Abstract
Soil bacteria are heavily consumed by protozoan predators, and many bacteria have evolved defense strategies such as the production of toxic exometabolites. However, the production of toxins is energetically costly and therefore is likely to be adjusted according to the predation risk to balance the costs and benefits of predator defense. We investigated the response of the biocontrol bacterium Pseudomonas fluorescens CHA0 to a common predator, the free-living amoeba Acanthamoeba castellanii. We monitored the effect of the exposure to predator cues or direct contact with the predators on the expression of the phlA, prnA, hcnA, and pltA genes, which are involved in the synthesis of the toxins, 2,4-diacetylphloroglucinol (DAPG), pyrrolnitrin, hydrogen cyanide, and pyoluteorin, respectively. Predator chemical cues led to 2.2-, 2.0-, and 1.2-fold increases in prnA, phlA, and hcnA expression, respectively, and to a 25% increase in bacterial toxicity. The upregulation of the tested genes was related to the antiprotozoan toxicity of the corresponding toxins. Pyrrolnitrin and DAPG had the highest toxicity, suggesting that bacteria secrete a predator-specific toxin cocktail. The response of the bacteria was elicited by supernatants of amoeba cultures, indicating that water-soluble chemical compounds were responsible for induction of the bacterial defense response. In contrast, direct contact of bacteria with living amoebae reduced the expression of the four bacterial toxin genes by up to 50%, suggesting that protozoa can repress bacterial toxicity. The results indicate that predator-prey interactions are a determinant of toxin production by rhizosphere P. fluorescens and may have an impact on its biocontrol potential.
Bacterial communities are heavily consumed by protozoan predators (30), and predation is a major force shaping the structure of microbial communities in both aquatic and terrestrial ecosystems (34, 35). The competitiveness of bacteria strongly depends on their ability to avoid predation (9, 22), and many species have developed defense mechanisms such as the production of toxins, which reduces mortality by repelling or killing predators (21, 24). Toxin production, however, is energetically costly, and defense theory predicts that prey species should optimize the investment in defense according to the resources available and the predation risk (40), for example, in response to predator-associated chemical cues (4, 15). In bacteria the production of defense traits is tightly regulated by various sensor cascades (11), and defense mechanisms, such as the formation of inedible morphotypes or microcolonies, can be elicited in the presence of predators (45).
Toxin production is one of the most powerful defense strategies, and in the present study we tested whether bacteria can also modulate the production of toxic secondary metabolites in response to protozoan predators. We investigated the chemical communication between the soil bacterium Pseudomonas fluorescens CHA0 and the bacterivorous amoeba Acanthamoeba castellanii, a ubiquitous soil protist feeding on a wide range of bacterial species (33). P. fluorescens CHA0 effectively colonizes the rhizosphere of plants and produces various extracellular toxins including pyrrolnitrin (PRN), 2,4-diacetylphloroglucinol (DAPG), hydrogen cyanide (HCN), and pyoluteorin (PLT) (18). These toxins reduce predation pressure and enhance the competitiveness of the bacteria in the rhizosphere (22) but also act antagonistically against plant pathogens, thereby promoting plant growth (11).
The production of secondary metabolites by P. fluorescens is a dynamic process that depends on environmental factors, such as nutrient availability (12), cell density (18), or the presence of phytopathogens (27). We hypothesized that P. fluorescens is also able to sense predators and responds by increasing the expression of toxin genes.
Predators or competitors susceptible to toxins can adopt counterstrategies to repress their production. For example, the fungal pathogen Fusarium can inhibit the production of DAPG by pseudomonads (26), and we hypothesized that A. castellanii can counteract prey defense by inhibiting bacterial toxin production.
We investigated the effects of predator-prey interactions on the regulation of the production of the extracellular toxins DAPG, PLT, PRN, and HCN using a set of autofluorescent green fluorescent protein (GFP)- and mCherry-based reporter fusions (2, 32). Autofluorescent reporter fusions allow in situ measurement of gene expression patterns and have been applied to monitor the expression of antifungal genes in the rhizosphere (10, 32). The response of the bacteria to predators or associated signal molecules was investigated in batch experiments and on barley roots.
MATERIALS AND METHODS
Organisms and plasmids used.
The organisms and plasmids used are listed in Table 1 . Bacterial strains were kept routinely on agar plates (blood agar base, 40 g liter−1; yeast extract, 5 g liter−1) amended, when required, with 100 μg of ampicillin sodium salt ml−1, 8 μg of gentamicin sulfate ml−1, and 50 μg of kanamycin sulfate ml−1 and, for the E. coli and P. fluorescens strains, 25 μg and 125 μg of tetracycline hydrochloride ml−1, respectively (all chemicals from Sigma-Aldrich, St. Louis, MO).
