Abstract
The genetic manageability of the biotechnologically important Bacillus licheniformis is hampered due to its poor transformability, whereas Bacillus subtilis efficiently takes up DNA during genetic competence, a quorum-sensing-dependent process. Since the sensor histidine kinase ComP, encoded by a gene of the quorum-sensing module comQXPA of B. licheniformis DSM13, was found to be inactive due to an insertion element within comP, the coding region was exchanged with a functional copy. Quorum sensing was restored, but the already-poor genetic competence dropped further. The inducible expression of the key regulator for the transcription of competence genes, ComK, in trans resulted in highly competent strains and facilitated the direct disruption of genes, as well as the conditional knockout of an essential operon. As ComK is inhibited at low cell densities by a proteolytic complex in which MecA binds ComK and such inhibition is antagonized by the interaction of MecA with ComS (the expression of the latter is controlled by cell density in B. subtilis), we performed an in silico analysis of MecA and the hitherto unidentified ComS, which revealed differences for competent and noncompetent strains, indicating that the reduced competence possibly is due to a nonfunctional coupling of the comQXPA-encoded quorum module and ComK. The obtained increased genetic tractability of this industrial workhorse should improve a wide array of scientific investigations.
Strains of the endospore-forming Gram-positive soil bacterium Bacillus licheniformis (a generally-regarded-as-safe [GRAS] species) serve as microbial workhorses, as they produce a number of valuable compounds (10), and they possess a high capacity for the secretion of exoenzymes, such as amylases and proteases, with yields of up to 25 g liter−1 (46). The availability of the genome sequences of strain B. licheniformis DSM13 (61) and the isogenic ATCC 14580 (44) already facilitated a number of developments aiming at strain improvement and the enhancement of biosafety (38, 63, 64).
However, the low frequency of transformation regularly observed with those strains is a drawback. Although a derivative (B. licheniformis MW3) was obtained by the deletion of the genes encoding type I restriction enzymes, allowing the routine creation of transformants by protoplast transformation, it still has a rather low transformation efficiency (65). Hence, the generation of mutants by homologous recombination is a rather time-consuming and cumbersome process, as is the case for other members of the genus (60).
During the part of their life cycle when they develop natural genetic competence, representatives of the genus Bacillus generally are capable of taking up exogenously supplied DNA. Such a DNA uptake mechanism was first described for B. subtilis (49) and frequently has been used to obtain transformants and generate deletions and conditional mutants with inducible gene expression (58). The establishment of such a system in B. licheniformis DSM13 would considerably improve genetic handling in this industrial workhorse.
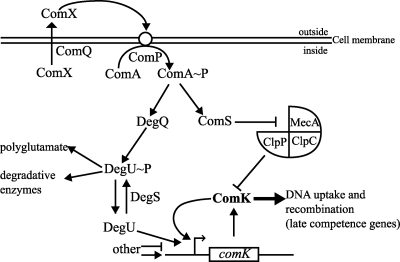
The key regulator responsible for the development of genetic competence is ComK, controlling the transcription of all genes involved in DNA binding, processing, uptake, and homologous recombination between the incoming and the host DNA (16, 20). ComK expression is turned down during exponential growth, and a number of stimuli must be integrated to control comK expression (Fig. 1) (14, 18, 23, 40). In B. subtilis 168 (27) and Bacillus amyloliquefaciens FZB42 (7), the quorum-sensing-dependent escape from ComK proteolysis depends on a regulatory operon containing four genes, comQ, comX, comP, and comA (20). ComQ is responsible for processing pre-ComX to generate the active ComX peptide pheromone (1). The accumulation of extracellular ComX is sensed by sensor histidine kinase ComP, which is capable of phosphorylating the cognate response regulator ComA. Phosphorylated ComA (ComA∼P) induces the transcription of the srf operon, which codes for the biosynthetic pathway of the biotenside surfactin, and which includes the 47 amino acids spanning ComS peptide (34). ComS prevents the proteolysis of ComK by competitively binding to MecA, thereby blocking the proteolytic MecA/ClpCP complex (13, 17, 31, 43, 57). Furthermore, ComA∼P also facilitates the transcription of degQ by promoting the phosphorylation of DegQ and increases the amount of DegU∼P, which facilitates the synthesis of extracellular enzymes and polyglutamate production (Fig. 1) (8, 35, 39, 50).
FIG. 1.
Regulation of comK expression (only proteins and loci addressed in this work are mentioned). ComK activates the transcription of its own gene, forming a positive autoregulatory loop (30, 47). The DegS/DegU two-component system influences competence development in a way that dephosphorylated DegU facilitates the binding of ComK to its cognate promoter, thus leading to the onset of the autoregulatory loop; phosphorylated DegU promotes the synthesis of extracellular enzymes and polyglutamate (19) for biofilm formation. ComK levels are reduced posttranscriptionally at low cell density when MecA recruits ComK to the proteolytic MecA-ClpCP complex, making competence development quorum-sensing dependent (for details, see the text). Arrows and T bars indicate positive and negative regulation, respectively. ComX, peptide pheromone; ComQ, modification and secretion of ComX; ComP, sensor histidine kinase; ComA, response regulator; ComS, inhibition of MecA/ClpCP; DegQ, facilitator of the phosphorylation of DegU; DegU, response regulator; DegS, sensor histidine kinase; MecA, adaptor-protein; ClpCP, protease; ComK, main transcriptional activator of natural competence.
However, B. licheniformis ATCC 14580 and the isogenic DSM13 carry an insertion element within the comP locus. Hence, poor genetic competence in B. licheniformis DSM13 may be due to the lack of a functional ComP. Since natural genetic competence has been reported to occur in B. licheniformis 9945A (32, 54), it was considered that understanding the lack of natural genetic competence in strain DSM13 and derivatives could lead to the development of competent strains and thus make efficient genetic tools available.
MATERIALS AND METHODS
Bioinformatic sequence analysis.
Sequence alignments were done with CLUSTALW (53). Visual alignments of GC contents were performed with Artemis (5).
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1 . Cultivation was done at 37°C in Luria-Bertani (LB) broth unless otherwise stated. Plasmid-carrying Escherichia coli strains were grown with ampicillin (100 μg/ml), and Bacillus transformants were grown with erythromycin (0.3 or 5 μg/ml) or tetracycline (12.5 to 37.5 μg/ml); the respective higher concentrations were applied for freely replicating plasmids (multicopy), and the lower concentrations were used for the selection of strains carrying an integrated plasmid (single copy).
