Abstract
Nuclear receptor estrogen receptor alpha (ERα) controls the expression of hundreds of genes responsible for target cell phenotypic properties, but the relative importance of direct versus tethering mechanisms of DNA binding has not been established. In this first report, we examine the genome-wide chromatin localization of an altered-specificity mutant ER with a DNA binding domain deficient in binding to estrogen response element (ERE)-containing DNA (DBDmut ER) versus wild-type ERα. Using high-throughput sequencing of ER chromatin immunoprecipitations (ChIP-Seq) and mRNA transcriptional profiling, we show that direct ERE binding is required for most of (75%) estrogen-dependent gene regulation and 90% of hormone-dependent recruitment of ER to genomic binding sites. De novo motif analysis of the chromatin binding regions in MDA-MB-231 human breast cancer cells defined unique transcription factor profiles responsible for genes regulated through tethering versus direct ERE binding, with Runx motifs enriched in ER-tethered sites. We confirmed a role for Runx1 in mediating ERα genomic recruitment and regulation of tethering genes. Our findings delineate the contributions of direct receptor ERE binding versus binding through response elements for other transcription factors in chromatin localization and ER-dependent gene regulation, paradigms likely to underlie the gene regulatory actions of other nuclear receptors as well.
Transcription in eukaryotes involves interactions between multiprotein complexes and chromosomal DNA to coordinately regulate gene expression in a stimulus-specific, temporal, and tissue-specific fashion (13, 19, 20). The polarity (stimulated versus repressed), the magnitude, and the duration of the response are the results of the combinatorial output of the receptor and coregulator proteins, histone-modifying enzymes, chromatin-remodeling complexes, and RNA polymerase II within the transcriptional complex (30, 38). Transcription factors have the ability to regulate gene expression by binding directly to DNA at sequence-specific response elements or by tethering to other response elements through protein-protein interactions with other DNA-bound factors (27). These response elements may be in the proximal promoter region (within 5 kb) of the target gene or at distal enhancer regulatory sites, or they may consist of a combination of proximal and distal elements (3, 5, 37). The combinatorial usage of these response elements drives the regulation of target genes and ultimately determines stimulus and tissue specificity.
Estrogen receptor alpha (ERα), a member of the nuclear hormone receptor family, is a ligand-activated transcription factor that controls the expression of hundreds of genes responsible for the diverse phenotypic properties of target cells, including growth, motility, and differentiation. The ER regulates genes such as TFF1, EBAG9, CASP7, and GREB1 through a classical regulatory mechanism involving its direct binding to DNA at estrogen response elements (EREs) through its zinc finger-containing DNA binding domain (22, 25, 41, 44, 56). Alternatively, the ER also has the ability to regulate gene expression through protein-protein interactions with other direct DNA binding transcription factors, such as Sp1, Ap1, CEBPβ, and Pitx1 (6, 7, 24, 28, 35, 40, 47, 53). However, the relative importance of this nonclassical, tethering paradigm in overall global gene expression and chromatin binding by the ER has not been extensively or systematically explored.
Recently, the genome-wide map for ER binding sites has been elucidated using human breast cancer cells (4, 26, 57). While these studies provide great insights into the binding sites for the ER, they do not distinguish between binding sites to which the ER binds through either the classical (direct ERE binding) versus nonclassical (tethering) paradigms, because the DNA in chromatin that is immunoprecipitated using ER antibodies may represent sites of direct DNA binding and/or tethering. These analyses have highlighted the importance of the ERE in ER DNA binding and gene regulation (2, 4, 26), yet a subset of ER binding sites contained no ERE or only an ERE half-site, suggestive of sites where the ER might be functioning through protein-protein interactions so that its activities would be mediated by indirect binding to other response elements through the agency of other transcription factors.
There is increasing evidence that gene regulation by the ER mediated through this nonclassical, tethering paradigm is likely to be of physiological significance. The selective estrogen receptor modulators (SERMs) tamoxifen and raloxifene are reported to function as agonists at least in part through the nonclassical mechanism (21, 24, 33, 42). Studies have also identified a subset of genes regulated by the ER through an ERE-independent mechanism in human breast tumors (12). Mice carrying an allele for an ER with mutations that eliminate DNA binding have deficiencies in ovarian function and in development of both the mammary gland and skeleton, and these mice exhibit enlarged uteri and altered patterns of gene expression, indicating tissue-selective effects for the classical versus nonclassical mechanisms of estrogen action (15, 17, 51, 52). In addition, chemical disruption of the DNA binding activities of the ER by electrophilic agents abrogated regulation of ERE-containing reporter genes and restored tamoxifen sensitivity in resistant breast cancer cells (54, 55). Therefore, characterizing estrogen regulation of gene expression through both the direct DNA binding and tethering paradigms could be important in directed therapeutics for hormone-dependent cancers.
In this report, we examine the genome-wide chromatin localization of a mutant nuclear hormone receptor, one in which point mutations in the DNA binding domain disable the receptor's ability to bind to its palindromic DNA response element. By using this altered specificity DNA (ERE) binding-deficient estrogen receptor (DBDmut ER), we were able to evaluate the roles of direct receptor DNA binding versus tethering as mechanisms in genome-wide recruitment of the ER to chromatin regulatory sites and in gene regulation in response to estradiol (E2) in human breast cancer cells. Through mRNA profiling and high-throughput sequencing of chromatin immunoprecipitations from cells with wild-type ER versus a DBD mutant ER that selectively regulates gene expression independent of ERE binding, we have defined subsets of genes regulated by this nuclear receptor through DNA binding and tethering modes. The studies also highlight novel enhancers and cooperating transcription factors, notably Runx1, having roles in estrogen-dependent chromatin localization and gene regulation by tethered ER.
MATERIALS AND METHODS
Cell culture.
MDA-MB-231 cells stably expressing the wild-type ERα or DBDmut were constructed and grown as previously described (1, 8, 45). The DBDmut ER is an altered specificity mutant that contains three point mutations that reduce binding of the ER to the ERE by greater than 95% while increasing its affinity for glucocorticoid receptor binding sites, thereby providing a positive control for the overall integrity of the DNA binding domain of this receptor (29). At 4 days prior to treatment, cells were switched to phenol red-free tissue culture medium containing 5% charcoal-dextran-treated calf serum. Medium was changed on days 2 and 4 of culture, and then cells were treated with control (0.1% ethanol) vehicle or 10 nM E2 (Sigma, St. Louis, MO).
Western blot analysis.
Cell protein lysates were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Blots were incubated in blocking buffer (5% milk in Tris-buffered saline with 0.5% Tween) and then with specific antibodies for ER (HC-20; Santa Cruz Biotechnology) and β-actin (AC-15; Sigma), followed by detection using horseradish peroxidase-conjugated secondary antibodies of the Supersignal West Femto detection kit (Pierce, Rockford, IL) as described by the manufacturer.
Plasmid preparation and transient transfections.
The 2ERE-pS2-luciferase vector has been described previously (36). The pGL3-promoter-TGFβ3 vector was constructed from a transforming growth factor β3-chloramphenicol acetyltransferase (TGFβ3-CAT) template using the primers forward_KpnI (GGGGTACCACAGCTGGCGAGAGGGCG) and reverse_XhoI (CCGCTCGAGCTTGGACTTGACTCTCTGCTTCCCTC) (14, 59). The identified ER binding sites were cloned by PCR into either pGL3_Promoter or pGL3-Basic luciferase vectors (Promega) using specific primers from human genomic DNA (Roche Molecular Biochemicals, Indianapolis, IN). Transient-transfection assays were performed as previously described (49).
RNA extraction and microarray analysis.
Preparation of RNA from cells and analysis of Affymetrix gene chip microarray data were as previously described (9). Affymetrix Hu133A GeneChips were used and analyzed using MicroArray Suite 5.0 (Affymetrix, Santa Clara, CA) and GeneSpring GX software (Silicon Genetics, Redwood City, CA) as described previously (1).
Quantitative real-time PCR.
RNA was processed, real-time PCRs were carried out in an ABI Prism 7700 sequence detection system (Applied Biosystems), and the fold change in expression for each gene was calculated as described previously, with the ribosomal protein 36B4 mRNA as an internal control (1, 45). Primer sequences will be provided upon request.
