Abstract
N-Acylhomoserine lactones (AHLs) are signaling molecules in many quorum-sensing (QS) systems that regulate interactions between various pathogenic bacteria and their hosts. Quorum quenching by the enzymatic inactivation of AHLs holds great promise in preventing and treating infections, and several such enzymes have been reported. In this study, we report the characterization of a novel AHL-degrading protein from the soil bacterium Ochrobactrum sp. strain T63. This protein, termed AidH, shares no similarity with any of the known AHL degradases but is highly homologous with a hydrolytic enzyme from Ochrobactrum anthropi ATCC 49188 that contains the alpha/beta-hydrolase fold. By liquid chromatography-mass spectrometry (MS) analysis, we demonstrate that AidH functions as an AHL-lactonase that hydrolyzes the ester bond of the homoserine lactone ring of AHLs. Mutational analyses indicate that the G-X-Nuc-X-G motif or the histidine residue conserved among alpha/beta-hydrolases is critical for the activity of AidH. Furthermore, the AHL-inactivating activity of AidH requires Mn2+ but not several other tested divalent cations. We also showed that AidH significantly reduces biofilm formation by Pseudomonas fluorescens 2P24 and the pathogenicity of Pectobacterium carotovorum, indicating that this enzyme is able to effectively quench QS-dependent functions in these bacteria by degrading AHLs.
Quorum sensing (QS) is a widespread phenomenon that facilitates intercellular communication in bacterial communities to regulate their behaviors in a cell density-dependent manner (5). In many Gram-negative bacteria, members of the community produce and release one or more derivatives of N-acylhomoserine lactone (AHL) into the environment. Structurally, these signaling compounds share the homoserine lactone ring but differ with respect to the length and the C3 substitution of the acyl side chain (49). The concentration of the QS signaling molecules in the environment rises in proportion to the increase in the bacterial population density. After being accumulated to a critical threshold concentration, AHLs bind and activate their cognate transcriptional regulators to control the expression of target genes (52).
QS systems in bacteria regulate a number of biological functions, including the production of degradative extracellular enzymes in Pseudomonas aeruginosa and Pectobacterium carotovorum (24), bioluminescence in Vibrio fischeri (51), plasmid transfer in Agrobacterium tumefaciens (43), swarming motility in Serratia liquefaciens (17), antibiotic production in P. carotovorum (2), and biofilm formation in P. aeruginosa and Pseudomonas fluorescens (11, 56). Some of these functions are key virulence factors during the interaction between pathogenic bacteria and their hosts (8, 12, 57, 59). Thus, the disruption of QS represents a potential strategy to intervene in infections caused by these pathogens, and some recent studies have successfully revealed several means to inhibit AHL-mediated QS systems. For example, S-adenosylmethionine (SAM) and 5′-methylthioadenosine (MTA) analogs are effective inhibitors of AHL synthesis by RhlI of P. aeruginosa (42), and halogenated furanones from Delisea pulchra inhibit AHL-mediated gene expression by promoting the degradation of the transcriptional activator (32, 33, 47). Some natural products and chemicals, such as garlic extracts, 4-nitro-pyridine-N-oxide (4-NPO), patulin, penicillic acid, and N-acyl cyclopentylamides, also inhibit QS systems but by less-clear mechanisms (22, 36, 45, 46). Enzymes of bacterial origin capable of degrading of AHLs represent another mechanism for inhibiting quorum sensing (10, 16, 54). Although such enzymes have been found in a wide range of bacteria, they can be divided into two families: AHL-lactonases and AHL-acylases. Members of the AHL-lactonase family inactivate AHLs by hydrolyzing the lactone bond. Noted examples of this family include AiiA from Bacillus sp. (13), AhlD from Arthrobacter sp. (40), AttM from A. tumefaciens strain A6 (60), AiiB from A. tumefaciens C58 (6), and QsdA from Rhodococcus erythropolis strain W2 (54). On the other hand, AHL-acylases degrade AHLs by hydrolyzing the amide linkage. Enzymes of this family are represented by AiiD from Ralstonia sp. strain XJ12B (30), PvdQ from P. aeruginosa PAO1 (21), AhlM from Streptomyces sp. (41), and an unknown protein from Comamonas sp. strain D1 (53). Finally, Leadbetter and Greenberg (29) previously reported that a strain of Variovorax paradoxus (VAI-C) is capable of using AHLs as the sole nutrient source. The presence of homoserine lactones in AHL metabolic products by V. paradoxus VAI-C suggests that the bacterium produces an AHL-acylase, but the gene coding for this enzyme remains unknown.