TABLE 1.
Organisms and plasmids used in this study
| Strains or plasmids | Propertiesa | Reference |
|---|---|---|
| Strains | ||
| Pseudomonas fluorescens | ||
| CHA0 | Biocontrol bacterium, wild-type strain | 43 |
| CHA19 | gacS isogenic mutant lacking extracellular toxins | 48 |
| Escherichia coli DH5α | Laboratory strain | 37 |
| Acanthamoeba castellanii | Soil-dwelling bacterivorous amoeba | 5 |
| Plasmids | ||
| pME7100 | phlA-gfp transcriptional fusion; reporter for 2,4-diacetylphloroglucinol biosynthetic gene expression; Tcr | 2 |
| pME7109 | pltA-gfp transcriptional fusion; reporter for pyoluteorin biosynthetic gene expression; Kmr | 2 |
| pME7116 | prnA-gfp transcriptional fusion; reporter for pyrrolnitrin biosynthetic gene expression; Tcr | 2 |
| pME7156 | hcnA-gfp transcriptional fusion; reporter for hydrogen cyanide biosynthetic gene expression; Tcr | 32 |
| pME9012 | phlA-mcherry transcriptional fusion; reporter for 2,4-diacetylphloroglucinol biosynthetic gene expression; Kmr | 32 |
| pSM1973 | Delivery plasmid for the mini Tn7 PrrnBP1 gfp-a cassette; reporter for rrnB ribosomal gene expression; Gmr | 23 |
| pUX-BF13 | Helper plasmid encoding the Tn7 transposition functions; Apr | 3 |
Apr, ampicillin resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; and Tcr, tetracycline resistant.
A. castellanii, isolated from a woodland soil (5) was cultivated axenically in PGY medium (peptone, 20 g liter−1; glucose, 10 g liter−1; yeast extract, 5 g liter−1) as described elsewhere (35). Prior to the experiment, the cells were harvested by gentle centrifugation (100 × g, 2 min) and washed three times in Page's Amoeba Saline (AS) (29). The cell density was measured in a Neubauer counting chamber and adjusted to 106 cells ml−1 in AS, and the cultures were incubated at 17°C in darkness for 3 days. The supernatant of the amoeba culture medium was obtained by separating the cells by gentle centrifugation. The supernatant was sterile filtered (0.22-μm pore size), snap-frozen in liquid nitrogen, and stored at −80°C until use.
Transformation of bacteria with transcriptional reporter fusions.
Midi-Prep extractions of plasmids from E. coli were carried out with a JetStar 2.0 Plasmid Midi-Prep kit (Genomed, Löhne, Germany). Plasmids pME7100, pME7109, pME7116, pME7156, and pME9012 containing the phlA-gfp, pltA-gfp, prnA-gfp, hcnA-gfp, and phlA-mcherry reporter fusions (Table 1), respectively, were electroporated into P. fluorescens CHA0 as described previously (2, 38). The rrnBP1-gfp cassette contained within the mini-Tn7 shuttle vector pSM1973 (23) was inserted at the neutral chromosomal Tn7 attachment site of strain CHA0 site after coelectroporation with the pUX-BF13 helper vector (3). Transformants were selected on NA plates with the appropriate antibiotics (Table 1).
Batch culture assay.
Assays to monitor expression of the GFP-based reporter fusions were performed in 96-well microtiter plates (Nunc, Langenselbold, Germany) using an Infinite M200 plate reader (Tecan, Männedorf, Switzerland). Bacteria from a single colony were grown at 26°C in 5 ml of LB medium on a rotary shaker (250 rpm) for 12 h. These precultures were centrifuged (8 krpm, 1 min), washed twice in 0.9% NaCl, and adjusted to an optical density at 600 nm (OD600) of 0.1 in minimal medium (OSG) (38) containing 0.5% (wt/vol) glycerol as sole carbon source. The effect of living amoebae or their cell-free supernatant on bacterial activity was assessed by mixing 90 μl of bacterial suspensions with 10 μl of freshly washed A. castellanii, (final concentration ranging from 101 to 106 amoebae ml−1) or 10 μl of supernatant (see above), respectively. The control treatments received 10 μl of AS. Eight replicates were set up per treatment. Plates were incubated with orbital agitation (1.5-mm amplitude) in the plate reader at 26°C for 10 h. At 10-min intervals the OD600 and the green fluorescence (excitation, 485 nm; emission, 518 nm; gain, 90) were recorded. Green fluorescence was expressed as relative fluorescence units (RFU) by dividing the total fluorescence signal by the OD600 (2). In treatments involving amoebae, the OD600 signal from A. castellanii suspension without bacteria was used as blank. The fluorescence signal of nontransformed P. fluorescens CHA0 was used for background correction.