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5αF′ | endA1 hsdR17 (rK−, mK+) supE44 thi-1 gyrA96 relA1 Δ(lacIZYA-argF)U169 deoR F′Φ80dlacZΔM15) | 67 |
| Bacillus subtilis DB104 | his npR2 nprE18 aprΔ3 | 24 |
| Bacillus licheniformis DSM13 | Wild type | DSMZ |
| Bacillus licheniformis DSM13ΔspoIV | Sporulation-deficient DSM13 derivative | S. Wemhoff, this laboratory |
| Bacillus licheniformis MW3 | ΔhsdR1, ΔhsdR2 | 65 |
| Bacillus licheniformis MW3.1 | ΔhsdR1, ΔhsdR2, ΔpyrE | This work |
| Bacillus licheniformis MW4.1 | ΔhsdR1, ΔhsdR2, ΔpyrE, Δpga | This work |
| Bacillus licheniformis MK1.1 | ΔhsdR1, ΔhsdR2, comP active, ΔpyrE | This work |
| Bacillus licheniformis MK2.1 | ΔhsdR1, ΔhsdR2, comP active, ΔpyrE, Δpga | This work |
| Bacillus licheniformis F11 | Wild type (ΔchiA) | 62 |
| Plasmids | ||
| pUCBM20 | E. coli cloning vector; Apr | Boehringer Mannheim, Germany |
| pSKE194 | E. coli/Bacillus shuttle vector, Apr, Emr, oriE.coli/orits | 38 |
| pE007 | pSKE194 derivative deprived of its E. coli fraction by XbaI restriction and religation of the Bacillus part and with mcs of pUCM20; Emr, orits | C. Borgmeier, this laboratory |
| pcomP | Bacillus vector Emr with comQXPA3/comQXPA4 amplicon of Bacillus licheniformis F11; Emr, orits | This work |
| pΔpyrE | pSKE194 derivate with deleted copy for Bacillus licheniformis pyrE; Emr, orits | This work |
| pΔpga | pSKE194 derivate with deleted copy for Bacillus licheniformis MW3 polyglutamate-operon; Emr, orits | This work |
| pMM1522 | Xylose-inducible expression vector for Bacillus | 60 |
| puc21uvrBABali2 | pUCBM20 derivative with deletion flanks for uvrBA | 63 |
| pUCuvrBAEm | pUCuvrBA derivative with pSKE194 erythromycin resistance cloned between the deletion flanks | This work |
| pMutpgk | pMutin2 derivative with pgk insertion flank | This work |
| pMMcomK | pMM1522 derivative, Bacillus licheniformis MW3 ComK expression vector | This work |
Apr, ampicillin resistance; Emr, erythromycin resistance; orits, temperature-sensitive Bacillus origin of replication; mcs, multiple cloning site; DSMZ, deutsche Sammlung für Mikroorganismen und Zellkulturen.
Molecular biological techniques.
Cloning in E. coli was performed essentially as described previously (45). Genomic DNA from B. licheniformis strains was isolated as previously described (9) or by using a commercial kit (MasterPure Gram-positive DNA purification kit; Epicentre Biotechnologies, Madison, WI). PCR and Southern blot analysis (48) were performed as previously described (37). Primers were deduced on the basis of the published B. licheniformis DSM13 (isogenic to ATCC 14580) genome sequence (44, 61) and are listed in Table S1 in the supplemental material. PCR mixtures (100 μl) consisted of 200 μM deoxynucleotides, 100 ng of template DNA, 1 pmol of each primer, and 1 U of Taq DNA polymerase or Vent DNA polymerase (New England Biolabs, Frankfurt, Germany). The purification of amplified fragments after gel electrophoresis was performed with a Qiaquick gel extraction kit (Qiagen, Hilden, Germany). Sequencing was done with fluorescence-labeled dideoxynucleotides of the BigDye Terminator v3.1 sequencing kit (Applied Biosystems, CA) and an ABI Prism capillary sequencer (model 3730). Sequences of B. licheniformis 9945A are part of an ongoing genome-sequencing project produced by the 454 technology (Roche, Brandford, CT). All fragments have been sequenced to a quality of phred45 or better.
Vector construction and gene deletion.
Vectors and plasmids used in this study are compiled in Table 1. For the deletion of the insertion sequence (IS) element in comP in B. licheniformis MW3, a fragment of an intact comP of B. licheniformis strain F11 was amplified by PCR with the primer pair comQXPA3/comQXPA4 and cloned into pE007, resulting in vector pcomP. For obtaining a stable uracil auxotrophic mutant, a small deletion of 129 bp comprising the α-d-5-P-ribosyl-pyrophosphate (PRPP) binding site of the pyrE gene product, which is responsible for the conversion of orotate and phosphoribosylpyrophosphate to orotidin-5-phosphate and PPi, was used. Respective recombination flanks were amplified by PCR; flank A was created with the primer pair BalipyrE1/BaliPyrESOE1, and flank B was obtained by using the primer pair BalipyrE2/BaliPyrESOE2 (see Table S1 and Fig. S2 in the supplemental material). The deletion cassette was constructed using splicing by overlap extensions (SOE) (22) and cloned into pUCBM20. The deletion cassette was excised from the pUCBM20 derivative by PstI restriction and cloned into the PstI-linearized pSKE194. XbaI restriction removed the E. coli part of the vector, and the remaining Bacillus part comprising the temperature-sensitive origin of replication, erythromycin resistance, and pyrE deletion cassette was religated; the resulting plasmid was termed pΔpyrE. For deletion, the vector pΔpga flank A was amplified with the primer pair pgaFlAf/pgaFlAr, flank B was constructed by using the primer pair pgaFlBf/pgaFlBr, and these were successively ligated into pUCBM20. Further cloning was the same as that for pΔpyrE. In PCRs for identifying mutants and for mutant screening, chromosomal DNA and the primers comQXPA1/comQXPAP2 specific for the comXQPA operon, the primers hsdR1a/hsdR1b specific for hsdR1, the primers hsdR2c/hsdR2d specific for hsdR2, the primers pga3/pgas20 specific for the pga region, and the primers pyrEscreenfor/pyrEscreenrev specific for the pyre region were used. ComK expression vectors were cloned by amplifying comK of B. licheniformis MW3 using the primer pair comKf_BsrGI/comKr_SphI and cutting with BsrGI and SphI, followed by cloning into the likewise-cut pMM1522 (Mobitec, Göttingen, Germany), a xylose-inducible expression vector (60). The ComK expression vector was designated pMMcomK.
For the direct disruption of the uvrBA genes, the suicide vector pUCuvrBAEm was constructed by amplifying the puc21uvrBABali2 vector (63) with the primer pair uvrABali1/uvrBBali2 and the subsequent ligation of the erythromycin resistance gene with the uvrAbali1/uvrBbali2 amplicon. The erythromycin resistance gene fragment was obtained by the PciI/ClaI restriction of pSKE194 by filling in the overlaps with Klenow polymerase (Roche Diagnostic GmbH, Mannheim, Germany) according to the manufacturer's instructions. The pMutpgk vector for the conditional inactivation of the pgk gene was constructed by the NotI/BamHI restriction of pMutin2 (58) and ligation with the NotI/BamHI-restricted pgkforNotI/pgkrevBamHI amplicon. The plasmids (pcomP, pΔpga, pΔpyrE, and pMMcomK) were transformed into Bacillus subtilis DB104 by the polyethylene glycol (PEG)-mediated transformation of protoplasts essentially as previously described (6). After reisolation, these were introduced into B. licheniformis MW3, and a PCR screening for deletion mutants was conducted as previously described (65). Mutants obtained are compiled in Table 1. Suicide vectors pUCuvrBAEm and pMutpgk were introduced into B. licheniformis MW3.1 carrying pMMcomK by transformation via induced competence.
Transformation with natural and induced competence.