The bacterium Ochrobactrum sp. strain A44 was previously reported to be capable of inactivating various synthetic AHL molecules and AHL produced by P. carotovorum subsp. carotovorum (23), although the gene and related mechanism responsible for degrading AHL were unknown. In this paper, we report the identification and characterization of a novel AHL-lactonase from the Gram-negative bacterium Ochrobactrum sp. strain T63 and demonstrate its quorum-quenching activity in two plant-associated bacteria.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Ochrobactrum sp. strain T63 was isolated from a soil sample collected in Yunnan Province, China. Pectobacterium carotovorum strain Z3-3 (laboratory stock), Pseudomonas fluorescens 2P24 (56), and Agrobacterium tumefaciens NTL4(pZLR4) (7) were grown in Luria-Bertani (LB) medium or ABM minimal medium (9) at 28°C. Ochrobactrum sp. strain T63 was similarly cultured. Unless otherwise specified, Escherichia coli strains were grown at 37°C in LB medium. When necessary, antibiotics were added at the following concentrations: ampicillin at 50 μg/ml, kanamycin at 50 μg/ml, gentamicin at 30 μg/ml, tetracycline at 20 μg/ml, and chloramphenicol at 20 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Ochrobactrum sp. | ||
| T63 | Apr; wild type | This study |
| T63ΔAidH | Apr; aidH deletion mutant | This study |
| T63-AidH | Apr; T63ΔAidH harboring pB8C-1 | This study |
| Agrobacterium tumefaciens NTL4(pZLR4) | Gmr; A. tumefaciens NT1 derivative harboring a traG::lacZ reporter fusion | 7 |
| Escherichia coli | ||
| DH5α | φ80 lacZΔM15 Δ(lacZYA-argF)U169 hsdR17 recA1 endA1 thi-1 | 18 |
| BL21(DE3) | Expression strain | Novagen |
| Pectobacterium carotovorum subsp. carotovorum | ||
| Z3-3 | Wild type | Laboratory stock |
| Z3-3(pBBR1MCS-2) | Kmr; strain Z3-3 harboring pBBR1MCS-2 | This study |
| Z3-3(pB8C-1) | Kmr; strain Z3-3 harboring pB8C-1 | This study |
| Pseudomonas fluorescens | ||
| 2P24 | Apr; wild type | 56 |
| 2P24(pRK415) | Apr Tcr; strain 2P24 harboring pRK415 | This study |
| 2P24(pR8C-1) | Apr Tcr; strain 2P24 harboring pR8C-1 | This study |
| Plasmids | ||
| pBluescript II SK(+) | Apr; ColE1 origin | Stratagene |
| p8C-1 | Apr; pBluescript harboring a 2.3-kb HindIII fragment with the aidH gene | This study |
| pBBR1MCS-2 | Kmr; Escherichia-Pseudomonas shuttle vector | 27 |
| pB8C-1 | Kmr; pBBR1MCS-2 harboring a 2.3-kb HindIII fragment with the aidH gene | This study |
| pSR47S | Kmr; mobRP4 oriR6K sacB | 1 |
| p47SΔAidH | Kmr; suicide plasmid pSR47S carrying a deleted aidH gene | This study |
| pLAFR5 | Tcr; oriV cosmid | 25 |
| pET-22b(+) | Apr; expression vector | Novagen |
| pET-AidH | Apr; pET-22b(+) carrying the aidH gene | This study |
| pHSG399 | Cmr; ColE1 origin | TaKaRa |
| p399-AidH | Cmr; pHSG399 carrying the aidH gene | This study |
| pRK415 | Tcr; IncP1 replicon, polylinker of pUC19; Mob+ | 25 |
| pR8C-1 | Tcr; pRK415 harboring a 2.3-kb HindIII fragment with the aidH gene | This study |
Apr, Cmr, Gmr, Kmr, and Tcr indicate resistance to ampicillin, chloramphenicol, gentamicin, kanamycin, and tetracycline, respectively.
Screening of AHL-degrading bacteria.
To isolate bacterial strains capable of inactivating AHLs, we collected soil samples from different geographical locations of China. For each sample, we suspended 1 g of soil sample in sterile water (10 ml) and spread serially diluted solutions onto ABM medium. After incubating the plates at 28°C for 1 to 2 days, colonies that appeared on the plates were struck to obtain single isolated colonies, which were then cultivated in 2-ml tubes at 28°C for 20 h in 270 μl LB broth with gentle shaking. N-(3-Oxooctanoyl)-l-homoserine lactone (OOHL) was then added to the bacterial cultures at a final concentration of 10 nM. After a further 2-h incubation, the OOHL present in the bacterial cultures was evaluated by spotting 2 μl of supernatant onto ABM medium seeded with A. tumefaciens strain NTL4(pZLR4) (7) and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The autoinducer detection plates were incubated at 28°C for 16 h, and the activity of OOHL was determined by the formation of a blue zone around the sample. Bacterial strains capable of significantly reducing the activity of OOHL within 2 h were retained for further study.