Effect of bacterial toxins on amoebae.
A. castellanii was grown and washed as described above. P. fluorescens CHA0 was grown at 26°C in OSG only or with addition of a cell-free amoebae supernatant as described above. Aliquots of 12-h-old bacterial cultures were then mixed 1:1 with A. castellanii (105 amoebae ml−1 in AS). Living and dead amoebae were counted after 6 h.
The dose-response curve of A. castellanii to cyanide, pyrrolnitrin, and DAPG was determined by mixing amoebae suspended in AS with a dilution series of KCN, pyrrolnitrin, and DAPG (50 μM to 5 mM). Living and dead organisms were counted after 6 h.
In situ assay on roots.
In addition to batch experiments, the effect of A. castellanii on the expression of biocontrol genes by P. fluorescens CHA0 was assessed in the rhizosphere of barley (Hordeum vulgare cv. Barke). In order to distinguish between variations of the global activity state of the bacteria and modulations of the secondary metabolism, two reporter fusions were used; a mCherry-based reporter fusion of the DAPG biosynthetic gene phlA (phlA-mcherry), i.e., the most strongly expressed toxin gene of P. fluorescens CHA0 in the rhizosphere (10), and a GFP-based reporter fusion of the expression of rRNA (rrnBP1-gfp). The diverging excitation and emission spectra of GFP and mCherry avoid cross excitation of the fluorophores, allowing the simultaneous detection of both signals by flow cytometry (32).
Barley seeds were surface sterilized by scarification in 50% H2SO4, immersion in 2% AgNO3 solution for 20 min, and five washing cycles in 1% NaCl and H2O (19). Seeds were germinated in the dark for 48 h on 1.5% water agar and transferred individually into autoclaved growth pouches made of clear plastic (Mega International, St. Paul, MN) containing 15 ml of a 1/5-strength Hoagland hydroponic solution. Bacteria and amoebae used for inoculation were washed and resuspended in AS as described above, and the pouches were inoculated with 500 μl of a 1:1 mixture of the two reporter strains (total of 108 bacteria) and 106 amoebae in the respective treatments. Ten replicates were set up for each treatment and time point. The root areas of the pouches were covered with aluminum foil, and plants were grown with 16 h of light (22°C, 500 μmol s−1 m−2), 8 h of darkness (18°C), and a 75% relative humidity.
After 4, 8, and 16 days the plants were harvested. Bacteria adhering to roots were collected and fixed by vigorously shaking the roots for 20 min in 6 ml of phosphate-buffered saline containing 0.8% formaldehyde. In situ expression of the reporter fusions for phlA and PrrnBP1 was analyzed simultaneously with a FACSCalibur flow cytometer (Becton and Dickinson, San Jose, CA) equipped with a 15-mW, air-cooled argon ion laser excitation light source (488 nm). GFP and mCherry fluorescence emissions were measured with FL1-H and FL3-H channels, respectively. Forward-scatter (FSC) signals were collected by using a photodiode with an amplification factor of 10, a threshold of 253, and a log gain. Side-scatter (SSC) signals were measured by using a photomultiplier tube set at 350 V, a threshold of 72, and a log gain. Green fluorescence was detected at 515 to 530 nm by the FL1-H detector set at 505 V and a log gain. Cherry fluorescence was detected at 670 nm by the FL3-H detector set at 690 V and a log gain.
Data were collected by using CellQuest software (Becton and Dickinson) and analyzed with WinMDI 2.8 (http://facs.scripps.edu/software.html). Selection of the counted events was done on the basis of the fetal calf serum and SSC signals. Fluorescence signals for each fluorophore were gated on the FL1-H and FL3-H histograms using root washes of plants inoculated with the complementary reporter strains as blank in order to avoid measuring potential unspecific signals from root particles or bacteria expressing the other fluorophore. FL1-H signals between 100 and 1023 and FL3-H signals between 50 and 1023 were taken into account. Bacterial colonization of barley roots was determined by epifluorescence counting to recheck fluorescence-activated cell sorting (FACS) counts (22).
Statistical analyses.
Geometric means from green and cherry fluorescence from the FACS measurement and the bacterial density were log transformed prior to analyses. The data were analyzed with a general linear model function and type III sum of squares, investigating the effect of the density of amoebae and the supernatant fraction on the relative fluorescence (RFU) in the batch experiment, and the presence of amoebae and the time postinoculation on the FACS data and the bacterial colonization in the root experiment. Analyses were performed with Statistica 7.1 (Statsoft, Tulsa, OK).
RESULTS
Batch culture experiment.