A two-step transformation protocol was performed essentially as described in reference 21. Cells were grown overnight on LB agar plates, and single colonies were inoculated into 3 ml HS medium, which contained 5 ml S base [2% (NH4)2SO4, 14% K2HPO4, 6% KH2PO4, 1% Na3 citrate × 2H2O, 0.2% MgSO4 × 7H2O (wt/vol)], 0.5 ml yeast extract (10% [wt/vol]), 0.5 ml Casamino Acids (2% [wt/vol]), 2 ml uracil solution (2 mg/ml), 5 ml arginin-histidine solution (8 and 0.4% [wt/vol], respectively), and 1.25 ml glucose (20% [wt/vol]). After overnight incubation at 37°C with vigorous shaking, 20 ml of prewarmed LS medium, containing 5 ml 10× S base, 0.5 ml yeast extract (10% [wt/vol]), 0.25 ml Casamino Acids (2% [wt/vol]), 2 ml uracil (2 mg/ml), 1.25 ml glucose (20% [wt/vol]), 0.125 ml 1 M MgCl2 was inoculated with 1 ml of the starter culture and cultivated at 37°C with vigorous shaking. Upon reaching an optical density at 546 nm (OD546) of 0.9 to 1 or at cultivation times of 1, 2, 3, or 4 h, 1 ml of competent cells was transferred to an Eppendorf cup containing 1 μg chromosomal DSM13ΔspoIV DNA for transformation and incubated for 2 to 4 h in a Thermomixer comfort (Eppendorf, Hamburg, Germany) at 37°C and 600 rpm. Cells were harvested (2 min at 16,000 × g) and washed three times with 1 ml of 15 mM NaCl to remove residual uracil and plated out on M9 minimal medium (45) supplemented with 0.1 mM CaCl2, 0.01% (wt/vol) yeast extract, 0.02% (wt/vol) Casamino Acids, and 0.2% (wt/vol) glucose as the carbon source without uracil to select for prototrophs.
The one-step transformation protocol was carried out as described previously (3). Cells were grown to the late stationary phase in competence medium. Transformation with chromosomal DSM13ΔspoIV-DNA was performed as described for B. subtilis 2, 3, and 4 h after the onset of stationary growth.
Induced competence with pMMcomK expression vectors.
A 10-ml LB-tet (12.5 μg/ml tetracycline [Tet], 8 μg/ml uracil) starter culture was inoculated with a single colony from a fresh LB plate (12.5 μg/ml Tet, 8 μg/ml uracil) and grown overnight at 37°C and 170 rpm in a New Brunswick Scientific shaker innova 4320 (Edison, NY). A 50-ml main culture (LB, 12.5 μg/ml Tet, 8.5 μg/ml uracil) was inoculated with 500 μl of the starter culture and grown in a four-baffle 300-ml Erlenmeyer flask at 37°C and 155 rpm until an OD546 of 0.5 was reached. The main culture was divided into two 20-ml cultures, and each was transferred into separate 100-ml Erlenmeyer flasks. In culture A, ComK expression was induced with 0.125 to 0.25% xylose, whereas culture B served as the uninduced negative control. Both of the cultures (A and B) were grown for 1 to 3 h. For the transformation, 0.5 ml culture (of A and B) was transferred to an Eppendorf cup containing 1 to 5 μg DNA. Transformation mixtures were incubated for 30 min in a Thermomixer comfort (Eppendorf, Hamburg, Germany) at 37°C and 700 rpm, after which the cells were harvested by centrifugation, resuspended in 500 μl M9 minimal medium supplemented with uracil (8 μg/ml), and incubated for another 30 min at 37°C and 700 rpm to stop comK induction. For the complementation of uracil auxotrophy, cells were washed three times with M9 minimal medium without uracil to remove residual uracil and finally plated on M9 minimal agar plates for the selection of prototrophs. For obtaining uvrBA-EmR mutants, washing was not necessary, as cells were directly plated on LB containing erythromycin (0.5 μg/ml). The construction and selection of conditional pgk mutant strains was done essentially as described for the uvrBA-EmR mutants, except that 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to growth media and selection plates.
RNA isolation.
For RNA isolation, cells were cultivated as described for the two-step transformation protocol; 1 ml was harvested after 1 h of cultivation in LS medium. The pellet was resuspended in 400 μl 10 mM Tris, transferred into Nalgene cryotubes (Nalgene Labware, Hereford, United Kingdom) filled with approximately 300 mg of glass beads (diameter, 150 to 212 μm; Sigma-Aldrich, Munich, Germany), and frozen in liquid nitrogen. Cell disruption was accomplished by using the Mikro Dismembrator S (Satorius AG, Goettingen, Germany) at 2,700 rpm for 4 min. RNA isolation was done according to the manufacturer's instructions with a high pure RNA isolation kit (Roche Diagnostics GmbH, Mannheim, Germany). The quality and the amount of the RNA samples were controlled using agarose gel electrophoresis and spectrophotometric analysis with a NanoDrop photometer (Peqlab Biotechnologie GmbH, Erlangen, Germany).
Reverse transcription-PCR (RT-PCR).
For cDNA synthesis, 0.5 μg of isolated RNA and a mixture of primers (rpsJ2, rpsE2, recA2, comK2, comGB1, and comA2) was employed in combination with a RevertAid H-minus first-strand cDNA synthesis kit (Fermentas, Leon-Rot, Germany) according to the manufacturer's instructions. The quantification of cDNA synthesis was accomplished by PCR with Taq polymerase from 18 to 25 cycles depending on the transcript. For the analysis of the genes, the following primer pairs were used: comK cDNA, primer pair comK1/comK2; comA cDNA, primer pair comA1/comA2; rpsE cDNA, primer pair rpsE1/rpsE2; recA cDNA, primer pair recA1/recA2; and comGB cDNA, primer pair comGB1/comGB2.
Enzyme assays.
Extracellular protease activities were determined using minimal medium agar plates as previously described (62). Extracellular glucanase activity was determined using LB plates containing 0.02% lichenin and subsequently stacked with Congo red. Clearing halos around the colony indicate enzyme activity.
UV survival measurements.
Cells of wild-type and randomly chosen uvrBA mutants (designated 1 to 4) were cultivated to the mid-exponential phase, and in several dilutions equal numbers of cells were spread on LB plates and UV irradiated (20 J/m2) or left nonirradiated as a control. Subsequently the plates were incubated overnight at 37°C.
Nucleotide sequence accession numbers.
Sequences determined in the course of this work have been submitted to GenBank under accession numbers GQ499198, GQ505078, GQ505079, GQ505080, GQ505081, and GQ505082.
RESULTS
Comparative analysis of genes involved in quorum sensing.
The only difference between the genome sequences of B. licheniformis DSM13 and ATCC14580 (except a negligible 370-bp difference within the 4.3-Mbp genome) is the orientation of the IS element accompanied by a breakdown in the GC content at the site of insertion in comP, possibly suggesting a flexible position within the genome (see Fig. S1 in the supplemental material). We have sequenced the comQXPA cluster of strain 9945A, which is known to be naturally competent, and also of the F11 strain for comparison. The corresponding genomic loci of the comQXPA clusters were aligned and compared to the genetic competent B. subtilis 168 and B. amyloliquefaciens FZB42. Differences from the sequence of DSM13 are observed in the first two large extracellular loops of the multimembrane-spanning protein ComP of the competent B. licheniformis 9945A. These loops are responsible for ComX recognition; however, as the sequence comparison revealed only 33% identities and 54% identities, respectively (42, 66), a replacement may not lead to the desired result. The GC plot of the sequence of comP of F11 almost perfectly matches that of DSM13 and ATCC 14580 (see Fig. S1 in the supplemental material), and the first two extracellular loops of ComP, which are responsible for ComX recognition, are almost identical (only one amino acid differs). The IS element is missing, making it a very promising candidate for the replacement of the interrupted gene. Furthermore, the functionality of ComP in B. licheniformis F11 can be derived from the polyglutamate formation of F11 (not shown), as B. subtilis strains with insertions in comP likewise lack polyglutamate synthesis (36, 55, 56), as is the case for B. licheniformis DSM13 (Fig. 2). Since different B. subtilis strains display genetic polymorphisms concerning the sensor histidine kinase ComP, the peptide pheromone ComX, and ComQ (55), the complete comQXPA cluster of Bacillus licheniformis F11 was compared to the respective B. licheniformis DSM13 sequence. ComQ and ComX of B. licheniformis F11 are identical to B. licheniformis DSM13. ComP of B. licheniformis F11 has few sequence deviations. These are composed of three silent mutations and two amino acid substitutions, one of which is at position 230 from serine in DSM13 to glycine in F11 (triplet change from AGT to GGT), and the other is at position 578 from aspartate in DSM13 to alanine in F11 (triplet change from GAT to GCT).
