, and P. R. Bohjanen. 2009. Tristetraprolin mediates interferon-gamma mRNA decay. J. Biol. Chem. 284:11216-11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paillard, L., F. Omilli, V. Legagneux, T. Bassez, D. Maniey, and H. B. Osborne. 1998. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 17:278-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng, S. S., C. Y. Chen, and A. B. Shyu. 1996. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol. Cell. Biol. 16:1490-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salisbury, E., K. Sakai, B. Schoser, C. Huichalaf, C. Schneider-Gold, H. Nguyen, G. L. Wang, J. H. Albrecht, and L. T. Timchenko. 2008. Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP1. Exp. Cell Res. 314:2266-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoecklin, G., S. A. Tenenbaum, T. Mayo, S. V. Chittur, A. D. George, T. E. Baroni, P. J. Blackshear, and P. Anderson. 2008. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J. Biol. Chem. 283:11689-11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi, N., N. Sasagawa, K. Suzuki, and S. Ishiura. 2000. The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem. Biophys. Res. Commun. 277:518-523. [DOI] [PubMed] [Google Scholar]
- 23.Tenenbaum, S. A., C. C. Carson, P. J. Lager, and J. D. Keene. 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. U. S. A. 97:14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenenbaum, S. A., P. J. Lager, C. C. Carson, and J. D. Keene. 2002. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods 26:191-198. [DOI] [PubMed] [Google Scholar]
- 25.Timchenko, L. T., J. W. Miller, N. A. Timchenko, D. R. DeVore, K. V. Datar, L. Lin, R. Roberts, C. T. Caskey, and M. S. Swanson. 1996. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 24:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timchenko, N. A., R. Patel, P. Iakova, Z. J. Cai, L. Quan, and L. T. Timchenko. 2004. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 279:13129-13139. [DOI] [PubMed] [Google Scholar]
- 27.Timchenko, N. A., A. L. Welm, X. Lu, and L. T. Timchenko. 1999. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPβ mRNA and regulates translation of C/EBPβ isoforms. Nucleic Acids Res. 27:4517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuda, K., K. Kuwasako, M. Takahashi, T. Someya, M. Inoue, T. Terada, N. Kobayashi, M. Shirouzu, T. Kigawa, A. Tanaka, S. Sugano, P. Guntert, Y. Muto, and S. Yokoyama. 2009. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 37:5151-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vlasova, I. A., and P. R. Bohjanen. 2008. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 5:201-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vlasova, I. A., N. M. Tahoe, D. Fan, O. Larsson, B. Rattenbacher, J. R. Sternjohn, J. Vasdewani, G. Karypis, C. S. Reilly, P. B. Bitterman, and P. R. Bohjanen. 2008. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell 29:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, L., J. E. Lee, J. Wilusz, and C. J. Wilusz. 2008. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J. Biol. Chem. 283:22457-22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.