Abstract
Shiga-toxigenic Escherichia coli (STEC) O157 occurrence was determined along the entire gastrointestinal tract (GIT) of each of four naturally shedding cattle and at three sites in 61 slaughter cattle. STEC O157 was distributed along the entire GIT, though interanimal distribution was variable. Neither feces nor rectoanal-junction samples accurately predicted the STEC O157-negative status of any particular animal.
Shiga-toxigenic Escherichia coli (STEC) O157:H7 colonizes the gastrointestinal tract (GIT) of healthy adult cattle, as evidenced by the natural and frequent occurrence of STEC O157 at the beginning (i.e., oral cavity) and end (i.e., rectal feces and rectoanal mucosa [RAM]) of the bovine GIT (7, 8, 12). Distribution of STEC O157 along the entire GIT has also been investigated in experimentally challenged livestock (4, 6, 9, 13), but these various challenge models may not accurately reflect natural bovine STEC O157 GIT infection or colonization sites. The objective of this study was to qualitatively determine viable STEC O157 occurrence along the entire GIT of naturally shedding adult cattle, including both mucosa and ingesta. We hypothesized that this distribution of STEC O157 would not be limited to the distal large intestine.
Four adult female mixed-breed beef cattle (Table 1) identified as shedding STEC O157 by fecal culture were humanely euthanized according to animal care and use guidelines. Sections at a total of 16 sites along the entire GIT (Fig. 1) were removed aseptically immediately posteuthanasia. Double-ligated sections were surface sterilized, a lateral incision was made in each section, and the solid or semisolid ingesta was gently removed while taking care not to scrape the underlying tissue. All remaining visible ingesta was removed using a stream of sterile water to rinse the tissue. The GIT tissues (10 g) and their adjacent luminal contents (ingesta, 10 g) were cultured for STEC O157 separately and in duplicate using selective Brilliant Green Bile broth enrichment, immunomagnetic separation (IMS), and selective agar plating on ChromAgar O157 with reduced tellurite as previously described (3). Suspect isolates were confirmed as STEC O157 by enzyme immunoassay for the O157 and H7 antigens (5, 14) and by PCR for Shiga toxin (stx1 and stx2), intimin (eae), hemolysin (hly), O-antigen (rfbEO157), and flagellar (fliCH7) genes (10).
TABLE 1.
Animals used in the necropsy study
| Animal no. | Age (yr) | Body wt (kg) | Description |
|---|---|---|---|
| 1 | 6.0 | 870 | Pastured cow, postpartum paresis |
| 2 | 1.9 | 422 | Pastured heifer, postpartum paresis |
| 3 | 1.5 | 498 | Healthy feedlot heifer |
| 4 | 1.5 | 484 | Healthy feedlot heifer |
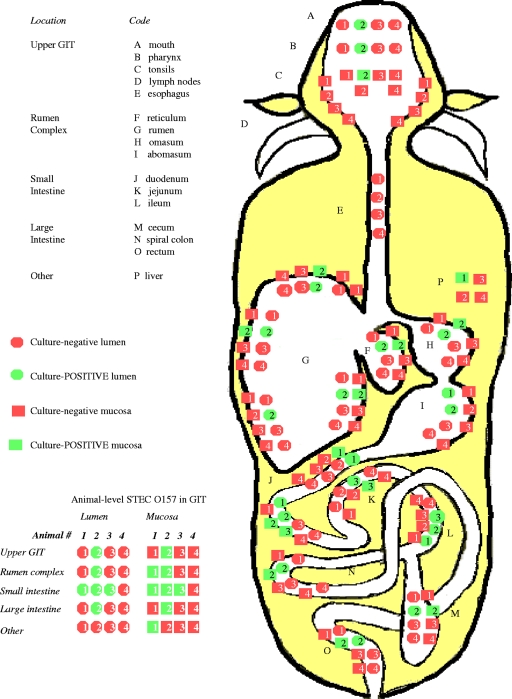
FIG. 1.
Composite schematic of the mouth-to-rectum distribution of STEC O157 in tissues and luminal contents at 16 sites in the GIT of four naturally infected cattle (frontal plane, dorsal view). Numbers refer to tested animals 1 to 4.
Culture results from the four euthanized animals are summarized in Fig. 1. Results for the rectal ingesta (feces) of these animals were considered to indicate the presence of STEC O157 by fecal culture alone. STEC O157 distribution in the GIT was highly variable among animals. STEC O157 was found only in the small intestines of animals 1 and 3 (ingesta and tissue positive). The liver of animal 1 was also culture positive. Animal 2 was STEC O157 positive along the entire GIT, including the upper GIT, the rumen complex, the small intestine, and the large intestine. Animal 4 was culture negative at all GIT locations.