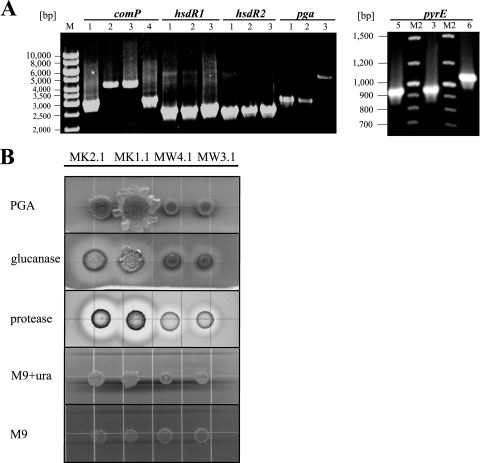
FIG. 2.
Genotypical and phenotypical examination of generated mutants. (A) PCR analysis of chromosomal DNA from B. licheniformis MK2.1 (ΔhsdR1, ΔhsdR2, comP active, ΔpyrE, Δpga) (1), MW4.1 (ΔhsdR1, ΔhsdR2, ΔpyrE, Δpga) (2), MW3.1 (ΔhsdR1, ΔhsdR2, ΔpyrE) (3), F11 (4), MK1.1 (ΔhsdR1, ΔhsdR2, comP active, ΔpyrE) (5), and MW3 (ΔhsdR1, ΔhsdR2) (6). As anticipated, mutants display smaller amplicons than the parental strain. (B) Equal amounts of cells of B. licheniformis MW3.1, MW4.1, MK1.1, and MK2.1 cultures were spotted onto agar plates containing LB only (PGA), LB plus lichenin (glucanase), and M9 plus skim milk (protease), respectively. Additionally, cells were dropped on M9 minimal medium supplemented with uracil (M9+ura) and on M9 minimal medium lacking uracil (M9). Note that the ComP-positive strain displays increased polyglutamate formation in either case.
Reconstitution of ComP in B. licheniformis MW3.
The disrupted comP of B. licheniformis MW3, the restriction-negative DSM13 derivative (65), was replaced by comP of B. licheniformis F11 (see Materials and Methods and Fig. S1 in the supplemental material). The generated strain B. licheniformis MK1 (ComP+) displayed polyglutamate synthesis, which was clearly discernible by slime formation (Fig. 2B). Furthermore, cells of the ComP-inactive strains B. licheniformis MW3.1 and MW4.1, as well as those of the ComP-active derivatives B. licheniformis MK1.1 and MK2.1, were spotted onto lichenin and skim milk agar plates for monitoring their extracellular glucanase and protease activity, respectively (Fig. 2B). B. licheniformis MK1.1 and MK2.1 displayed enhanced extracellular glucanase and protease activities compared to those of the ComP-inactive MW3.1 and MW4.1. The strains also were analyzed with respect to their genetic background (Fig. 2A) via PCR to ensure that all carried the deletions in the two loci encoding HsdR1/HsdR2, the restriction endonucleases (65). Based on the knowledge that ComX-dependent quorum sensing enhances polyglutamate slime formation and extracellular enzyme synthesis in B. subtilis (35, 50), ComP of B. licheniformis F11 appears to be functional when inserted into B. licheniformis MW3, as ComX-mediated quorum sensing apparently is reactivated. Additionally, we have added a His6 tag to ComP to verify the expression of ComP from the xylose-inducible promoter. Signals obtained by Western blotting proved expression (data not shown).
To analyze genetic competence, uracil auxotrophs of MW3 and MK1 were generated by deleting sequences encoding the phosphoribosyl-binding site of PyrE, the orotate phosphoribosyltransferase, which is essential for pyrimidine synthesis (see Fig. S2B in the supplemental material). The respective deletion and concomitant uracil auxotrophy is documented in Fig. 2A and B. Since a negative effect on transformation by excessive polyglutamate production cannot be excluded, mutants with a defect in PGA synthesis were constructed by the almost-complete deletion of the polyglutamate (pga) synthetase operon (see Fig. S2A in the supplemental material) in both MW3.1 and MK1.1 (ComP inactive and active, respectively). In the pga deletion mutant B. licheniformis MK2.1 (ComP active, Δpga, ΔpyrE), slime formation was completely abolished (Fig. 2B), providing further evidence that the slime formation of MK1.1 is indeed due to polyglutamate synthesis. As expected, no change in colony morphology was observed after pga deletion in strain MW4.1 (ComP inactive, Δpga, ΔpyrE), as the disruption of comP already sufficed to prevent polyglutamate synthesis (36, 56). The successful removal of the IS element was demonstrated by PCR. B. licheniformis MK2.1 and MK1.1 (not shown) display the same comP amplicon size of 2,965 bp as that of B. licheniformis F11. MK2.1 also carries the deletions of the genes for PGA synthesis and uracil auxotrophy (PyrE) (amplicons of 2,646 and 938 bp, respectively), whereas B. licheniformis MW4.1 is characterized by the 4,253-bp comP amplicon, which includes the IS element.
Transformation via natural competence.
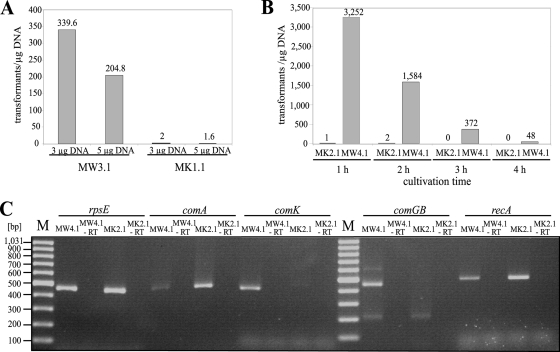
For the first experiments, B. licheniformis MW3.1 and MK1.1 were transformed to prototrophy by applying chromosomal DNA of a sporulation-negative but uracil-prototrophic B. licheniformis strain (DSM13ΔspoIV) via a two-step transformation protocol (Fig. 3A). Experiments with DNA from uracil-auxotrophic B. licheniformis MW3.1 and MK1.1 served as negative controls and, as expected, no uracil prototrophic transformants could be obtained. B. licheniformis MW3.1 carrying the inactive comP was only poorly transformable via natural competence with approximately 340 transformants/μg DNA. However, MK1.1, encoding an active ComP, exhibited a more-than hundredfold decrease in transformation efficiency, i.e., 2 and 1.6 transformants/μg DNA, respectively. Transformants were routinely checked by PCR analysis for their genetic background (data not shown). To rule out that the polyglutamate slime formation of the ComP-active strain negatively influenced transformation efficiencies, such experiments again were carried out with the Δpga mutant strains B. licheniformis MW4.1 and MK2.1. Furthermore, possible effects due to extended cultivation times (1 to 4 h within the exponential phase) on transformation efficiencies after the change from the HS to the LS medium were investigated for both strains (Fig. 3B). In general, transformation efficiencies were higher immediately after medium change from the starter culture to the main culture, with maximum transformation efficiencies of 3,250 transformants/μg DNA for MW4.1 cultivation for 1 h. The pga deletion did not enhance the transformation of the ComP active strain MK2.1, as efficiencies of B. licheniformis MK2.1 remained low, with 1 and 2 transformants/μg DNA after 1 and 2 h of cultivation. Thus, poor transformation efficiencies are not due to polyglutamate synthesis. To further investigate the reason for the poor transformation efficiency in the ComP-active strain B. licheniformis MK2.1 in contrast to that of MW4.1, a quantitative PCR (qPCR) analysis was conducted with RNA isolated from cultures after 1 h of cultivation (Fig. 3C). Negative controls without reverse transcriptase showed no amplicon. The mRNA of a ribosomal protein (RpsE) served as an internal control. In the ComP-active strain MK2.1, the transcription levels of the genes for the main activator of natural competence, ComK, and also for ComGB, a protein of the DNA uptake machinery, were significantly lower than that in the ComP-inactive strain MW4.1. Furthermore, the transcription of comA and recA are enhanced in B. licheniformis MK2.1 (Fig. 3C). Hence, the reactivation of the sensor histidine kinase ComP in B. licheniformis MW3 diminished transformation efficiencies during natural competence due to reduced comK expression in the ComP-active strain MK2.1, as shown here with a two-step transformation protocol. A one-step procedure for transformation during natural competence (3, 54) also was applied for B. licheniformis MW4.1 and MK2.1, but no prototrophic transformants could be obtained. As the reactivation of ComP did not enhance transformation efficiencies via natural competence, another strategy that focused on inducible ComK expression was tested.