ChIP assays and Solexa ChIP-Seq analysis.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described before (2, 31), and the antibodies used were against ERα (HC-20; Santa Cruz Biotechnology) and Runx1 (PC384; Calbiochem). DNA was purified using QiaQuick columns (Qiagen, Valencia, CA) and was subjected to quantitative PCR or Solexa analysis using the Genome Analyzer system according to the manufacturer's recommendations (Illumina, San Diego, CA). Single-end sequences were trimmed to 25 bp and mapped to the human genome assembly hg18 using Eland (Illumina), keeping only tags that mapped uniquely. The 3′ ends of the tags were adjusted by 75 bp, corresponding to half the recommended fragment length for Illumina sequencing. Only one tag from each unique position was considered, to eliminate peaks resulting from clonal amplification of fragments during the ChIP-Seq protocol. Peaks (binding sites) were identified by searching for clusters of tags within a sliding 200-bp window, requiring adjacent clusters to be at least 500 bp away from each other. The threshold for the number of tags that determined a valid peak was selected for a false discovery rate of 0.001, as determined empirically by peak finding using randomized tag positions. Peaks were required to have 4-fold more tags relative to the local background region (10 kb), to avoid identifying regions with genomic duplications or nonlocalized binding. Peaks that overlapped more than 20% with repeat regions of the genome were eliminated from the analysis.
Computational motif analysis.
De novo motif analysis was performed as previously described (32), and the results are available at http://biowhat.ucsd.edu/homer/. Sequences corresponding to the ERα binding sites were extracted from the March 2006 human genome assembly hg18. Sequences were divided into target and background sets for each application of the algorithm as described below. Background sequences were then selectively weighted to equalize the distributions of CpG content in target and background sequences to avoid comparing sequences of different general sequence content. Motifs of lengths 8, 10, 12, and 14 bp were identified separately by first exhaustively screening all oligos for enrichment in the target set compared to the background set by using the cumulative hypergeometric distribution to score enrichment. Up to two mismatches were allowed in each oligonucleotide sequence to increase the sensitivity of the method. The top 200 oligonucleotides of each length with the lowest P values were then converted into probability matrices and heuristically optimized to maximize hypergeometric enrichment of each motif in the given data set. As optimized motifs were found, they were removed from the data set to facilitate the identification of additional motifs. Sequence logos were generated using WebLOGO (http://weblogo.berkeley.edu/).
Microarray sequence accession numbers.
The microarray expression data have been deposited in the Gene Expression Omnibus database and assigned accession number GSE 22593. Also, the ChIP-Seq data have been deposited in the Gene Expression Omnibus database and assigned accession number GSE 22609. The Gene Expression Omnibus superseries containing both the microarray expression and ChIP-Seg data is accession number GSE 22610.
RESULTS
The ERα DNA binding domain mutant DBDmut ER selectively regulates gene expression independent of ERE binding.
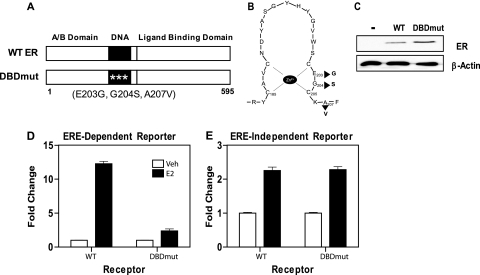
The ERα DNA binding domain contains two nonequivalent zinc fingers, CI and CII, which interact with the major groove and phosphate backbone of DNA, respectively (43). Three mutations, E203G/G204S/A207V, in CI of the human ER (DBDmut) have been shown to disrupt binding of the ER to EREs by more than 95% while not affecting hormone binding (29). Of note, this is an altered specificity mutant that exhibits substantially reduced binding to EREs and has increased affinity for the hormone response element (HRE), which is recognized by the glucocorticoid, androgen, and progesterone nuclear hormone receptors, thereby providing a positive control for the overall integrity of the DNA binding domain. The positions of the three mutations are shown in Fig. 1A, and the relative locations of these mutations in the P-box are diagramed in Fig. 1B. We used this DBDmut ER as a tool to discriminate genes where regulation by the ER required direct DNA (ERE) binding from those regulated independent of ERE binding, through tethering. To do so, human MDA-MB-231 breast cancer cells were stably transfected with either the wild type (WT) or the DBDmut receptors, creating 231ER+ cells. The stably expressing ER cell lines chosen for these studies expressed quite similar receptor levels, with expression of the DBDmut being slightly higher than that of the WT ER (Fig. 1C).
FIG. 1.
The estrogen receptor DNA binding domain mutant selectively activates ERE binding-independent estrogen signaling. (A) Schematic diagram of the WT ERα and DBDmut ER, which contains three alterations (E203G, G204S, and A207V) in the first zinc finger of the ER DNA binding domain that greatly diminishes binding to the ERE. (B) Detailed diagram for the locations of the three mutations in the DBDmut receptor. (C) Western blot analysis of ERα in MDA-MB-231 breast cancer cells stably expressing the WT or DBDmut ER. (D and E) Regulation of the ERE-containing reporter (2ERE-pS2-luciferase) (D) or the ERE-independent reporter (TGFβ3-luciferase) (E) by the WT or DBDmut receptor in MDA-MB-231 cells treated with vehicle (Veh) or 10 nM E2. Values from triplicate experiments ± standard errors of the means are expressed as normalized light units (firefly luciferase/Renilla luciferase).
We first examined the activities of these ERs on an ERE-dependent and an ERE-independent reporter gene. These reporter genes were transfected into the 231 cells stably expressing either the WT or DBDmut ER. As expected, in the presence of 10 nM E2, the ERE-dependent reporter (2ERE-pS2-luciferase) was activated by the WT ER to a much higher level than by the DBDmut receptor (Fig. 1D). In contrast, the ERE-independent reporter gene, TGFβ3-luciferase, which contains the proximal promoter of the TGFβ3 gene but no ERE or ERE half-sites, was activated very well by both the WT and DBDmut receptors (Fig. 1E). Hence, the DBDmut ER has lost most of the ability of the receptor to regulate transcription by an ERE-dependent mechanism, while it retains the ability of the ER to regulate gene expression independent of ERE interaction.
Identification of genes regulated by the ER through direct DNA (ERE) binding versus non-ERE-mediated tethering mechanisms.
To determine the relative importance of the direct DNA binding versus tethering mechanisms in E2-dependent gene regulation, we performed global microarray gene expression analyses for the 231ER+ cell lines expressing the WT or DBDmut receptors. The WT and DBDmut ER-containing cells were treated with E2 for 1, 2, 4, 8, and 24 h. We hypothesize that genes regulated much more robustly by the WT ER than by DBDmut ER are those that require ERE binding (either alone or in possible combination with tethering), while genes regulated equally by the WT ER and DBDmut ER represent genes regulated principally by tethering. Genes were considered to be significantly regulated by hormone if RNA levels changed by ≥l.5-fold with a P value of <0.05.
This analysis identified 420 E2-regulated genes in the 231 WT ER-expressing cells and 402 regulated genes in the 231 DBDmut ER-expressing cells, with 109 genes regulated by both receptors (see Fig. S1A in the supplemental material). We chose to focus on the genes regulated by E2 in the 231WT ER-expressing cells (420 genes, constituting the ERα transcriptome), which includes those that overlap with the 109 genes regulated in the DBDmut ER-expressing cells, since these represent genes also regulated by the WT ER. We believe that genes regulated only by the DBDmut ER most likely represent genes regulated through estrogen receptor-independent mechanisms, including binding to additional response elements or proteins, and hence were not considered further.