In a separate set of experiments examining mucosal and ingesta samples from one site each in the distal small intestine (ileum), distal large intestine (mid-rectum), and rectoanal junction (RAJ) of 61 fed cattle at processing whose STEC O157 shedding status was unknown, fecal STEC O157 culture underestimated the prevalence of STEC O157 present along the bovine GIT (Table 2). Whereas 19/61 animals (31%) were STEC O157 positive at any site, only 5 (26%) of these were fecal culture positive. These fecal culture results, taken alone, indicate a prevalence of only 8% (5/61). Additionally, 15/19 STEC O157-positive samples (78.9%) had positive tissue cultures and 10/19 (52.6%) ingesta samples had positive cultures. There were six animals (31.6%) that were both ingesta and tissue positive at any of the three sample sites, while nine animals (47.4%) yielded a STEC O157-positive culture only from tissue samples and four animals (21.1%) yielded a STEC O157-positive culture only from ingesta samples (Table 2). STEC O157 isolation from the ileum, mid-rectum, and RAJ of the 61 animals showed that STEC was present at all locations, with prevalences of 11.5% (95% confidence interval [CI], 4.7 to 22.2), 16.4% (95% CI, 8.2 to 28.1), and 21.3% (95% CI, 11.9 to 33.7), respectively (Table 3).
TABLE 2.
STEC O157 isolation from mucosa and ingesta samples from one site each in the ileum, mid-rectum, and RAJ of 19 of 61 cattle of unknown infection status at processinga
| Animal no. | Ileum mucosal tissue | Ileum ingesta | Rectal mucosa tissue | Rectal ingesta (feces) | Rectoanal mucosa tissue | Rectoanal mucosa swab | Total no. positive | No. positive/3 sitesb |
|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 2 |
| 5 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 6 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 7 | 0 | 0 | 0 | 1 | 1 | 1 | 3 | 2 |
| 8 | 0 | 0 | 1 | 1 | 1 | 1 | 4 | 2 |
| 15 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 17 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 2 |
| 19 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 2 |
| 20 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 |
| 21 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 2 |
| 23 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 2 |
| 28 | 1 | 1 | 0 | 0 | 1 | 0 | 3 | 2 |
| 29 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| 34 | 0 | 1 | 1 | 1 | 1 | 1 | 5 | 3 |
| 37 | 1 | 0 | 1 | 0 | 0 | 1 | 3 | 3 |
| 44 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| 45 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| 51 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 58 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 |
| Total | 4 | 4 | 8 | 5 | 9 | 7 | NA | NA |
Animals that were culture negative at all sites (n = 42) are not shown. 0, negative; 1, positive; NA, not applicable.
Number of sites (ileum, rectal mucosa, or RAM) that contained a positive sample.
TABLE 3.
Prevalence of STEC O157 in the ileum, mid-rectum, and RAJ of 61 fed cattle at processing
| GIT location | No. of cattle |
% Prevalence (95% CI) | |
|---|---|---|---|
| STEC O157 positive | STEC O157 negative | ||
| Ileum | 7 | 54 | 11.5 (4.7-22.2) |
| Rectum | 10 | 51 | 16.4 (8.2-28.1) |
| RAJ | 13 | 48 | 21.3 (11.9-33.7) |
| Any of 3 sites | 19 | 42 | 31.3 (19.9-44.3) |
Our data show that STEC O157 can be isolated from tissues throughout the cattle GIT, including the tonsils, reticulum, rumen, omasum, abomasum, duodenum, jejunum, cecum, spiral colon, rectum, and even the liver, suggesting that STEC O157 is broadly adapted to many cattle GI microhabitats. It is not known whether the bacteria we detected using enrichment methods represented resident or transient strains. Recent studies debate the role of the bovine gallbladder as a focus of STEC O157 colonization in cattle (6, 13,11). While gallbladders were not examined as part of this study, our results showed that STEC O157 can be found throughout the bovine GIT. Like others, we also isolated STEC O157 from tonsils and cecums (1, 2).