FIG. 3.
Influence of the ComP status (active/inactive) on the transformation efficiencies of B. licheniformis MW3 derivatives and on gene expression during transformation. (A) Transformation efficiencies of B. licheniformis MW3.1 (ComP−, ΔpyrE) and of MK1.1 (ComP+, ΔpyrE) using different amounts (3 or 5 μg) of chromosomal DSM13ΔspoIV-DNA to regain uracil prototrophy. (B) Transformation efficiencies of polyglutamate-negative strains B. licheniformis MW4.1 (ComP−, ΔpyrE, Δpga) and B. licheniformis MK2.1 (ComP+, ΔpyrE, Δpga) after different cultivation times after medium exchange. (C) RT-PCR analyses of isolated RNA from B. licheniformis MW4.1 (ComP−, ΔpyrE, Δpga) and B. licheniformis MK2.1 (ComP+, ΔpyrE, Δpga). −RT, negative controls without reverse transcriptase; M, 100-bp DNA ladder (gene ruler; Fermentes, St-Leon-Roth, France); RpsE, ribosomal protein; ComA, response regulator; ComK, main transcriptional activator of natural competence; ComGB, DNA transport protein; RecA, multifunctional protein involved in homologous recombination and DNA repair.
Inducible genetic competence.
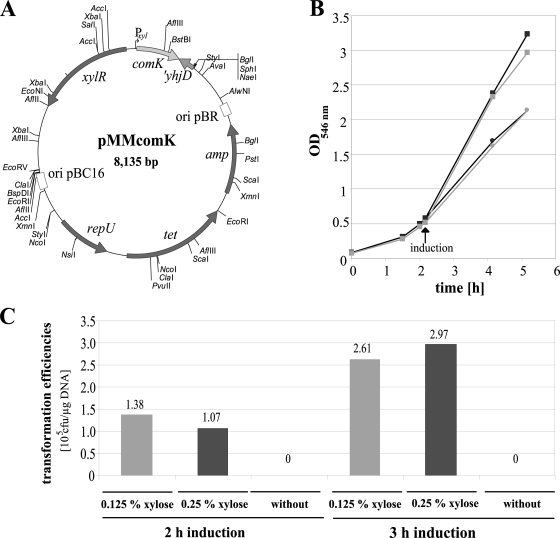
The comK gene of B. licheniformis DSM13 was placed under the control of a xylose-inducible promoter in the pMM1522 vector to give pMMcomK (60). Strain B. licheniformis MW3.1 (ComP−, ΔpyrE) transformed with the ComK expression vector (pMMcomK) subsequently was used in transformation experiments (Fig. 4A). For the determination of the optimal transformation conditions, the strain described above was transformed to prototrophy by using chromosomal DSM13ΔspoIV DNA and applying different xylose concentrations (0.125 and 0.25%) for comK induction along with several cultivation times after induction (1, 2, and 3 h). The highest transformation efficiency, 2.97 × 105 CFU/μg DNA, was obtained with 0.25% xylose and a 3-h cultivation time postinduction; induction with 0.125% xylose and a 3-h cultivation time yielded 2.61 × 105 prototrophic transformants/μg DNA (Fig. 4C). Shorter induction times (for example, 2 h) lowered the transformation efficiency to 1.38 × 105 CFU/μg DNA for induction with 0.125% xylose and 1.07 × 105 CFU/μg DNA with 0.25% xylose. As artificial ComK induction in B. licheniformis MW3 resulted in high transformation efficiencies, the regulatory pathway downstream of ComK activation apparently is functional. Growth retardation observed in the induced cultures provided the basis for the curing of the pMMcomK vector (Fig. 4B). The obtained prototrophic strains were grown overnight in LB supplemented with 0.5% xylose but without tetracycline, which are conditions that favor the growth of transformants that have lost the vector. Cells subsequently were plated out on LB medium with 0.5% xylose, and single colonies were used to inoculate LB medium with 0.5% xylose. Such cultures plated on LB medium with 0.5% xylose containing 12.5 μg/ml tetracycline revealed no growth, indicating the loss of the tetracycline resistance carried on the vector pMMcomK. In a control experiment in which the overnight culture was supplemented with tetracycline, up to 1.14 × 108 colonies were observed on LB medium with 0.5% xylose containing 12.5 μg/ml tetracycline.
FIG. 4.
Induced natural competence by xylose-driven ComK expression. (A) pMMcomK, vector for xylose-inducible ComK expression. Open reading frames are depicted as arrows. Restriction enzymes are abbreviated. ComK, the main activator for natural competence; ′YhjD, gene downstream of comK, part of a hypothetical protein; XylR, xylose repressor; RepU, replication protein; Tet, tetracycline resistance (Bacillus); Amp, ampicillin resistance (E. coli); ori pBC16, Bacillus origin of replication; ori pBR, ColE1 origin of replication. (B) Growth curves of B. licheniformis MW3.1 pMMcomK with or without the induction of ComK expression. Black circles, induction with 0.25% xylose; gray circles, induction with 0.125% xylose; black and gray squares correspond to uninduced controls. (C) Prototrophic transformants per microgram of DNA of induced (0.125% xylose and 0.25% xylose) and not induced (without) cultures for 2- and 3-h induction times.
Direct gene disruption.
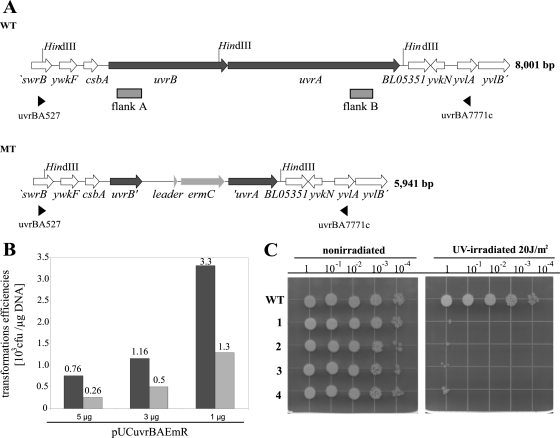
As the target for direct gene disruption experiments, the uvrBA operon was chosen, as such mutants previously were shown to be UV sensitive, rendering phenotypic identification possible (63). A suicide vector comprising the erythromycin resistance gene flanked by the recombination sequences was used (Fig. 5A). Approximately 3.3 × 103 erythromycin-resistant colonies per μg of DNA were obtained (Fig. 5B). Thirty-four to 43% of the latter were uvrBA mutants with the expected genotype (PCR data not shown). The remaining erythromycin-resistant transformants had ectopic integrations of the erythromycin resistance gene (revealed by Southern blot analysis; data not shown). The highest transformation efficiencies were obtained with 1 μg of pUCuvrBAEmR, which yielded 1.3 × 103 uvrBA mutants/μg DNA (Fig. 5B). Increasing the amount of DNA from 1 μg pUCuvrBAEmR to 3 or 5 μg did not improve the observed efficiency. Randomly chosen uvrBA mutants were tested for UV sensitivity in a drop dilution assay as shown in Fig. 5C. Hence, by applying the induced competence method, it could be shown that the direct gene disruption of the uvrBA operon was successful and rapidly achievable in B. licheniformis MW3.