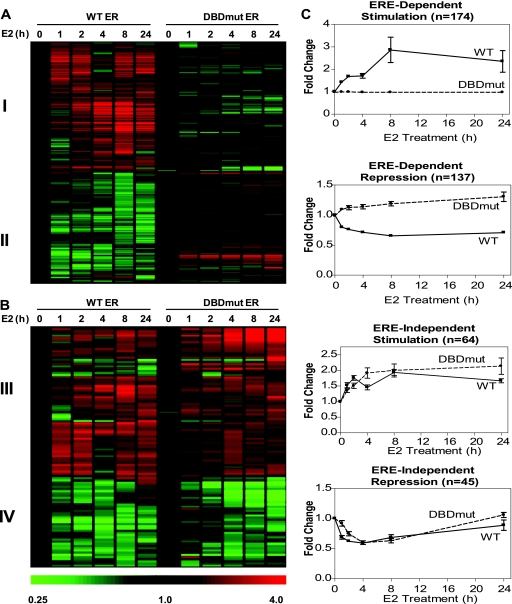
Hierarchical clustering of these E2-regulated genes using the microarray expression data from the WT or DBDmut ER-expressing cells segregated the E2-regulated genes into two major classes: (i) genes that were regulated only by the WT ER (Fig. 2A) and (ii) genes that were regulated by both the WT ER and DBDmut ER (Fig. 2B). These two major groups could be further subdivided based on whether the genes were stimulated (I and III) or repressed (II and IV) by E2. The microarray data for these four regulation patterns are shown in Fig. 2C. This analysis identified 311 genes (174 stimulated and 137 repressed) that were regulated by the WT ER only (ERE binding dependent), while 109 genes (64 stimulated and 45 repressed) were regulated equally by the WT ER and DBDmut ER (ERE binding independent). Therefore, approximately 75% of the genes that are stimulated by E2 or repressed by E2 appear to require ERE binding of the receptor in their regulation, with 25% regulated independently of ERE binding. That the majority (75%) of genes repressed by E2 also require the ER with a functional DNA binding domain emphasizes the importance of DNA binding in both gene stimulation and repression activities of the ER in these cells.
FIG. 2.
ER gene regulation is predominantly through ERE binding-dependent mechanisms. (A and B) Hierarchal gene tree clustering for genes regulated by 10 nM E2 over time (0, 1, 2, 4, 8, and 24 h) through an ERE binding-dependent mechanism (n = 311) (A) or an ERE binding-independent mechanism (n = 109) (B) in MDA-MB-231 cells stably expressing either the WT ER or DBDmut ER. Upregulated genes are shown in red, and genes downregulated by E2 are shown in green. Groups I, II, III, and IV are described in the text. (C) Average microarray gene expression fold change ± the standard error of the mean for genes regulated through an ERE binding-dependent mechanism (stimulation [n = 174] or repression [n = 137]) or through an ERE binding-independent mechanism (stimulation [n = 64] or repression [n = 45]) in MDA-MB-231 cells containing either the WT ER or DBDmut ER.
To ensure that the identified ERE-independent tethering genes were specifically regulated by the estrogen receptor, 231ER+ cells were treated with agonists for the estrogen and glucocorticoid receptors. As expected, the mRNA for TFF1 was stimulated by E2 treatment but not dexamethasone (DEX), while GILZ mRNA was stimulated by DEX treatment but remained unresponsive to E2 treatment (see Fig. S1B in the supplemental material). Examination of the mRNA regulation of five representative genes identified to be activated by ER through the ERE-independent tethering mechanism displayed E2-dependent activation; however, all five mRNAs were unchanged by dexamethasone treatment, demonstrating that these genes are specifically targeted by estrogen signaling (see Fig. S1B, right).
Genome-wide localization of binding sites for the DBDmut ER and WT ER.
Recent studies have identified binding sites for the ER upon E2 treatment of MCF-7 human breast cancer cells on a genome-wide basis (4, 26, 57). In our previous studies, we demonstrated that E2 regulated common as well as distinct sets of genes in MCF-7 cells and 231 cells expressing WT ER (45), suggesting that the ER might have a partly distinct profile of genomic ER binding sites in 231ER+ cells compared to MCF-7 cells.
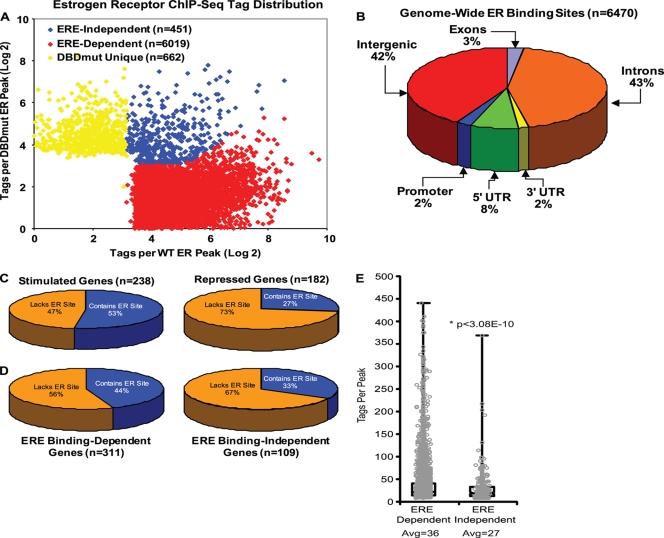
To compare the genome-wide binding of the DBDmut ER and the wild-type ER, we performed ChIP assays for these two ERs in E2-treated 231WT ER and DBDmut ER cells followed by Solexa high-throughput sequencing. This ChIP-Seq analysis provided 7,008,583 and 6,971,823 unique DNA fragments for the WT and DBDmut ER, respectively, that mapped to the human genome and corresponded to 6,470 and 662 unique peaks, respectively (Fig. 3A ). Of these 6,470 identified WT ER binding sites in 231ER+ cells, 3,638 (56%) overlapped with the high-confidence binding sites identified in MCF-7 cells (cf. Fig. 6D), demonstrating that ERα partially shares its binding profile within different breast cancer cell lines; however, each cell line also contains unique genomic positions where ERα localizes.
FIG. 3.
ChIP-Seq identified ER binding sites that involve ERE-dependent or ERE-independent binding of the ER. (A) ER ChIP-Seq tag distribution. The scatter plot analysis shows peaks identified in WT ER-containing and DBDmut ER-containing cells. Peaks preferential for WT ER recruitment (n = 6,019) are denoted in red, while peaks common for both WT ER and DBDmut ER (n = 451) are blue. Peaks unique for the DBDmut ER (n = 662) are shown in yellow. (B) Genomic distribution of the 6,470 unique ER binding sites in 231 WT ER cells treated with 10 nM E2 for 45 min. (C) Pie graphs showing the percentages of stimulated genes (left) or repressed genes (right) that are in proximity to wild-type ER binding sites. (D) Pie graphs showing the percentages of ERE binding-dependent genes (left) or ERE binding-independent (tethering) genes (right) that are in proximity to ER binding sites. (E) Box plot of tags for WT ER binding to ERE binding-dependent or ERE binding-independent peaks.
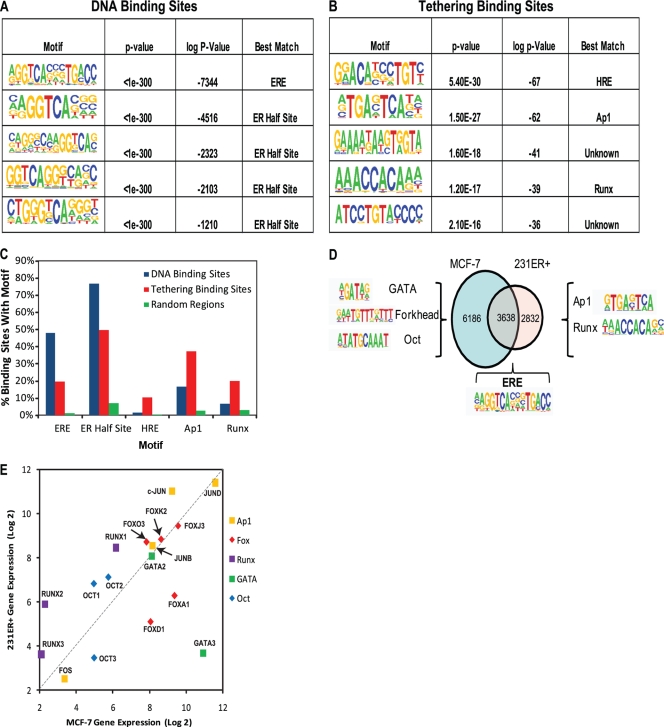
FIG. 6.
Analysis of motifs enriched for the WT ER or DBDmut ER binding sites. (A and B) De novo identification of enriched motifs in DNA binding sites (A) and tethering binding sites (B) relative to the genomic background. Nucleotide sequences were visualized by using WebLogo (http://weblogo.berkeley.edu/). (C) Distribution of Ap1 and Runx motifs in WT ER binding sites and in DBDmut binding sites. (D) Venn diagram demonstrating the overlap of binding sites (peaks) between the ERα ChIP-Seq data from MCF-7 and 231ER+ cells. The sequence logo for the top enriched motifs from MCF-7 (GATA, Forkhead, and Oct), 231ER+ (Ap1 and Runx), and common (ERE) peaks are also shown. (E) Microarray expression data for the indicated transcription factors in MCF-7 and 231ER+ cells.