Research with artificially challenged animals suggests that the RAJ is the principal site of STEC O157 colonization in cattle (9). These conclusions were based, in part, on a failure to isolate STEC from rumen and ileum mucosa samples. We obtained samples from a larger number of GIT sites and were able to detect STEC O157 in a larger proportion of non-RAJ samples than Naylor et al. (9). Lim et al. (8) also concluded that the RAJ was the site of colonization because of an inability to culture STEC O157 from non-RAJ samples. Since we were working with naturally occurring STEC O157 strains, we were able to incorporate an IMS step into our protocol, which likely increased the sensitively of our detection method. Naylor et al. (9) used a marked STEC O157 strain that prohibited the use of an IMS step, and Lim et al. (8) also did not use IMS. Unlike Naylor et al. (9), we did not quantify STEC O157 or use immunochemistry to try to determine colonization status. Our results, based on culture of STEC O157 from 65 feedlot beef cattle that were naturally exposed to STEC O157, suggest that the dynamics of actual STEC O157 infections in the cattle GIT may involve more sites than just the RAJ. While no single sample site accounted for all of the positive animals, RAM tissue samples from cattle presented for slaughter were the most likely of the sites sampled to yield a positive result.
Based on STEC O157 culture and isolation from multiple sites along the entire GIT of naturally shedding cattle, fecal shedding greatly underestimates the true extent and prevalence of STEC O157 occurrence in the bovine reservoir. While a positive STEC O157 culture from any bovine GIT site indicates the presence of the pathogen, no single sample type, including feces or the RAM, can accurately predict the STEC O157-negative status of any particular animal. Our data suggest that there is high animal-to-animal variability in the location of STEC O157 in the GIT and that the small intestine may support STEC O157 colonization.
Acknowledgments
We thank Sandy Fryda-Bradley, Ron Mlejnek, Tammy Sorensen, Stacy Bierman, Liz Ossian, Scott Schroetlin, Halah Bajaber, Josh Soucek, and Jenna Williams for microbiological and technical contributions and Joan Rosch for secretarial assistance.
Footnotes
Published ahead of print on 11 June 2010.
REFERENCES
- 1.Bonardi, S., E. Foni, C. Chiapponi, A. Salsi, and F. Brindani. 2007. Detection of verocytotoxin-producing Escherichia coli serogroups O157 and O26 in the cecal content and lymphatic tissue of cattle at slaughter in Italy. J. Food Prot. 70:1493-1497. [DOI] [PubMed] [Google Scholar]
- 2.Cray, W. C., Jr., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durso, L. M., K. Reynolds, N. Bauer, Jr., and J. E. Keen. 2005. Shiga-toxigenic Escherichia coli O157:H7 infections among livestock exhibitors and visitors at a Texas county fair. Vector Borne Zoonotic Dis. 5:193-201. [DOI] [PubMed] [Google Scholar]
- 4.Grauke, L. J., I. T. Kudva, J. W. Yoon, C. W. Hunt, C. J. Williams, and C. J. Hovde. 2002. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He, Y., J. E. Keen, R. B. Westerman, E. T. Littledike, and J. Kwang. 1996. Monoclonal antibodies for detection of the H7 antigen of Escherichia coli. Appl. Environ. Microbiol. 62:3325-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong, K. C., M. Y. Kang, C. Heimke, J. A. Shere, I. Erol, and C. W. Kaspar. 2007. Isolation of Escherichia coli O157:H7 from the gall bladder of inoculated and naturally-infected cattle. Vet. Microbiol. 119:339-345. [DOI] [PubMed] [Google Scholar]
- 7.Keen, J. E., and R. O. Elder. 2002. Isolation of Shiga-toxigenic Escherichia coli O157 from hide surfaces and the oral cavity of finished beef feedlot cattle. J. Am. Vet. Med. Assoc. 220:756-763. [DOI] [PubMed] [Google Scholar]
- 8.Lim, J. Y., J. Li, H. Sheng, T. E. Besser, K. Potter, and C. J. Hovde. 2007. Escherichia coli O157:H7 colonization at the rectoanal junction of long-duration culture-positive cattle. Appl. Environ. Microbiol. 73:1380-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, D. G. E. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinstein, S., J. T. Fox, X. Shi, and T. G. Nagaraja. 2007. Prevalence of Escherichia coli O157:H7 in gallbladders of beef cattle. Appl. Environ. Microbiol. 73:1002-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice, D. H., H. Q. Sheng, S. A. Wynia, and C. J. Hovde. 2003. Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. J. Clin. Microbiol. 41:4924-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoffregen, W. C., J. F. L. Pohlenz, and E. A. Dean-Nystrom. 2004. Escherichia coli O157:H7 in the gallbladders of experimentally infected calves. J. Vet. Diagn. Invest. 16:79-83. [DOI] [PubMed] [Google Scholar]
- 14.Westerman, R. B., Y. He, J. E. Keen, E. T. Littledike, and J. Kwang. 1997. Production and characterization of monoclonal antibodies specific for the lipopolysaccharide of Escherichia coli O157. J. Clin. Microbiol. 35:679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]