FIG. 5.
Direct disruption of the uvrBA operon by induced competence. (A) Schematic representation of the uvrBA operon from wild-type B. licheniformis (WT) and the uvrBA mutant (MT). SwrB, motility/swarming protein; YwkF, hypothetical protein; CsbA, conserved uncharacterized protein; UvrB, excinuclease ABC subunit B; UvrA, excinuclease ABC subunit A; BL05351, pseudogene; YvkN and YvlA, hypothetical proteins; YvlB, uncharacterized conserved protein; ErmC, erythromycin resistance. (B) Transformation efficiencies via induced competence with suicide vector pUCuvrBAEmR. Black bars, total amounts of obtained erythromycin-resistant transformants; gray bars, fraction of the uvrBA mutants. (C) UV sensitivity of the uvrBA mutant strains (1, 2, 3, and 4) compared to that of the wild type.
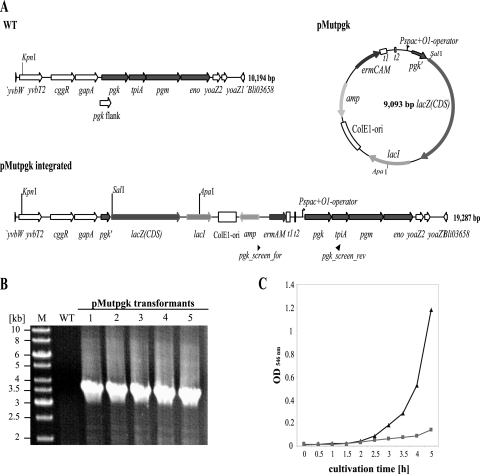
Conditional knockout of essential genes.
A vector based on pMutin2 was used (58) to determine whether transformation efficiencies suffice for the conditional inactivation of an essential gene. The vector (pMutin2) allows the expression of the target gene with the IPTG-inducible Pspac promoter by integrating pMutin at the position of the gene in the genome via homologous recombination. Due to promoter exchange, the target gene is transcribed only in the presence IPTG. Thus, the growth of the culture is IPTG dependent, given that the target gene is essential for the organism. For the conditional gene inactivation approach, an operon was chosen that includes four putative essential genes: pgk, tpiA, pgm, and eno. The products of these genes are thought to be instrumental in the conversion of trioses to pyruvate (26). Transformants having integrated the respective suicide vector were obtained (Fig. 6A), as shown by PCR with the primer pair pgk_screen_for/pgk_screen_rev (Fig. 6B). It also was shown that the growth of the transformants with an integrated vector (pMutpgk) is IPTG dependent (see Fig. 6C), showing that this operon comprises at least one essential gene.
FIG. 6.
Conditional inactivation of essential genes in B. licheniformis MW3 by induced competence. (A) Schematic representation of the genetic organization of the pgk operon of the wild type (WT), conditional mutant (pMutpgk integrated), and vector for inactivation (pMutpgk). Pgk, phosphoglyceratkinase; TpiA, triosephoshatisomerase; Pgm, phosphoglyceratmutase; Eno, enolase. Pspac+O1, IPTG-inducible promoter with operator 1; t1 and t2, strong transcriptional terminators. For detailed information, see reference 58). (B) Verification of obtained transformants by PCRs. (C) IPTG growth of transformants. Black curve marked with triangles, transformants grown in LB with IPTG; gray curve with boxes and curve with triangles, transformants grown without IPTG.
DISCUSSION
The surmised negative impact on the development of genetic competence due to the insertion of an IS element into the comP locus is not unique to B. licheniformis DSM13/ATCC 14580 (44, 61). In B. subtilis natto NAF4, an element belonging to the IS4 family (IS4Bsu1) frequently integrates into comP (36, 56), thereby not only rendering the strain less competent but also causing the complete loss of PGA formation and a decrease in extracellular protease secretion (36, 52, 66), as shown here for B. licheniformis DSM13.
Surprisingly, the replacement of the IS3-interrupted comP with an intact copy did not restore full genetic competence but did indeed effect the opposite. PGA formation, known to be detrimental for competence (12, 29, 54), was excluded as the cause of the observed impairment. As the transcription of comK as well as that of downstream genes, such as comGB, was significantly lowered, no direct link between ComP and ComK via ComS, as observed in B. subtilis, could be shown for B. licheniformis. The quantitative conversion of DegU into its phosphorylated state, DegU∼P, via the ComP/ComA-activated DegQ may additionally provide a clue to the lowered competence development, as unphosphorylated DegU is crucial to start the autoregulatory comK loop (19, 20, 25, 35). The enhanced expression of recA and, in particular, comA again probably is due to the lack of the insertion element, as comA transcription is possible not only from its cognate promoter but also from the promoter of comP by a readthrough mechanism (66). When ComK was inducibly expressed on a plasmid, the achieved transformation frequencies are in good agreement with those of the known naturally competent strain B. licheniformis 9945A (54), demonstrating not only a functional DNA uptake machinery but also its regulation by ComK.
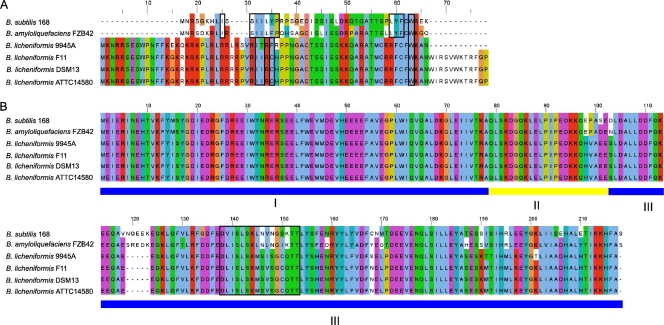
The link between ComP and competence development via ComS may not exist in B. licheniformis DSM13, as an open reading frame encoding a potential ComS could not be identified due to poor sequence conservation along with the short sequence length of ComS (28). Thus, the identification was attempted by comparing the N-terminal parts of the B. subtilis 168 surfactin synthetase B with the corresponding protein lichenysin synthetase B of the B. licheniformis strains, followed by a search for conserved peptides of ComS. A significant C-terminal conservation of the B. subtilis ComS compared to that of the considered B. licheniformis ComS was identified (Fig. 7A).
FIG. 7.
ClustalW alignments of ComS (A) and MecA (B) colored with JalView. The same colors in all rows mark conserved amino acids. (A) Black boxes show ComS amino acids essential for the competence development (40, 43). (B) The domain structure of MecA is highlighted. The roman numeral I corresponds to the ComK and ComS binding domain, II indicates a predicted loop region, and III indicates the ClpC binding domain (41). The black box marks a second ComS binding domain (43).