The majority of the peaks identified in 231ER+ cells were located either in the introns of known genes (43%) or in intergenic regions (42%) located more than 1 kb away from genes (Fig. 3B). Only 10% of the peaks were localized to the proximal promoter or 5′ untranslated region of known genes, consistent with two previous genome-wide ER binding site data sets (4, 26). Therefore, the ability of ERα to regulate many genes through long-range distal sites most likely remains a common mechanism in ER-positive breast cancer cells despite each having some unique binding profiles and regulation programs. Of the 420 E2-regulated genes we identified in 231ER+ cells, 174 (41%) contained WT ER binding sites within 50 kb of the transcription start site, and there was a higher association of WT ER binding sites with E2-stimulated genes than with repressed genes (53% versus 27%) (Fig. 3C). Also, more ERE binding-dependent genes had ER binding sites associated with them than did ERE binding-independent genes (44% versus 33%) (Fig. 3D).
To differentiate ER peaks that are ERE dependent or independent for binding, we compared high-throughput Solexa sequencing results for the DBDmut to that of the WT ER (Fig. 3A). The DBDmut colocalized to only 451 (7%) (blue dots) of the 6470 WT binding peaks (red dots plus blue dots), which indicates that the majority of ER recruitment to ER binding sites requires a fully functional DNA binding domain. Notably, use of the DBDmut allowed for the identification of a subset of ER sites that are bound by ER in an ERE-independent mode. It is interesting that the high-throughput sequencing revealed, as expected, that the altered specificity DBDmut ER also bound to some sites different from those to which WT ER bound (DBDmut unique sites [yellow dots]), due to its binding to hormone response elements, and indeed the HRE was an enriched motif in this subset of binding sites (see Fig. S2 in the supplemental material), thereby providing a positive control for the overall integrity of the DNA binding domain. Interestingly, the ERα binding sites that require ERE binding of the receptor contained more tags/peaks than did the ERE binding-independent sites (36 compared to 27; P < 3.08E-10) (Fig. 3E), which is consistent with our observation (see below) that gene stimulations involving direct receptor ERE binding are often more robust.
Differential recruitment of WT ER and DBDmut ER to binding sites of estrogen-regulated genes.
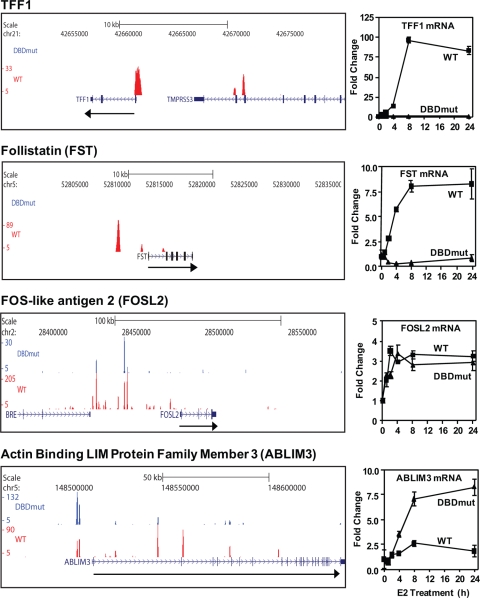
Treatment of WT ER cells with E2 promoted rapid receptor recruitment to the promoter and enhancer regions of the TFF1 gene and marked stimulation of TFF1 gene expression (Fig. 4 top, TFF1); however, no recruitment of receptor to these TFF1 regions was observed in DBDmut cells, consistent with the lack of induction of this mRNA by the DBDmut ER (Fig. 4, top left and right panels). Likewise, the WT receptor, but not the DBDmut ER, was recruited to the Follistatin (FST) promoter and an enhancer approximately 3 kb upstream of the transcription start site (Fig. 4, FST). In contrast, the WT and the DBDmut ERs were both recruited to binding sites in the FOSL2 and ABLIM3 gene loci (Fig. 4, left), and these genes showed E2-dependent upregulation in both 231WT ER and 231 DBDmut cells (Fig. 4, right). Therefore, only the WT ER, but not the DBDmut ER, was recruited to genes that are regulated through ERE binding (TFF1 and FST), while both WT and DBDmut receptors were recruited to genes regulated through non-ERE-dependent tethering (FOSL2 and ABLIM3). Validation of the ChIP-Seq findings for representative genes is shown in Fig. S3 of the supplemental material.
FIG. 4.
Recruitment of the WT ER and DBDmut ER to specific gene targets. (Left) UCSC Genome Browser images of TFF1, FST, FOSL2, and ABLIM3. FST, FOSL2, and ABLIM3 are shown in the natural 5′-to-3′ orientation, while TFF1 is shown in the 3′-to-5′ orientation. Black arrows indicate the direction of transcription. Red peaks represent peaks where the WT ER is recruited, while blue peaks represent peaks where the DBDmut ER is recruited. (Right) Microarray mRNA expression is shown for TFF1, FST, FOSL2, and ABLIM3 in 231 cells containing WT or DBDmut ERs treated with 10 nM E2 for periods of up to 24 h.
Estradiol stimulates genes requiring DNA binding of the ER to a higher magnitude than genes regulated by ER tethering only.
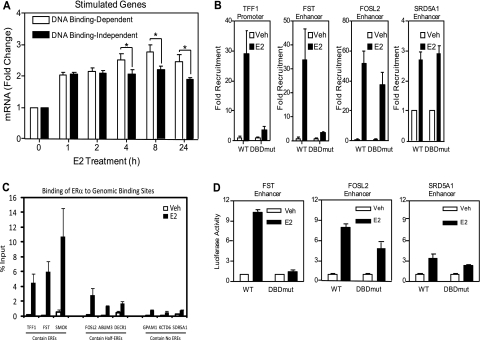
The overall gene regulation patterns for ERE binding-dependent and ERE binding-independent paradigms suggested that genes might be activated more robustly through direct DNA binding than tethering (Fig. 1D and E and 2C). This prompted us to compare the average change in gene expression for the genes regulated through ERE-dependent and ERE-independent modes. As seen in Fig. 5A, the average stimulation for the ERE-binding versus ERE-independent genes was similar at 1 and 2 h of E2 treatment, whereas the magnitude of stimulation for the ERE binding-dependent genes was significantly higher after 4, 8, and 24 h of hormone treatment (Fig. 5A). These data demonstrate that gene expression requiring DNA binding of the ER is, on average, increased more robustly over time than is gene expression activated through indirect ER-DNA tethered interactions, suggesting that direct ERE binding might function to stabilize a complex between ER and the transcriptional machinery in a manner that results in more efficient transcription.
FIG. 5.
Correlation of magnitude of gene expression and ER recruitment to chromatin binding sites. (A) Average Affymetrix microarray gene expression (± standard errors of the means) for genes stimulated through ERE binding-dependent or ERE binding-independent mechanisms in MDA-MB-231 cells containing the WT or DBDmut ER. *, P < 0.05. (B) Differential recruitment of the WT ER and/or DBDmut ER, determined by chromatin immunoprecipitation assays, to known ER binding sites located in the TFF1 promoter, FST enhancer, FOSL2 enhancer, or SRD5A1 enhancer in MDA-MB-231 cells containing either the WT or DBDmut ER exposed to control vehicle or 10 nM E2 for 45 min. (C) ERα binding to the indicated enhancers in 231 WT ER cells following treatment with vehicle or 10 nM E2 for 45 min. (D) Regulation of representative ER binding sites in luciferase reporter gene assays. 231 cells stably containing the WT ER or DBDmut ER were transfected with the designated gene enhancer-Luc reporter construct and then treated with vehicle (Veh) or 10 nM E2 for 24 h. Values are normalized luciferase units ± standard deviations of triplicate experiments.