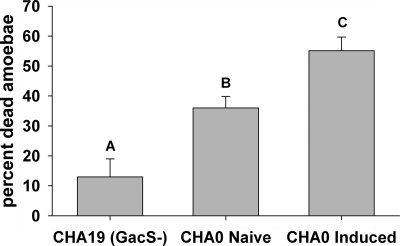
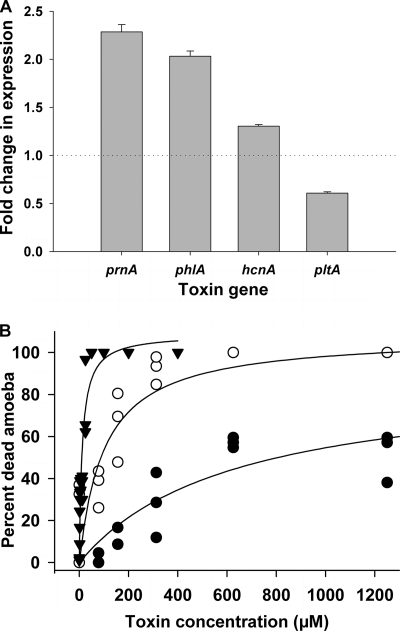
Incubation of P. fluorescens CHA0 with cell-free A. castellanii supernatant increased the acute toxicity toward the amoebae by ca. 25% compared to “naive” bacteria never put in contact with the amoebae (F2,18 = 18.1, P < 0.001; Fig. 1), indicating that bacteria responded to predator cues by upregulating defense mechanisms. At the gene level, supernatant elicited rapid changes in expression of each of the four tested extracellular toxin genes compared to the control treatment mixed with fresh AS (F3,101 = 32.0, P < 0.001). However, the four genes responded differently (F3,101 = 202.2, P < 0.001) to the supernatant. The expression of the DAPG and PRN biosynthetic genes phlA and prnA, respectively, exhibited the highest increase in response to signals of amoebae (Fig. 2 A). The supernatant of A. castellanii cultures also induced a moderate increase in the expression of the cyanide biosynthetic gene hcnA, and the PLT biosynthetic gene pltA was even downregulated in the presence of the supernatant (Fig. 2A). Interestingly, the changes in the expression levels of the tested genes in response to the amoeba supernatant were correlated with the toxicity of the resulting toxins. Amoebae were insensitive to cyanide and survived to up to 5 mM KCN, while being more sensitive to PRN and DAPG, which already killed most of the cells at concentrations of about 13 and 100 μM, respectively (Fig. 2B).
FIG. 1.
Toxicity of overnight cultures of P. fluorescens CHA0 grown alone (“naive”) or with cell-free supernatant of A. castellanii (“induced”). The isogenic toxin-deficient mutant CHA19 was used as control. Different letters indicate significant differences between the treatments (Tukey's HSD, α = 0.05).
FIG. 2.
(A) Induction of the extracellular toxin genes prnA, phlA, hcnA, and pltA of P. fluorescens CHA0 after coincubation with supernatant of A. castellanii. Bacteria were incubated for 5 h in OSG minimal medium, and toxin gene expression was monitored with GFP-based reporters. Effects are expressed as the fold change (induction/repression) relative to the control treatment (OSG medium without A. castellanii). Error bars show the standard error (SE; n = 8). (B) Dose-response curve of A. castellanii to pyrrolnitrin (▾), DAPG (○), and KCN (•) measured as the percentage of dead organisms after incubation at 20°C for 6 h. Curves were fitted with Michaelis-Menten kinetics (R2 = 0.94, 0.81 and 0.74 for pyrrolnitrin, DAPG, and KCN, respectively).
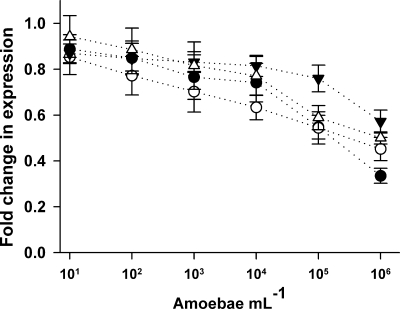
Cocultivation of P. fluorescens CHA0 with living A. castellanii cells resulted in contrasting gene expression patterns compared to the effect of the supernatant. Increased density of amoebae reduced the expression of phlA, hcnA, prnA, and pltA genes after 5 h (F6,177 = 25.1, P < 0.001; Fig. 3). The different genes were distinctly inhibited (F3,177 = 3.3, P = 0.02), with the expression of phlA and pltA being the most affected. At a concentration of 106 ml−1, amoebae reduced the specific fluorescence of the the phlA-gfp and pltA-gfp reporter fusions by up to 66 and 55%, respectively.
FIG. 3.