Identification of bacterial strain T63.
We identified bacterial strains exhibiting the phenotypes of inactivating OOHL by analyzing their 16S rRNA gene sequences. For Ochrobactrum sp. strain T63, described in this study, we amplified its 16S rRNA genes by PCR with primers 63SF (5′-TGTCGACAGGCCTAACACATGCAAGTC-3′) and 1494SR (5′-TGTCGACGGYTACCTTGTTACGACTT-3′) (the SalI sites are underlined) (34, 38). After cloning into pBluescript II SK(+) (Stratagene), the PCR products were sequenced (SinoGenoMax Company Limited) and analyzed by using the BLAST programs of the National Center for Biotechnology Information (NCBI).
Cloning and sequencing of the DNA fragment harboring the aidH gene.
To clone the gene responsible for the AHL-inactivating function in strain T63, we first constructed a cosmid library that represents its genome with the broad-host-range vector pLAFR5 using a previously described method (25). Escherichia coli strains harboring individual cosmid clones were screened for the AHL-inactivating property by using the A. tumefaciens reporter system with a procedure similar to the one employed to identify strain T63. Subcloning was carried out with the vector pBluescript II SK(+) by standard molecular techniques (48).
Construction of an aidH in-frame deletion mutant of strain T63.
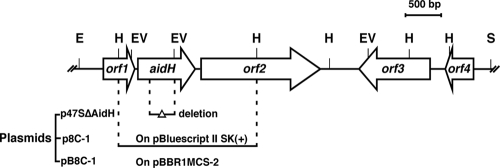
To create an aidH gene deletion mutant of strain T63, two fragments flanking the aidH gene were amplified by PCR. One was generated by primers A01 (5′-CAGGAATTCCGCACCGATCCG-3′) and A1065 (5′-ATGGGATCCATCGAATAGCTGCGGTCG-3′) (EcoRI and BamHI sites are underlined), and the second was created with primers A1307 (5′-ATGGATCCTACGCTCGCAGCACCTG-3′) and A2354 (5′-ATGAGCTCAGACCCATTGAGAATGTC-3′) (BamHI and SacI sites are underlined). After being digested with the relevant restriction enzymes, the two DNA fragments were ligated into suicide plasmid pSR47S (1) digested with EcoRI and SacI to generate p47SΔAidH. The deletion mutant T63ΔAidH was created with p47SΔAidH according to a standard procedure (31) (for detailed procedures, see the supplemental material). To construct a complementation plasmid, a 2.3-kb HindIII fragment harboring the aidH gene was inserted into vector pBBR1MCS-2 (27) to create pB8C-1 (Fig. 1).
FIG. 1.
Physical map of the aidH gene locus. The single-headed arrows represent the locations and orientations of the genes in the region of the Ochrobactrum sp. T63 chromosome that carries the aidH gene. A 2.3-kb HindIII fragment encoding the AHL inactivation function was inserted into pBluescript II SK(+) and pBBR1MCS-2 to create plasmids p8C-1 and pB8C-1, respectively. The construction of vector p47SΔAidH for deleting the aidH gene was described in Materials and Methods. Abbreviations: E, EcoRI; H, HindIII; EV, EcoRV; S, SalI.
Expression and purification of the AidH protein.
The predicted open reading frame (ORF) of the aidH gene was amplified by PCR with primers AidH-F (5′-GACCATATGACAATCAATTATCACGAACTTG-3′) and AidH-R (5′-GAGCTCGAGTTGTGTGCAATCGCGGATAAAG-3′) (NdeI and XhoI sites are underlined). The amplified DNA fragment was digested with NdeI and XhoI and was inserted into similarly digested pET-22b(+) (Novagen) to give pET-AidH for the production of His6-AidH. Protein expression was performed with E. coli BL21(DE3) (Novagen). To induce protein expression, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to E. coli cultures grown to an optical density at 600 nm (OD600) of 0.6 at a final concentration of 0.8 mM. The induction was allowed to proceed for 8 h at 28°C. His6-AidH was purified by using Ni2+-nitrilotriacetic acid (NTA) superflow according to the manufacturer's instructions (Amersham). After elution with 200 mM imidazole, fractions containing active His6-AidH were pooled and dialyzed in a Tris-Cl buffer (50 mM Tris-Cl and 400 mM NaCl [pH 7.0]), and the purity of the protein was assessed by SDS-PAGE.
Site-directed mutagenesis.
To introduce specific mutations into the aidH gene, we first cloned the predicted ORF into pHSG399 (TaKaRa) to give p399-AidH, which was used to mutate the specific amino acids according to the protocol of the TaKaRa MutanBEST system. Each mutation was introduced into aidH on p399-AidH by PCR using a pair of primers that contain the desired nucleotide changes (Table 2). The PCR products were phosphorylated and ligated with T4 DNA ligase (TaKaRa). All mutations were verified by double-strand DNA sequencing. To express mutant proteins, each mutated allele was inserted into the expression vector pET-22b(+) in a manner similar to that for the wild-type gene.