To understand possible mechanisms for these observations, we analyzed the pattern of recruitment of the WT and DBDmut ERs to binding sites for a few genes (4, 57) with enhancers representing each regulation paradigm. As shown in Fig. 5B, only WT ER was recruited to the TFF1 (trefoil factor 1) promoter and the FST (follistatin) enhancer sites, whereas both WT and DBDmut ERs were recruited equally to the FOSL2 (FOS-like 2) and SRD5A1 (steroid reductase 5A1) enhancers. Time-course ChIP assays to follow the dynamics of receptor recruitment to three representative binding sites demonstrated similar kinetics for recruitment of both the WT and DBDmut receptors, with recruitment as early as 5 min of E2 treatment (data not shown). The magnitude of E2-dependent recruitment of WT ER to binding sites containing canonical full EREs was stronger than recruitment to sites containing half-EREs or lacking EREs (Fig. 5C). These data suggest that ER may be recruited more robustly to ER binding sites that contain full ERE sequences than to binding sites containing either a half-ERE or lacking an ERE or half-ERE site.
Because ER recruitment to a genomic binding site need not always result in gene regulation, we examined gene expression regulation for representative genes. Genomic DNA encompassing representative ER binding sites for the FST, FOSL2, and SRD5A1 genes was cloned into luciferase reporter vectors and transfected into 231 cells containing the WT or DBDmut receptors, and the magnitude of estrogen response was determined (Fig. 5D). The FST enhancer was activated 10-fold by E2 in WT ER-containing cells, whereas the DBDmut ER was very ineffective (Fig. 5D). In contrast, both the WT and DBDmut receptors activated the FOSL2 and SRD5A1 enhancers. These results confirm that the identified ER binding regions are indeed responsive to E2 and that the relative recruitment of the two receptors observed in the ChIP assays correlates well with the magnitude of E2-dependent regulation of these genes.
Determinants of ER direct DNA binding and tethering.
To identify cooperating factors that might be responsible for ERα recruitment to tethering sites, we performed an unbiased search for DNA motifs enriched in the identified ER binding sites. The DNA sequences corresponding to direct ER binding sites were searched for enriched motif sequences relative to genomic background sequence selected at random (Fig. 6). The five most enriched DNA motifs in the WT ER binding sites are listed in Fig. 6A, and the five most enriched motifs in the tethered binding sites are given in Fig. 6B. As expected, the ERE was the most enriched motif for WT ER and was present in approximately 50% of the sites (Fig. 6C). In addition, motifs corresponding to half EREs were also very highly enriched in direct binding sites.
To identify motifs that are specifically involved in tethering, we searched for motifs in tethered binding sites while using direct WT ER binding sites as a background set. In contrast to direct binding sites, the most enriched motifs for the tethering sites included Ap1, Runx, and HRE (Fig. 6B). The Ap1 motif was present in 37% of the binding sites of the DBDmut ER, while being present in only 16% of the WT ER DNA binding sites (Fig. 6C). In addition, the Runx motif was present in 20% of DBDmut sites, while only 7% of the WT ER binding sites contained a Runx motif. These data suggest that members of the Ap1 and the Runx families may be potential candidate tethering factors involved in mediating ERα-dependent gene regulation. Notably, ERE and half-ERE motifs were found at lower frequencies in the DBDmut ER sites (Fig. 6C). Binding sites for glucocorticoid/androgen/progesterone receptors (HRE; GR, AR, and PR, respectively) were present in less than 10% of tethering binding sites.
Although we cannot rule out the possibility that the identified tethering sites are ones that interact with both the ER and the DNA binding domains of other nuclear receptors (GR, PR, and AR), our data support that ∼90% of these sites are true ER tethering sites, based on the following: (i) the recruitment of wild-type ERα and DBDmut ER to these sites (see Fig. S3, right panel, in the supplemental material), (ii) the presence of an HRE in less than 10% of the tethering sites (Fig. 6C), and (iii) the responsiveness of tethering genes to E2 treatment but not to treatment by the GR agonist dexamethasone (see Fig. S1B, right).
Comparison of the genome-wide binding programs of ERα in 231 WT ER versus MCF-7 cells.
As mentioned above, the genome-wide binding program for WT ERα in 231ER+ cells shared a significant overlap of sites (56%) with that previously observed for ERα in MCF-7 cells (Fig. 6D); however, ERα also bound preferentially to several thousand enhancers in either MCF-7 or 231ER+ breast cancer cells. To gain further insights into the cell-specific binding programs for ERα, we performed unbiased de novo motif analysis on sites bound by ERα in both cell lines and also those bound by ERα preferentially in either cell type. The top motif identified among the common binding sites was a canonical ERE compared to random genomic background, as expected (Fig. 6D). Of note, however, sites bound by WT ERα preferentially in the MCF-7 cells were highly enriched for GATA, Forkhead and Octamer (Oct) motifs, while Ap1 and Runx motifs were specifically enriched in sites preferentially bound by WT ERα in 231ER+ cells (Fig. 6D).
Examination of the expression levels of these transcription factors in 231ER+ cells and in MCF-7 cells revealed that members of the Forkhead (e.g., FOXA1) and GATA (e.g., GATA3) families were expressed at much higher levels in MCF-7 cells than in 231ER+ cells (Fig. 6E), whereas expression of Runx family members was much higher in 231ER+ cells, which is of interest because the Runx motif was preferentially enriched in the 231ER+ data set (Fig. 6E). Some members of the Octamer and Ap1 families exhibited similar expression levels in MCF-7 and 231ER+ cells and therefore might not be involved in directing cell-type-specific ERα binding. Our data suggest a role for GATA3 and FOXA1 in coordinating cell-type-specific binding of ERα in MCF-7 cells, with members of the Runx family facilitating ERα binding in 231ER+ cells.
ERα tethers to Runx1 to regulate gene expression.
A role for Runx family transcription factors in mediating ER-dependent gene activation has not previously been described; however, the enrichment of the Runx motif in the ER tethering sites suggested that Runx family members might mediate the recruitment of ERα to some chromatin sites and, therefore, be necessary for estrogen regulation of a subset of tethering genes. Of the three known Runx family members, mRNA expression for Runx1 was highest in the 231ER+ cells by Affymetrix microarray analysis and by quantitative real-time PCR. Runx2 was present at a lower level (Fig. 6E), and Runx3 expression was very low. Knockdown of Runx1 or Runx2 by small interfering (siRNA) resulted in efficient and specific depletion of each mRNA (see Fig. S4A and B in the supplemental material) and protein (see Fig. S4C).
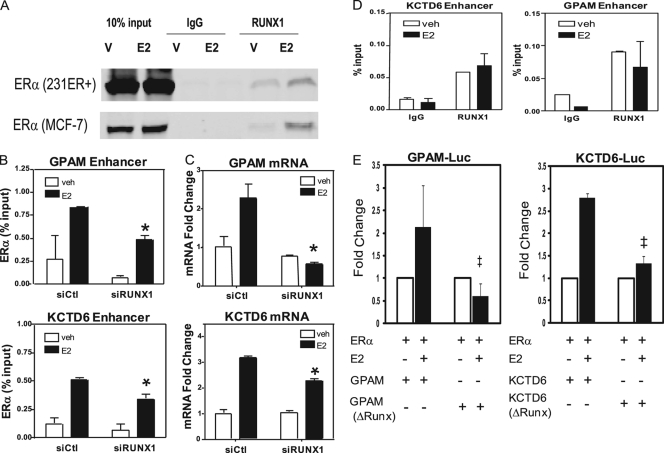
The observation that the Runx motif was specifically enriched in a subset of ER tethering sites suggested the possibility that Runx1 might bind to and serve as a tethering protein for ERα at distinct chromosomal locations. Indeed, coimmunoprecipitation experiments in 231ER+ cells (Fig. 7A) and MCF-7 cells (Fig. 7A) demonstrated interaction of ERα and Runx1 under basal conditions and that E2 treatment enhances this interaction. In addition, depletion of Runx1 by siRNA resulted in diminished E2-dependent recruitment of ERα to enhancer sites in GPAM and KCTD6, which contain consensus Runx motifs (Fig. 7B), while not affecting the recruitment of ERα to binding sites in the enhancer of HEG1 that lacks Runx binding motifs (see Fig. S5 in the supplemental material). In addition, knockdown of Runx1 eliminated the stimulation of GPAM mRNA expression by estrogen (Fig. 7C) and reduced estrogen stimulation of KCTD6 mRNA (Fig. 7C), but had little effect on the E2 stimulation of HEG1, which lacks Runx motifs (see Fig. S5). In contrast to the impact of Runx1 loss on ERα recruitment and gene regulation, we found that knockdown of Runx2, which is less abundant in 231 cells, had no impact on the gene regulation by estrogen, and knockdown of both Runx1 and Runx2 gave ERα recruitment and effects on gene stimulations by E2 identical to those observed with the Runx1-only knockdown (data not shown). Interestingly, we detected Runx1 occupancy by ChIP assays at both the GPAM and KCTD6 ER binding sites in the presence and absence of E2 treatments, demonstrating that Runx1 is present at these sites in vivo (Fig. 7D).