The transcription of the lichenysin operon is known to be activated by B. subtilis ComA∼P; consistently with this finding, a putative ComA Box in the promoter region of the lichenysin operon has been proposed (68, 69). However, ComS9945A differs in several aspects from its counterparts of B. subtilis and B. amyloliquefaciens: (i) there is an N-terminal extension of 16 amino acids, (ii) there are four amino acids inserted at position 27, and (iii) the potential core sequence for MecA binding (ITRFRP) in B. licheniformis 9945A differs from those of B. subtilis (ILLYPR) and B. amyloliquefaciens (IILFPQ) (7, 40, 43). To prove the functionality of the putative ComS9945A further, experiments are being performed and will be reported later.
Interestingly, the putative ComS proteins of the B. licheniformis strains F11, DSM13, and ATCC 14580 are identical, whereas ComS of the only known naturally competent B. licheniformis 9945A has some differences (Fig. 7). The C-terminal extension and the differences in the putative binding site of MecA could be responsible for the lack of function in B. licheniformis DSM13 and thus also will be the focus of further experiments.
Upon ComK induction, very high transformation efficiencies were achieved. As the growth rate of cultures with induced ComK expression was reduced compared to that of the uninduced control, an elegant means for vector curing is provided, as competent cells of B. subtilis are known to block chromosome replication and cell division via ComGA, which is governed by ComK (13, 15, 51). Additionally, genes encoding inhibitors of cell division such as maf, minC, and minD are upregulated by ComK (2, 4, 11).
For Bacillus cereus, a vector-based comK induction was used to obtain mutants by natural competence; however, efficiencies were rather poor: the use of chromosomal DNA of B. cereus FM1400, which contains an erythromycin resistance gene (ermC) in sigB, yielded 1 to 9 CFU/μg DNA (33). Possibly, the heterologous ComK of B. subtilis or detrimental effects of permanent ComK induction by IPTG (13, 14, 59) caused the low number of obtainable transformants, which are three orders of magnitude less than that for B. licheniformis. As the high transformation efficiencies facilitated not just an easy direct disruption of the uvrBA genes but also the conditional inactivation of essential genes by using the pMutin system (58), the developed method is suitable for the alteration of any chromosomal locus, irrespective of function, making further studies of regulators and essential genes possible.
Supplementary Material
Acknowledgments
We thank Carla Denschlag for help with the sequencing of DNA fragments from strain F11 and Timothy O'Connell from Henkel AG & Co. KGaA for reading the manuscript.
The work was supported by Federal Ministry of Education and Research (BMBF, Bonn-Bad Godesberg, Germany) grant no. 0315283 for the Westfälische Wilhelms-Universität Münster and grant no. 0313917D for the Georg-August Universität Göttingen.
Footnotes
Published ahead of print on 11 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bacon Schneider, K., T. M. Palmer, and A. D. Grossman. 2002. Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis. J. Bacteriol. 184:410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 3.Bott, K. F., and G. A. Wilson. 1967. Development of competence in the Bacillus subtilis transformation system. J. Bacteriol. 94:562-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, Y. X., Y. Abhayawardhane, and G. C. Stewart. 1993. Amplification of the Bacillus subtilis maf gene results in arrested septum formation. J. Bacteriol. 175:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carver, T., M. Berriman, A. Tivey, C. Patel, U. Bohme, B. G. Barrell, J. Parkhill, and M. A. Rajandream. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, S., and S. N. Cohen. 1979. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol. Gen. Genet. 168:111-115. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X. H., A. Koumoutsi, R. Scholz, A. Eisenreich, K. Schneider, I. Heinemeyer, B. Morgenstern, B. Voss, W. R. Hess, O. Reva, H. Junge, B. Voigt, P. R. Jungblut, J. Vater, R. Sussmuth, H. Liesegang, A. Strittmatter, G. Gottschalk, and R. Borriss. 2007. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25:1007-1014. [DOI] [PubMed] [Google Scholar]
- 8.Comella, N., and A. D. Grossman. 2005. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 57:1159-1174. [DOI] [PubMed] [Google Scholar]
- 9.Gärtner, D., M. Geissendörfer, and W. Hillen. 1988. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J. Bacteriol. 170:3102-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gherna, R., P. Pienta, and R. Cote. 1989. Catalogue of bacteria and bacteriophages, 17th ed. American Type Culture Collection, Rockville, MD.
- 11.Gregory, J. A., E. C. Becker, and K. Pogliano. 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 22:3475-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gwinn, D. D., and C. B. Thorne. 1964. Transformation of Bacillus licheniformis. J. Bacteriol. 87:519-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, J., J. Bylund, M. Haines, M. Higgins, and D. Dubnau. 1995. Inactivation of mecA prevents recovery from the competent state and interferes with cell division and the partitioning of nucleoids in Bacillus subtilis. Mol. Microbiol. 18:755-767. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, J., L. Kong, and D. Dubnau. 1994. The regulation of competence transcription factor synthesis constitutes a critical control point in the regulation of competence in Bacillus subtilis. J. Bacteriol. 176:5753-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haijema, B. J., J. Hahn, J. Haynes, and D. Dubnau. 2001. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 40:52-64. [DOI] [PubMed] [Google Scholar]
- 16.Haijema, B. J., D. van Sinderen, K. Winterling, J. Kooistra, G. Venema, and L. W. Hamoen. 1996. Regulated expression of the dinR and recA genes during competence development and SOS induction in Bacillus subtilis. Mol. Microbiol. 22:75-85. [DOI] [PubMed] [Google Scholar]
- 17.Hamoen, L. W., H. Eshuis, J. Jongbloed, G. Venema, and D. van Sinderen. 1995. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol. Microbiol. 15:55-63. [DOI] [PubMed] [Google Scholar]
- 18.Hamoen, L. W., D. Kausche, M. A. Marahiel, D. van Sinderen, G. Venema, and P. Serror. 2003. The Bacillus subtilis transition state regulator AbrB binds to the −35 promoter region of comK. FEMS Microbiol. Lett. 218:299-304. [DOI] [PubMed] [Google Scholar]
- 19.Hamoen, L. W., A. F. Van Werkhoven, G. Venema, and D. Dubnau. 2000. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 97:9246-9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamoen, L. W., G. Venema, and O. P. Kuipers. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9-17. [DOI] [PubMed] [Google Scholar]
- 21.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley and Sons, Ltd., Chichester, West Sussex, England.