Effect of cocultivation of P. fluorescens CHA0 with A. castellanii on the expression of phlA (•), hcnA (▾), prnA (▵), and pltA (○) genes. Bacteria were grown for 5 h in OSG minimal medium; inhibition is expressed as the relative reduction of the GFP fluorescence (RFU) compared to the control treatment. Error bars show ± the SE (n = 8).
Plant microcosms.
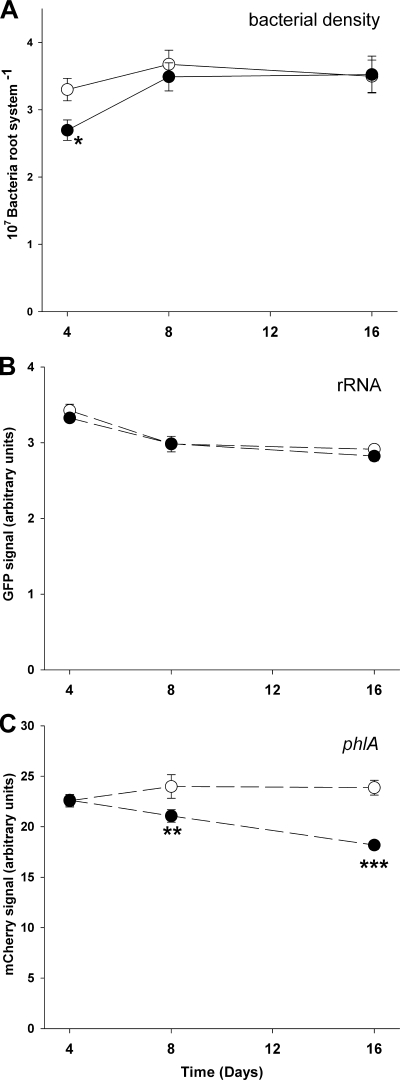
P. fluorescens CHA0 cell numbers increased during the experiment from 2.99 × 107 bacteria per root system on day 4 to 3.58 × 107 and 3.51 × 107 bacteria per root system on days 8 and 16, respectively (F2,54 = 4.51, P = 0.015). The bacterial density was not significantly affected by amoebae (F1,54 = 2.28, P = 0.16), indicating low consumption of bacteria by the amoebae (Fig. 4A). The green fluorescence signal, coupled with the expression of the growth-dependent ribosomal operon rrnB, decreased by 16% during the experiment (F2,54 = 29.66, P<0.001) but was not affected by the presence of amoebae (F1,54 = 1.20, P = 0.277; Fig. 4B), suggesting that amoebae did not affect the basal metabolism of P. fluorescens. During the experiment, however, amoebae increasingly affected the mCherry signal reporting phlA gene expression (F2,54 = 7.43, P = 0.0015 for the amoeba × time interaction) until reducing it by 25% after 16 days (Fig. 4C).
FIG. 4.
Bacterial density on barley roots (A), expression of the reporter fusions rrnBP1-gfp (B) and phlA-mcherry (C) by P. fluorescens CHA0 in the absence (○) or the presence of A. castellanii (•). Reporter activity was measured by flow cytometry 4, 8, and 16 days after inoculation and is expressed as the geometric mean of the respective fluorescence signal per bacterial cell. Error bars show ± the SE (n = 10). Asterisks indicate significant differences compared to the control (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [Tukey's HSD]).
DISCUSSION
The results of the present study show that the production of toxins by rhizosphere bacteria depends on signals from microfaunal predators. No direct contact was required for this effect, suggesting that bacteria sensed predator-associated chemical cues and readily responded by enhancing the expression of toxicity-related genes. Interestingly, direct contact between amoebae and bacteria resulted in an inhibition of the defense gene expression, suggesting that amoebae could interfere with toxin production.
Induced defense in bacteria.
The addition of amoeba culture supernatant led to an increase in bacterial toxicity against amoebae. Toxicity is an important defense strategy for P. fluorescens significantly reducing consumption by protozoa (22, 25). Bacteria responded to amoeba cues within minutes by increasing the expression of most of the studied genes regulating the biosynthesis of extracellular toxins in P. fluorescens with the exception of pltA. The response in the absence of amoebae suggests that predator perception by bacteria is mediated by water-soluble chemical compounds, as is the case in aquatic systems (31).
Since bacteria are consumed as whole cells, early perception of predators and fast upregulation of secondary metabolite production is crucial for effective defense. Expression of antipredator traits often is costly for the producer and is only advantageous at certain conditions, i.e., at high predator density or high nutrient availability (14). The advantage of defense mechanisms depends on the feeding behavior of predators and prey population density (20), and modulating the expression of defense traits according to their costs and benefits is advantageous for prey species (1).