FIG. 7.
Runx1 is required for ERα localization to, and regulation of, a subset of tethering genes. (A) Coimmunoprecipitation of ERα and Runx1 in 231ER+ cells and in MCF-7 cells. Cells were treated with vehicle (V; 0.1% ethanol) or 10 nM E2 for 45 min prior to immunoprecipitation with Runx1 antibody or IgG followed by Western immunoblotting for ERα. (B) ChIP assays assessing the recruitment of ERα to the GPAM or KCTD6 enhancers in 231ER+ cells treated with siControl (siCtl) or siRunx1 and then exposed to vehicle or 10 nM E2 for 45 min. (C) Quantitative PCR analysis of GPAM and KCTD6 mRNA in 231ER+ cells treated with siCtl or siRunx1 and then with vehicle or 10 nM E2 for 4 h. (D) ChIP assays assessing the recruitment of Runx1 to the GPAM or KCTD6 enhancers in 231ER+ cells treated with vehicle or 10 nM E2 for 45 min. (E) Regulation of luciferase reporters containing the ER binding site near the GPAM gene, GPAM-luc (left), or the GPAM(ΔRunx1)-luc (right), in which the Runx1 motif has been deleted, in 231ER+ cells treated with vehicle or 10 nM E2 for 24 h. Values from triplicate experiments ± standard errors of the means are expressed as normalized light units. *, P < 0.05 compared to siCtl or E2 treatment; ‡, P < 0.05 compared to luciferase reporter with intact Runx1 motifs and E2 treatment.
To further confirm a role for Runx1 in ERα gene regulation, the enhancer sites for ER in GPAM and KCTD6 were cloned directly upstream of a luciferase reporter gene. In transient cell transfections, we observed E2-dependent stimulation of both reporter genes by ERα (Fig. 7E), and deletion of the Runx motifs in these enhancers abrogated the E2 stimulation (Fig. 7E). Collectively, these findings demonstrate the involvement of Runx1 in ERα regulation of a subset of tethering genes in breast cancer cells.
DISCUSSION
In this study we have investigated the roles of direct DNA binding and tethering in gene regulation by ERα by analyzing the recruitment of WT ERα and an ERE binding-deficient ER to chromatin binding sites across the genome and their comparative gene regulations in response to hormone in breast cancer cells. By parsing the contributions of direct ERE binding versus tethering in receptor activities we found, interestingly, that the majority of gene regulations and ERα recruitment to chromatin binding sites require a functional ER DNA binding domain; however, a subset of E2-regulated genes were regulated by ERα in the absence of ERE binding. In addition, our findings demonstrate the importance of Runx1 as a cooperating factor in ER-mediated gene regulation and chromatin binding at tethered sites. Our ChIP-Seq analyses, the first to be done in ER-containing 231 human breast cancer cells, have also enabled important comparisons to be made with another ER-positive breast cancer cell line, MCF-7, in which all prior ER chromatin localization analyses have been conducted. The use of an ERα mutant in which activation of gene expression is restricted to non-ERE-mediated responses facilitated the identification of this subset of genes and allowed us to uncover properties of the ERα tethering mechanism that would not have been otherwise possible.
Traditionally, the ER has been characterized as a transcription factor that binds to specific DNA sequences in the regulatory regions of genes. Our data support this as the dominant mechanism of gene transcriptional activation, since approximately 75% of the genes regulated by the ERα in these breast cancer cells required an intact DNA binding domain and 50% of the ER binding sites for the WT ER contained a full ERE. It is important to note, however, that ER-regulated genes often contain multiple ER binding sites, as already described for several genes, such as GREB1 (29, 50) and pS2 (34) in MCF-7 breast cancer cells, and that genes may be regulated by ER through a combination of direct DNA binding and indirect tethering binding modes. Hence, the overall increased magnitude of gene regulation by the WT ER versus DBDmut ER could reflect the combined actions of DNA binding and tethering mechanisms. While we expected, and indeed found, that the regulation of gene expression by the WT ER, which represents the combined output from direct DNA binding and/or tethering modes, was usually equal to or greater than that of the DBDmut ER, it is noteworthy that at least one gene, ABLIM3, showed higher regulation by the DBDmut ER than the WT ER (Fig. 4). Hence, in this case, gene expression might be stimulated by the ER via tethering and repressed by ER via a direct DNA binding mechanism. With the DBDmut ER, where direct DNA binding to EREs is lacking, this repression would be removed, enabling gene stimulation by the DBDmut ER to exceed that of the WT ER.
In our studies, binding sites containing a full ERE demonstrated, on average, higher recruitment of ER than binding sites containing only a half-ERE or lacking EREs entirely, and the WT ER activated gene expression to a greater extent, on average, than did the DBDmut ER. However, the overall time course of gene stimulation through DNA binding-dependent or tethering modes appeared indistinguishable, with the ER in both cases becoming rapidly (by 5 min) associated with binding sites. We have observed equally rapid recruitment of the ER to chromatin binding sites in MCF-7 breast cancer cells after hormone treatment (48).
Recent studies with breast cancer cells that have identified the genome-wide binding sites for ER upon E2 treatment have all been done in MCF-7 human breast cancer cells (4, 11, 26, 57). These ChIP-chip, ChIP-Seq, and ChIA-PET approaches have established that ERα regulates many genes through distal enhancers (2, 11, 29, 50, 57). Interestingly, the majority of the ER genomic binding sites identified in the 231ER+ cells were also located several kb away from regulated genes, supporting a mechanism of long-range control by the ERα in these cells as well. Our comparison of the genome-wide ER binding programs between 231ER+ cells and MCF-7 cells identified an overlap of 56%, suggesting that cell-specific factors, along with probable differences in chromatin architecture, may constitute major determinants in ERα genome binding, with considerable similarities but also distinct differences being evident in different ER-positive human breast cancer cell lines.
A notable difference between the two breast cancer cell lines studied was that the binding site for FOXA1, which facilitates ER binding in MCF-7 cells, was not enriched in the genome-wide 231WT ER binding site set, suggesting that additional factors such as Runx1 drive cell-specific ERα binding and hormonal regulation of gene expression in these cells. While studies by us (46) and others (23, 39) have revealed marked differences in the genes regulated by the estrogen-occupied ER in breast cancer (MCF-7) and osteosarcoma (U2OS) cells and only limited overlap of ER binding sites in these two estrogen target cells (23), our work extends these observations to reveal that there are also pronounced differences not only in the genes regulated by ER (45) but also in ER binding site utilization between different human breast cancer cell lines.
The previous genome-wide ERα localization studies in MCF-7 cells have provided a very useful map of binding sites for ERα (4, 11, 26, 57), but they do not distinguish between genes regulated by the ER through direct DNA binding versus tethering, because WT ER interacts with DNA by both modes. The advantage of our using the DBDmut ERα is that it has specifically enabled the identification of sites where ERα localizes in the absence of direct binding to EREs. This approach identified several hundred novel enhancers at which ERα binds through tethering to other transcription factors, and the de novo motif analysis revealed enrichment of Ap1 and Runx binding motifs in sites binding the DBDmut ER. Several previous reports have implicated ERα tethering to c-Jun in the regulation of genes through Ap1 sites (6, 24, 53); however, this report is the first to our knowledge to implicate the involvement of the Runx transcription factor in supporting ERα regulation of gene expression by tethering.
Runx transcription factors are critical regulators of cell growth and differentiation and function as cell context-dependent tumor suppressors or oncogenes (10, 16). Previous studies have documented that Runx1 exists in a complex with chromatin-modifying enzymes, including the histone acetyltransferases p300, CBP, and MOZ, implicating a role for Runx1 in modulating the epigenetics of ERα targeted enhancers (58). Our ChIP data for Runx1 suggest that Runx1 occupies ERα binding sites prior to E2 stimulation and, therefore, Runx1 may function to establish a permissive chromatin structure for ERα binding. Interestingly, it was recently reported that high-grade breast tumors have reduced Runx1 expression and that progressive loss of Runx1 expression correlates with increasingly advanced stages of breast cancer (18). Therefore, the regulation of gene expression by ERα and Runx1 may serve to restrict progression of breast cancer in ER-positive tumors. Although the Runx motif is not the most highly enriched motif in MCF-7 cells, its binding site is enriched in genome-wide ERα binding sites in MCF-7 cells compared to genomic background (P < 9.9E−8) (57). Therefore, the observed interaction between Runx1 and ERα in MCF-7 and in ERα-containing 231 cells suggests that at least a subset of genes within the ERα gene regulation program in ER-positive breast cancer cells could be controlled through Runx1-ERα interactions.