- 22.Heckman, K. L., and L. R. Pease. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924-932. [DOI] [PubMed] [Google Scholar]
- 23.Hoa, T. T., P. Tortosa, M. Albano, and D. Dubnau. 2002. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 43:15-26. [DOI] [PubMed] [Google Scholar]
- 24.Kawamura, F., and R. H. Doi. 1984. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J. Bacteriol. 160:442-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66:395-409. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, N. Ogasawara, et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U. S. A. 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 28.Lapidus, A., N. Galleron, J. T. Andersen, P. L. Jorgensen, S. D. Ehrlich, and A. Sorokin. 2002. Co-linear scaffold of the Bacillus licheniformis and Bacillus subtilis genomes and its use to compare their competence genes. FEMS Microbiol. Lett. 209:23-30. [DOI] [PubMed] [Google Scholar]
- 29.Leonard, C. G., D. K. Mattheis, M. J. Mattheis, and R. D. Housewright. 1964. Transformation to prototrophy and polyglutamic acid synthesis in Bacillus licheniformis. J. Bacteriol. 88:220-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maamar, H., and D. Dubnau. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magnuson, R., J. Solomon, and A. D. Grossman. 1994. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77:207-216. [DOI] [PubMed] [Google Scholar]
- 32.McCuen, R. W., and C. B. Thorne. 1971. Genetic mapping of genes concerned with glutamyl polypeptide production by Bacillus licheniformis and a study of their relationship to the development of competence for transformation. J. Bacteriol. 107:636-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mironczuk, A. M., Á. Kovács, and O. Kuipers. 2008. Induction of natural competence in Bacillus cereus ATCC14579. Microb. Biotechnol. 1:226-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Msadek, T. 1999. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 7:201-207. [DOI] [PubMed] [Google Scholar]
- 35.Msadek, T., F. Kunst, A. Klier, and G. Rapoport. 1991. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J. Bacteriol. 173:2366-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagai, T., L. S. Phan Tran, Y. Inatsu, and Y. Itoh. 2000. A new IS4 family insertion sequence, IS4Bsu1, responsible for genetic instability of poly-gamma-glutamic acid production in Bacillus subtilis. J. Bacteriol. 182:2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nahrstedt, H., and F. Meinhardt. 2004. Structural and functional characterization of the Bacillus megaterium uvrBA locus and generation of UV-sensitive mutants. Appl. Microbiol. Biotechnol. 65:193-199. [DOI] [PubMed] [Google Scholar]
- 38.Nahrstedt, H., J. Waldeck, M. Gröne, R. Eichstadt, J. Feesche, and F. Meinhardt. 2005. Strain development in Bacillus licheniformis: construction of biologically contained mutants deficient in sporulation and DNA repair. J. Biotechnol. 119:245-254. [DOI] [PubMed] [Google Scholar]
- 39.Nakano, M. M., and P. Zuber. 1991. The primary role of comA in establishment of the competent state in Bacillus subtilis is to activate expression of srfA. J. Bacteriol. 173:7269-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogura, M., L. Liu, M. Lacelle, M. M. Nakano, and P. Zuber. 1999. Mutational analysis of ComS: evidence for the interaction of ComS and MecA in the regulation of competence development in Bacillus subtilis. Mol. Microbiol. 32:799-812. [DOI] [PubMed] [Google Scholar]
- 41.Persuh, M., K. Turgay, I. Mandic-Mulec, and D. Dubnau. 1999. The N- and C-terminal domains of MecA recognize different partners in the competence molecular switch. Mol. Microbiol. 33:886-894. [DOI] [PubMed] [Google Scholar]
- 42.Piazza, F., P. Tortosa, and D. Dubnau. 1999. Mutational analysis and membrane topology of ComP, a quorum-sensing histidine kinase of Bacillus subtilis controlling competence development. J. Bacteriol. 181:4540-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prepiak, P., and D. Dubnau. 2007. A peptide signal for adapter protein-mediated degradation by the AAA+ protease ClpCP. Mol. Cell 26:639-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rey, M. W., P. Ramaiya, B. A. Nelson, S. D. Brody-Karpin, E. J. Zaretsky, M. Tang, A. Lopez de Leon, H. Xiang, V. Gusti, I. G. Clausen, P. B. Olsen, M. D. Rasmussen, J. T. Andersen, P. L. Jorgensen, T. S. Larsen, A. Sorokin, A. Bolotin, A. Lapidus, N. Galleron, S. D. Ehrlich, and R. M. Berka. 2004. Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol. 5:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1 to 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schallmey, M., A. Singh, and O. P. Ward. 2004. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50:1-17. [DOI] [PubMed] [Google Scholar]
- 47.Smits, W. K., C. C. Eschevins, K. A. Susanna, S. Bron, O. P. Kuipers, and L. W. Hamoen. 2005. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56:604-614. [DOI] [PubMed] [Google Scholar]
- 48.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 49.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. U. S. A. 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanley, N. R., and B. A. Lazazzera. 2005. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-gamma-δl-glutamic acid production and biofilm formation. Mol. Microbiol. 57:1143-1158. [DOI] [PubMed] [Google Scholar]
- 51.Süel, G. M., J. Garcia-Ojalvo, L. M. Liberman, and M. B. Elowitz. 2006. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440:545-550. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi, K., Y. Sekine, T. Chibazakura, and H. Yoshikawa. 2007. Development of an intermolecular transposition assay system in Bacillus subtilis 168 using IS4Bsu1 from Bacillus subtilis (natto). Microbiology 153:2553-2559. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorne, C. B., and H. B. Stull. 1966. Factors affecting transformation of Bacillus licheniformis. J. Bacteriol. 91:1012-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tortosa, P., L. Logsdon, B. Kraigher, Y. Itoh, I. Mandic-Mulec, and D. Dubnau. 2001. Specificity and genetic polymorphism of the Bacillus competence quorum-sensing system. J. Bacteriol. 183:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran, L. S., T. Nagai, and Y. Itoh. 2000. Divergent structure of the ComQXPA quorum-sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol. 37:1159-1171. [DOI] [PubMed] [Google Scholar]
- 57.Turgay, K., L. W. Hamoen, G. Venema, and D. Dubnau. 1997. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11:119-128. [DOI] [PubMed] [Google Scholar]
- 58.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 59.van Sinderen, D., and G. Venema. 1994. comK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J. Bacteriol. 176:5762-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vary, P. S., R. Biedendieck, T. Fuerch, F. Meinhardt, M. Rohde, W. D. Deckwer, and D. Jahn. 2007. Bacillus megaterium-from simple soil bacterium to industrial protein production host. Appl. Microbiol. Biotechnol. 76:957-967. [DOI] [PubMed] [Google Scholar]
- 61.Veith, B., C. Herzberg, S. Steckel, J. Feesche, K. H. Maurer, P. Ehrenreich, S. Baumer, A. Henne, H. Liesegang, R. Merkl, A. Ehrenreich, and G. Gottschalk. 2004. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J. Mol. Microbiol. Biotechnol. 7:204-211. [DOI] [PubMed] [Google Scholar]
- 62.Waldeck, J., G. Daum, B. Bisping, and F. Meinhardt. 2006. Isolation and molecular characterization of chitinase-deficient Bacillus licheniformis strains capable of deproteinization of shrimp shell waste to obtain highly viscous chitin. Appl. Environ. Microbiol. 72:7879-7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waldeck, J., H. Meyer-Rammes, H. Nahrstedt, R. Eichstädt, S. Wieland, and F. Meinhardt. 2007. Targeted deletion of the uvrBA operon and biological containment in the industrially important Bacillus licheniformis. Appl. Microbiol. Biotechnol. 73:1340-1347. [DOI] [PubMed] [Google Scholar]
- 64.Waldeck, J., H. Meyer-Rammes, S. Wieland, J. Feesche, K. H. Maurer, and F. Meinhardt. 2007. Targeted deletion of genes encoding extracellular enzymes in Bacillus licheniformis and the impact on the secretion capability. J. Biotechnol. 130:124-132. [DOI] [PubMed] [Google Scholar]
- 65.Waschkau, B., J. Waldeck, S. Wieland, R. Eichstädt, and F. Meinhardt. 2008. Generation of readily transformable Bacillus licheniformis mutants. Appl. Microbiol. Biotechnol. 78:181-188. [DOI] [PubMed] [Google Scholar]
- 66.Weinrauch, Y., R. Penchev, E. Dubnau, I. Smith, and D. Dubnau. 1990. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 4:860-872. [DOI] [PubMed] [Google Scholar]
- 67.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yakimov, M. M., and P. N. Golyshin. 1997. ComA-dependent transcriptional activation of lichenysin A synthetase promoter in Bacillus subtilis cells. Biotechnol. Prog. 13:757-761. [DOI] [PubMed] [Google Scholar]
- 69.Yakimov, M. M., A. Kroger, T. N. Slepak, L. Giuliano, K. N. Timmis, and P. N. Golyshin. 1998. A putative lichenysin A synthetase operon in Bacillus licheniformis: initial characterization. Biochim. Biophys. Acta 1399:141-153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.