Interestingly, the studied bacterial toxin genes differentially responded to chemical cues of predators, and the response was related to the efficiency of the toxins against the predator. The expression of phlA, responsible for the production of DAPG, was strongly induced in the presence of amoeba supernatant, and DAPG toxicity proved to kill amoebae at a comparatively low threshold concentration. Hydrogen cyanide is a broad-spectrum toxin inhibiting cytochrome c oxidase activity and, for example, causes paralytic death of nematodes (16). Interestingly, Acanthamoeba species possess cyanide-resistant terminal oxidases (13), and cyanide was only marginally toxic to our strain, even at high doses. The low response of hcnA to predator cues may thus be linked to the low efficiency of cyanide against A. castellanii. The expression of pltA was reduced in the presence of amoebae, and this might be linked to the low antiprotozoan activity of this toxin (21). The production of DAPG and PLT is negatively coregulated by P. fluorescens (2, 38), and reduced expression of pltA probably results from a tradeoff in favor of DAPG. Similar to phlA, prnA was strongly upregulated in the presence of chemical cues of A. castellanii. Pyrrolnitrin is also a potent inhibitor of the respiratory pathway (41) and of osmotic regulation (28), affecting a wide spectrum of fungi and protists (6, 8), and was lethal to the amoebae at very low concentration. Overall, the results indicate that in P. fluorescens, and potentially other soil bacteria, the investment in defense compounds is adjusted to secrete a toxin cocktail efficiently killing predators. Characterization of the predator associated signal cues may help to better understand the molecular mechanisms involved in predator recognition.
Counteraction of bacterial defense by amoebae.
The presence of living amoebae had a contrasting effect on bacterial gene expression. They reduced the expression of all of the investigated genes of P. fluorescens in vitro and negatively affected the expression of phlA on plant roots. Since amoebae did not reduce bacterial density in the short-term batch experiment and did not affect bacterial basal metabolism on barley roots, the observed effect suggest a specific inhibition of bacterial secondary metabolism. Interference with toxin production has been described by soil fungi such as Fusarium sp., which can detoxify DAPG (39) or inhibit its synthesis (26). Moreover, both prokaryotes and eukaryotes can interfere with bacterial signals (17), which regulate toxicity by P. fluorescens (18). The observed inhibition of phlA expression by A. castellanii suggests that the amoebae also evolved defense mechanisms to reduce prey toxicity. A. castellanii produces extracellular enzymes that may interfere with bacterial metabolism and communication (44). Detailed investigations on the effect of amoebae on the different regulatory cascades, such as the Gac/Rsm pathway, involved in the control of secondary metabolism (18), may help elucidate the mechanisms of chemical interaction between protozoan predators and their bacterial prey.
Since supernatant of the culture medium of amoebae resulted in increased bacterial secondary metabolism, the inhibitory effect of amoebae cocultured with bacteria on bacterial gene expression is probably induced by prey signals. Eukaryotes use conserved receptors, such as Toll-like receptors or mannose-binding receptors, to detect bacteria based on general molecular patterns, such as flagellin or lipopolysaccharides (46). Moreover, microfauna predators secrete opsonins (36) to detect and localize bacterial prey based on diffusible chemical cues (47). The ability to detect bacterial cues appeared early in the evolution of eukaryotes and permitted the evolution of selective phagocytosis (7). Our results suggest that recognition mechanisms may not only control phagocytosis but also help the protozoa respond to prokaryotic prey defense mechanisms.
Induced defense is one of the major mechanisms alleviating predator impact on prey populations and has received considerable attention in aquatic and aboveground systems (40, 42). Predator-prey interactions also are of paramount importance in belowground systems, and we have shown that induced defense also governs the interaction between bacteria and protozoa, one of the most widespread and functionally important predator-prey systems in soil. The production of extracellular toxins by rhizosphere pseudomonads appears as a dynamic process adjusted according to the risk of predation by protozoa. Moreover, our results indicate that microfaunal predators are able to partially disarm their prey by inhibiting the production of these toxins. This mutual perception and response indicates intricate chemical warfare between bacterivorous protozoa and bacterial prey. Since the same toxins function as biocontrol agents by improving the resistance of plants against fungal pathogens, manipulating predator-prey interactions may allow improving biocontrol of soilborne plant diseases.
Acknowledgments
We are grateful to Lotte Lambertsen and Søren Molin (DTU, Denmark) for providing the PrrnB-gfp reporter fusion on pSM1973.
This study was funded by the fellowship program of the German Federal Foundation for the Environment (DBU). We gratefully acknowledge support from the Swiss National Science Foundation (projects 3100A0-105881 and 31003A-120121).
Footnotes
Published ahead of print on 4 June 2010.