Our findings document the importance of both direct DNA binding and tethering modes in ERα-dependent gene regulation and localization to chromatin binding sites, and they highlight novel enhancers and transcription factors that cooperate in ER-mediated control of gene expression in breast cancer cells. It is likely that the paradigm of receptor tethering through Runx1 or related transcription factors we have observed for ERα may likewise be involved more generally in the actions of other members of the nuclear receptor superfamily.
Supplementary Material
Acknowledgments
This research was supported by NIH grants 5R01 CA18119 and P01 AG 024387 (B.S.K.), T32 HD07028 (J.D.S.), T32 CA009523 (J.D.S.), R01 CA52599 (C.B. and C.K.G.), and R01 DK 058110 (W.L.K.) and by a grant from the Breast Cancer Research Foundation (B.S.K.). T.H.C. was supported by an A*STAR graduate fellowship from the Singapore Agency for Science, Technology and Research. Sequencing was supported by P30 DK063491 (UCSD Core Facility).
Footnotes
Published ahead of print on 14 June 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Acevedo, M. L., K. C. Lee, J. D. Stender, B. S. Katzenellenbogen, and W. L. Kraus. 2004. Selective recognition of distinct classes of coactivators by a ligand-inducible activation domain. Mol. Cell 13:725-738. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, D. H., S. Sheng, T. H. Charn, A. Waheed, W. S. Sly, C. Y. Lin, E. T. Liu, and B. S. Katzenellenbogen. 2008. Estrogen receptor regulation of carbonic anhydrase XII through a distal enhancer in breast cancer. Cancer Res. 68:3505-3515. [DOI] [PubMed] [Google Scholar]
- 3.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33-43. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38:1289-1297. [DOI] [PubMed] [Google Scholar]
- 5.Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, E., M. L. Acevedo, P. A. Cole, and W. L. Kraus. 2005. Altered pharmacology and distinct coactivator usage for estrogen receptor-dependent transcription through activating protein-1. Proc. Natl. Acad. Sci. U. S. A. 102:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, J., C. H. Tsai-Morris, and M. L. Dufau. 2006. A novel estradiol/estrogen receptor alpha-dependent transcriptional mechanism controls expression of the human prolactin receptor. J. Biol. Chem. 281:18825-18836. [DOI] [PubMed] [Google Scholar]
- 8.Ediger, T. R., W. L. Kraus, E. J. Weinman, and B. S. Katzenellenbogen. 1999. Estrogen receptor regulation of the Na+/H+ exchange regulatory factor. Endocrinology 140:2976-2982. [DOI] [PubMed] [Google Scholar]
- 9.Frasor, J., J. M. Danes, B. Komm, K. C. Chang, C. R. Lyttle, and B. S. Katzenellenbogen. 2003. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144:4562-4574. [DOI] [PubMed] [Google Scholar]
- 10.Friedman, A. D. 2009. Cell cycle and developmental control of hematopoiesis by Runx1. J. Cell. Physiol. 219:520-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fullwood, M. J., M. H. Liu, Y. F. Pan, J. Liu, H. Xu, Y. B. Mohamed, Y. L. Orlov, S. Velkov, A. Ho, P. H. Mei, E. G. Chew, P. Y. Huang, W. J. Welboren, Y. Han, H. S. Ooi, P. N. Ariyaratne, V. B. Vega, Y. Luo, P. Y. Tan, P. Y. Choy, K. D. Wansa, B. Zhao, K. S. Lim, S. C. Leow, J. S. Yow, R. Joseph, H. Li, K. V. Desai, J. S. Thomsen, Y. K. Lee, R. K. Karuturi, T. Herve, G. Bourque, H. G. Stunnenberg, X. Ruan, V. Cacheux-Rataboul, W. K. Sung, E. T. Liu, C. L. Wei, E. Cheung, and Y. Ruan. 2009. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glidewell-Kenney, C., J. Weiss, E. J. Lee, S. Pillai, T. Ishikawa, E. A. Ariazi, and J. L. Jameson. 2005. ERE-independent ERα target genes differentially expressed in human breast tumors. Mol. Cell. Endocrinol. 245:53-59. [DOI] [PubMed] [Google Scholar]
- 13.Hall, J. M., J. F. Couse, and K. S. Korach. 2001. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J. Biol. Chem. 276:36869-36872. [DOI] [PubMed] [Google Scholar]
- 14.Harrington, W. R., S. Sheng, D. H. Barnett, L. N. Petz, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2003. Activities of estrogen receptor alpha- and beta-selective ligands at diverse estrogen responsive gene sites mediating transactivation or transrepression. Mol. Cell. Endocrinol. 206:13-22. [DOI] [PubMed] [Google Scholar]
- 15.Hewitt, S. C., J. E. O'Brien, J. L. Jameson, G. E. Kissling, and K. S. Korach. 2009. Selective disruption of ERα DNA-binding activity alters uterine responsiveness to estradiol. Mol. Endocrinol. 23:2111-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, Y. 2008. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 99:33-76. [DOI] [PubMed] [Google Scholar]
- 17.Jakacka, M., M. Ito, F. Martinson, T. Ishikawa, E. J. Lee, and J. L. Jameson. 2002. An estrogen receptor (ER)α deoxyribonucleic acid-binding domain knock-in mutation provides evidence for nonclassical ER pathway signaling in vivo. Mol. Endocrinol. 16:2188-2201. [DOI] [PubMed] [Google Scholar]
- 18.Kadota, M., H. H. Yang, B. Gomez, M. Sato, R. J. Clifford, D. Meerzaman, B. K. Dunn, L. M. Wakefield, and M. P. Lee. 2010. Delineating genetic alterations for tumor progression in the MCF10A series of breast cancer cell lines. PLoS One 5:e9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katzenellenbogen, B. S., and J. A. Katzenellenbogen. 2002. Biomedicine. Defining the “S” in SERMs. Science 295:2380-2381. [DOI] [PubMed] [Google Scholar]
- 20.Katzenellenbogen, B. S., M. M. Montano, T. R. Ediger, J. Sun, K. Ekena, G. Lazennec, P. G. Martini, E. M. McInerney, R. Delage-Mourroux, K. Weis, and J. A. Katzenellenbogen. 2000. Estrogen receptors: selective ligands, partners, and distinctive pharmacology. Recent Prog. Horm Res. 55:163-193. [PubMed] [Google Scholar]
- 21.Kim, K., N. Thu, B. Saville, and S. Safe. 2003. Domains of estrogen receptor alpha (ERα) required for ERα/Sp1-mediated activation of GC-rich promoters by estrogens and antiestrogens in breast cancer cells. Mol. Endocrinol. 17:804-817. [DOI] [PubMed] [Google Scholar]
- 22.Klinge, C. M. 2001. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 29:2905-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krum, S. A., G. A. Miranda-Carboni, M. Lupien, J. Eeckhoute, J. S. Carroll, and M. Brown. 2008. Unique ERα cistromes control cell type-specific gene regulation. Mol. Endocrinol. 22:2393-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kushner, P. J., D. A. Agard, G. L. Greene, T. S. Scanlan, A. K. Shiau, R. M. Uht, and P. Webb. 2000. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74:311-317. [DOI] [PubMed] [Google Scholar]
- 25.Lin, C. Y., A. Strom, V. B. Vega, S. L. Kong, A. L. Yeo, J. S. Thomsen, W. C. Chan, B. Doray, D. K. Bangarusamy, A. Ramasamy, L. A. Vergara, S. Tang, A. Chong, V. B. Bajic, L. D. Miller, J. A. Gustafsson, and E. T. Liu. 2004. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 5:R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, C. Y., V. B. Vega, J. S. Thomsen, T. Zhang, S. L. Kong, M. Xie, K. P. Chiu, L. Lipovich, D. H. Barnett, F. Stossi, A. Yeo, J. George, V. A. Kuznetsov, Y. K. Lee, T. H. Charn, N. Palanisamy, L. D. Miller, E. Cheung, B. S. Katzenellenbogen, Y. Ruan, G. Bourque, C. L. Wei, and E. T. Liu. 2007. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonard, D. M., and B. W. O'Malley. 2006. The expanding cosmos of nuclear receptor coactivators. Cell 125:411-414. [DOI] [PubMed] [Google Scholar]