REFERENCES
- 1.Agrawal, A. A. 2007. Macroevolution of plant defense strategies. Trends Ecol. Evol. 22:103-109. [DOI] [PubMed] [Google Scholar]
- 2.Baehler, E., M. Bottiglieri, M. Pechy-Tarr, M. Maurhofer, and C. Keel. 2005. Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 99:24-38. [DOI] [PubMed] [Google Scholar]
- 3.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 4.Bergkvist, J., E. Selander, and H. Pavia. 2008. Induction of toxin production in dinoflagellates: the grazer makes a difference. Oecologia 156:147-154. [DOI] [PubMed] [Google Scholar]
- 5.Bonkowski, M., and F. Brandt. 2002. Do soil protozoa enhance plant growth by hormonal effects? Soil Biol. Biochem. 34:1709-1715. [Google Scholar]
- 6.Carlone, N., and S. Scannerini. 1974. Scanning and transmission electron microscopy evidence of the cytological effect of pyrrolnitrin on Microsporon audouinii Gruby CBS 313-54 ‘in vitro’. Mycopathologia 53:111-123. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith, T. 2009. Predation and eukaryote cell origins: a coevolutionary perspective. Int. J. Biochem. Cell Biol. 41:307-322. [DOI] [PubMed] [Google Scholar]
- 8.Chernin, L., A. Brandis, Z. Ismailov, and I. Chet. 1996. Pyrrolnitrin production by an Enterobacter agglomerans strain with a broad spectrum of antagonistic activity toward fungal and bacterial phytopathogens. Curr. Microbiol. 32:208. [Google Scholar]
- 9.Corno, G., and K. Jurgens. 2006. Direct and indirect effects of protist predation on population size structure of a bacterial strain with high phenotypic plasticity. Appl. Environ. Microbiol. 72:78-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Werra, P., E. Baehler, A. Huser, C. Keel, and M. Maurhofer. 2008. Detection of plant-modulated alterations in antifungal gene expression in Pseudomonas fluorescens CHA0 on roots by flow cytometry. Appl. Environ. Microbiol. 74:1339-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubuis, C., C. Keel, and D. Haas. 2007. Dialogues of root-colonizing biocontrol pseudomonads. Eur. J. Plant Pathol. 119:311-328. [Google Scholar]
- 12.Duffy, B. K., and G. Defago. 1999. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65:2429-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, S. W., and D. Lloyd. 1978. Properties of mitochondria isolated from cyanide-sensitive and cyanide-stimulated cultures of Acanthamoeba castellanii. Biochem. J. 174:203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friman, V. P., T. Hiltunen, J. Laakso, and V. Kaitala. 2008. Availability of prey resources drives evolution of predator-prey interaction. Proc. R. Soc. B Biol. Sci. 275:1625-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fyda, J., A. Warren, and J. Wolinska. 2005. An investigation of predator-induced defence responses in ciliated protozoa. J. Nat. Hist. 39:1431-1442. [Google Scholar]
- 16.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez, J. E., and N. D. Keshavan. 2006. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 70:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 19.Henkes, G. J., M. R. Thorpe, P. E. H. Minchin, U. Schurr, and U. S. R. Rose. 2008. Jasmonic acid treatment to part of the root system is consistent with simulated leaf herbivory, diverting recently assimilated carbon toward untreated roots within an hour. Plant Cell Environ. 31:1229-1236. [DOI] [PubMed] [Google Scholar]
- 20.Jeschke, J. M. 2006. Density-dependent effects of prey defenses and predator offenses. J. Theor. Biol. 242:900-907. [DOI] [PubMed] [Google Scholar]
- 21.Jousset, A., E. Lara, L. G. Wall, and C. Valverde. 2006. Secondary metabolites help biocontrol strain Pseudomonas fluorescens CHA0 to escape predation. Appl. Environ. Microbiol. 72:7083-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jousset, A., S. Scheu, and M. Bonkowski. 2008. Secondary metabolite production facilitates establishment of rhizobacteria by reducing both protozoan predation and the competitive effects of indigenous bacteria. Funct. Ecol. 22:714-719. [Google Scholar]
- 23.Lambertsen, L., C. Sternberg, and S. Molin. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6:726-732. [DOI] [PubMed] [Google Scholar]
- 24.Matz, C., P. Deines, J. Boenigk, H. Arndt, L. Eberl, S. Kjelleberg, and K. Jürgens. 2004. Impact of violacein-producing bacteria on survival and feeding of bacterivorous nanoflagellates. Appl. Environ. Microbiol. 70:1593-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazzola, M., I. de Bruijn, M. F. Cohen, and J. M. Raaijmakers. 2009. Protozoan-induced regulation of cyclic lipopeptide biosynthesis is an effective predation defense mechanism for Pseudomonas fluorescens. Appl. Environ. Microbiol. 75:6804-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notz, R., M. Maurhofer, H. Dubach, D. Haas, and G. Defago. 2002. Fusaric acid-producing strains of Fusarium oxysporum alter 2,4-diacetylphloroglucinol biosynthetic gene expression in Pseudomonas fluorescens CHA0 in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 68:2229-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notz, R., M. Maurhofer, U. Schnider-Keel, B. Duffy, D. Haas, and G. Defago. 2001. Biotic factors affecting expression of the 2,4-diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology 91:873-881. [DOI] [PubMed] [Google Scholar]
- 28.Okada, A., S. Banno, A. Ichiishi, M. Kimura, I. Yamaguchi, and M. Fujimura. 2005. Pyrrolnitrin interferes with osmotic signal transduction in Neurospora crassa. J. Pesticide Sci. 30:378-383. [Google Scholar]
- 29.Page, F. C. 1988. A new key to freshwater and soil gymnaboeae. Freshwater Biological Association, Ambleside, United Kingdom.
- 30.Pernthaler, J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3:537-546. [DOI] [PubMed] [Google Scholar]
- 31.Pohnert, G., M. Steinke, and R. Tollrian. 2007. Chemical cues, defence metabolites and the shaping of pelagic interspecific interactions. Trends Ecol. Evol. 22:198. [DOI] [PubMed] [Google Scholar]
- 32.Rochat, L., M. Péchy-Tarr, E. Baehler, M. Maurhofer, and C. Keel. 2010. Combination of fluorescent reporters for simultaneous monitoring of root colonization and antifungal gene expression by biocontrol agent Pseudomonas fluorescens CHA0 with flow cytometry. Mol. Plant-Microbe Interact. 23:949-961. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Zaragoza, S. 1994. Ecology of free-living amebas. Crit. Rev. Microbiol. 20:225-241. [DOI] [PubMed] [Google Scholar]
- 34.Rønn, R., A. McCaig, B. Griffiths, and J. Prosser. 2002. Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl. Environ. Microbiol. 68:6094-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg, K., J. Bertaux, S. Scheu, and M. Bonkowski. 2009. Soil amoeba rapidly change bacterial community composition in Arabidopsis thaliana rhizosphere. ISME J. 3:675-684. [DOI] [PubMed] [Google Scholar]
- 36.Sakaguchi, M., H. Murakami, and T. Suzaki. 2001. Involvement of a 40-kDa glycoprotein in food recognition, prey capture, and induction of phagocytosis in the protozoon Actinophrys sol. Protist 152:33-41. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, and C. Gigot-Bonnefoy. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schouten, A., G. van den Berg, V. Edel-Hermann, C. Steinberg, N. Gautheron, C. Alabouvette, C. H. de Vos, P. Lemanceau, and J. M. Raaijmakers. 2004. Defense responses of Fusarium oxysporum to 2,4-diacetylphloroglucinol, a broad-spectrum antibiotic produced by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 17:1201-1211. [DOI] [PubMed] [Google Scholar]
- 40.Steiner, U. K. 2007. Investment in defense and cost of predator-induced defense along a resource gradient. Oecologia 152:201-210. [DOI] [PubMed] [Google Scholar]
- 41.Tripathi, R. K., and D. Gottlieb. 1969. Mechanism of action of antifungal antibiotic pyrrolnitrin. J. Bacteriol. 100:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Donk, E. 2007. Chemical information transfer in freshwater plankton. Ecol. Informatics 2:112-120. [Google Scholar]
- 43.Voisard, C., C. T. Bull, C. Keel, J. Laville, M. Maurhofer, and M. Schnider. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67-89. In F. O'Gara, D. N. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH Verlagsgesellschaft GmbH, Berlin, Germany.
- 44.Weekers, P. H. H., A. M. W. Engelberts, and G. D. Vogels. 1995. Bacteriolytic activities of the free-living soil amebas, Acanthamoeba castellanii, Acanthamoeba polyphaga, and Hartmannella vermiformis. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 68:237-243. [DOI] [PubMed] [Google Scholar]
- 45.Weitere, M., T. Bergfeld, S. A. Rice, C. Matz, and S. Kjelleberg. 2005. Grazing resistance of Pseudomonas aeruginosa biofilms depends on type of protective mechanism, developmental stage and protozoan feeding mode. Environ. Microbiol. 7:1593-1601. [DOI] [PubMed] [Google Scholar]
- 46.Wildschutte, H., D. M. Wolfe, A. Tamewitz, and J. G. Lawrence. 2004. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 101:10644-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willard, S. S., and P. N. Devreotes. 2006. Signaling pathways mediating chemotaxis in the social amoeba, Dictyostelium discoideum. Eur. J. Cell Biol. 85:897-904. [DOI] [PubMed] [Google Scholar]
- 48.Zuber, S., F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]