- 28.Luo, M., M. Koh, J. Feng, Q. Wu, and P. Melamed. 2005. Cross talk in hormonally regulated gene transcription through induction of estrogen receptor ubiquitylation. Mol. Cell. Biol. 25:7386-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mader, S., V. Kumar, H. de Verneuil, and P. Chambon. 1989. Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature 338:271-274. [DOI] [PubMed] [Google Scholar]
- 30.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 31.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa, S., J. Lozach, C. Benner, G. Pascual, R. K. Tangirala, S. Westin, A. Hoffmann, S. Subramaniam, M. David, M. G. Rosenfeld, and C. K. Glass. 2005. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 122:707-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paech, K., P. Webb, G. G. Kuiper, S. Nilsson, J. Gustafsson, P. J. Kushner, and T. S. Scanlan. 1997. Differential ligand activation of estrogen receptors ERαand ERβ at AP1 sites. Science 277:1508-1510. [DOI] [PubMed] [Google Scholar]
- 34.Pan, Y. F., K. D. Wansa, M. H. Liu, B. Zhao, S. Z. Hong, P. Y. Tan, K. S. Lim, G. Bourque, E. T. Liu, and E. Cheung. 2008. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J. Biol. Chem. 283:32977-32988. [DOI] [PubMed] [Google Scholar]
- 35.Porter, W., B. Saville, D. Hoivik, and S. Safe. 1997. Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol. Endocrinol. 11:1569-1580. [DOI] [PubMed] [Google Scholar]
- 36.Rajendran, R. R., A. C. Nye, J. Frasor, R. D. Balsara, P. G. Martini, and B. S. Katzenellenbogen. 2003. Regulation of nuclear receptor transcriptional activity by a novel DEAD box RNA helicase (DP97). J. Biol. Chem. 278:4628-4638. [DOI] [PubMed] [Google Scholar]
- 37.Razin, S. V., O. V. Iarovaia, N. Sjakste, T. Sjakste, L. Bagdoniene, A. V. Rynditch, E. R. Eivazova, M. Lipinski, and Y. S. Vassetzky. 2007. Chromatin domains and regulation of transcription. J. Mol. Biol. 369:597-607. [DOI] [PubMed] [Google Scholar]
- 38.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20:1405-1428. [DOI] [PubMed] [Google Scholar]
- 39.Rudnik, V., A. Sanyal, F. A. Syed, D. G. Monroe, T. C. Spelsberg, M. J. Oursler, and S. Khosla. 2008. Loss of ERE binding activity by estrogen receptor-alpha alters basal and estrogen-stimulated bone-related gene expression by osteoblastic cells. J. Cell Biochem. 103:896-907. [DOI] [PubMed] [Google Scholar]
- 40.Safe, S. 2001. Transcriptional activation of genes by 17 beta-estradiol through estrogen receptor-Sp1 interactions. Vitam. Horm. 62:231-252. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez, R., D. Nguyen, W. Rocha, J. H. White, and S. Mader. 2002. Diversity in the mechanisms of gene regulation by estrogen receptors. Bioessays 24:244-254. [DOI] [PubMed] [Google Scholar]
- 42.Saville, B., M. Wormke, F. Wang, T. Nguyen, E. Enmark, G. Kuiper, J. A. Gustafsson, and S. Safe. 2000. Ligand-, cell-, and estrogen receptor subtype (alpha/beta)-dependent activation at GC-rich (Sp1) promoter elements. J. Biol. Chem. 275:5379-5387. [DOI] [PubMed] [Google Scholar]
- 43.Schwabe, J. W., L. Chapman, J. T. Finch, and D. Rhodes. 1993. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell 75:567-578. [DOI] [PubMed] [Google Scholar]
- 44.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 45.Stender, J. D., J. Frasor, B. Komm, K. C. Chang, W. L. Kraus, and B. S. Katzenellenbogen. 2007. Estrogen regulated gene networks in human breast cancer cells: Involvement of E2F1 in the regulation of cell proliferation. Mol. Endocrinol. 21:2112-2123. [DOI] [PubMed] [Google Scholar]
- 46.Stossi, F., D. H. Barnett, J. Frasor, B. Komm, C. R. Lyttle, and B. S. Katzenellenbogen. 2004. Transcriptional profiling of estrogen-regulated gene expression via estrogen receptor (ER) α or ERβ in human osteosarcoma cells: distinct and common target genes for these receptors. Endocrinology 145:3473-3486. [DOI] [PubMed] [Google Scholar]
- 47.Stossi, F., V. S. Likhite, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2006. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J. Biol. Chem. 281:16272-16278. [DOI] [PubMed] [Google Scholar]
- 48.Stossi, F., Z. Madak-Erdogan, and B. S. Katzenellenbogen. 2009. Estrogen receptor alpha represses transcription of early target genes via p300 and CtBP1. Mol. Cell. Biol. 29:1749-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, J., M. J. Meyers, B. E. Fink, R. Rajendran, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 1999. Novel ligands that function as selective estrogens or antiestrogens for estrogen receptor-alpha or estrogen receptor-beta. Endocrinology 140:800-804. [DOI] [PubMed] [Google Scholar]
- 50.Sun, J., Z. Nawaz, and J. M. Slingerland. 2007. Long-range activation of GREB1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Mol. Endocrinol. 21:2651-2662. [DOI] [PubMed] [Google Scholar]
- 51.Syed, F. A., D. G. Fraser, T. C. Spelsberg, C. J. Rosen, A. Krust, P. Chambon, J. L. Jameson, and S. Khosla. 2007. Effects of loss of classical estrogen response element signaling on bone in male mice. Endocrinology 148:1902-1910. [DOI] [PubMed] [Google Scholar]
- 52.Syed, F. A., U. I. Modder, D. G. Fraser, T. C. Spelsberg, C. J. Rosen, A. Krust, P. Chambon, J. L. Jameson, and S. Khosla. 2005. Skeletal effects of estrogen are mediated by opposing actions of classical and nonclassical estrogen receptor pathways. J. Bone Miner. Res. 20:1992-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Umayahara, Y., R. Kawamori, H. Watada, E. Imano, N. Iwama, T. Morishima, Y. Yamasaki, Y. Kajimoto, and T. Kamada. 1994. Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. J. Biol. Chem. 269:16433-16442. [PubMed] [Google Scholar]
- 54.Wang, L. H., X. Y. Yang, X. Zhang, P. An, H. J. Kim, J. Huang, R. Clarke, C. K. Osborne, J. K. Inman, E. Appella, and W. L. Farrar. 2006. Disruption of estrogen receptor DNA-binding domain and related intramolecular communication restores tamoxifen sensitivity in resistant breast cancer. Cancer Cell 10:487-499. [DOI] [PubMed] [Google Scholar]
- 55.Wang, L. H., X. Y. Yang, X. Zhang, K. Mihalic, Y. X. Fan, W. Xiao, O. M. Howard, E. Appella, A. T. Maynard, and W. L. Farrar. 2004. Suppression of breast cancer by chemical modulation of vulnerable zinc fingers in estrogen receptor. Nat. Med. 10:40-47. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe, T., S. Inoue, H. Hiroi, A. Orimo, H. Kawashima, and M. Muramatsu. 1998. Isolation of estrogen-responsive genes with a CpG island library. Mol. Cell. Biol. 18:442-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welboren, W.-J., M. A. van Driel, E. M. Janssen-Megens, S. J. van Heeringen, F. C. G. J. Sweep, P. N. Span, and H. G. Stunnenberg. 2009. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 28:1418-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamagata, T., K. Maki, and K. Mitani. 2005. Runx1/AML1 in normal and abnormal hematopoiesis. Int. J. Hematol. 82:1-8. [DOI] [PubMed] [Google Scholar]
- 59.Yang, N. N., M. Venugopalan, S. Hardikar, and A. Glasebrook. 1996. Identification of an estrogen response element activated by metabolites of 17β-estradiol and raloxifene. Science 273:1222-1225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.