TABLE 2.
Primers for site-directed mutagenesis
| Primera | Amino acid substitution | Sequenceb |
|---|---|---|
| G100V-F | Gly100Val | 5′-GTCTGGTCGCTCGGCGGACAT-3′ |
| G100V-R | 5′-GAAAACCACCGCATCGGCAAT-3′ | |
| S102V-F | Ser102Val | 5′-GTGCTCGGCGGACATATCGGCATC-3′ |
| S102V-R | 5′-CCAGCCGAAAACCACCGCATC-3′ | |
| G104V-F | Gly104Val | 5′-GTCGGACATATCGGCATCGAG-3′ |
| G104V-R | 5′-GAGCGACCAGCCGAAAACCAC-3′ | |
| E219T-F | Glu219Thr | 5′-ACGCCTTTTGTTGAACTCGATTTC-3′ |
| E219T-R | 5′-GTCACGGCCATTGACGACCGC-3′ | |
| H248S-F | His248Ser | 5′-TCTGCGCCATTCCGTGAAGCA-3′ |
| H248S-R | 5′-ACCTGCATTGTCGATAACGTG-3′ |
F, forward primer; R, reverse primer.
The bases changed are shown in boldface type in each primer.
Determining the mechanism of AidH catalysis.
To determine the chemical structures of products from the reaction between AidH and AHLs, N-hexanoyl-l-homoserine lactone (HHL) was subjected to digestion by AidH, and the resulting products were analyzed by using high-performance liquid chromatography (HPLC) and electrospray ionization (ESI)-mass spectrometry (MS). We mixed purified AidH (50 μg) with HHL (1 μmol) in 1 ml of reaction buffer (50 mM Tris-Cl and 400 mM NaCl [pH 7.0]). After a 30-min incubation at 37°C, the mixture was extracted three times with ethyl acetate, and the combined organic phase was evaporated in a rotary evaporator. For HPLC analysis, the sample was redissolved in 0.1 ml methanol and analyzed by using a symmetry C18 reverse-phase column (4.6 by 150 mm) (Agilent TC-18). Fractions were separated by elution isocratically with 40:60 methanol-water (vol/vol) at a flow rate of 0.25 ml min−1. ESI-MS was performed by using an LCMS2010 instrument at the Institute of Chemistry, Chinese Academy of Sciences. Samples were dissolved in methanol and ionized by negative-ion electrospray. N-Hexanoyl-l-homoserine, the lactonolysis product of HHL, was prepared by using a method described previously (14). HHL (0.2 μmol) was incubated in a solvent containing 200 μl dimethyl sulfoxide and 200 μl NaOH (1 M) for 30 min at 37°C. The mixture was then adjusted to pH 6 with H3PO4 and was extracted three times with ethyl acetate. The combined organic fractions were evaporated to dryness, and the product was purified by HPLC under the conditions described above.
Extraction and detection of AHLs.
To evaluate AHL production, strains of P. fluorescens and P. carotovorum were grown in LB broth overnight at 28°C, 0.5 ml of culture sampled at different time points was extracted with an equal volume of ethyl acetate, and the mixture was then brought to dryness by vacuum evaporation and was resuspended in 50 μl methanol. One microliter of the sample was cocultured with 0.3 ml of the AHL biosensor A. tumefaciens NTL4(pZLR4) (OD600 of 0.8) at 28°C for 3 h, and the β-galactosidase expressed by biosensor cells was determined by using the Miller method (35). The thin-layer chromatography (TLC) assays were performed according to a previously described method (50). Briefly, 2 μl of methanol samples was applied to C18 reversed-phase TLC plates (catalog no. 1.15389; Merck, Germany) and air dried. The TLC plates were developed by using 60:40 (vol/vol) methanol-water as the mobile phase. The detection of AHLs was performed by overlaying the TLC plate with a 3-mm thin film of 0.6% (wt/vol) ABM agar (100 ml) containing 5 ml of exponentially grown A. tumefaciens NTL4(pZLR4) and X-Gal (40 μg/ml). The overlaid TLC plates were incubated at 28°C for 24 h. AHL activities were determined by the appearance of blue spots on the plate.
Biofilm formation by P. fluorescens 2P24 and virulence of P. carotovorum subsp. carotovorum.
For biofilm formation, P. fluorescens 2P24 and its derivatives harboring plasmid pR8C-1 or the empty vector (pRK415) were used according to a previously described procedure (56). P. carotovorum subsp. carotovorum strain Z3-3 and its derivatives carrying pB8C-1 or the vector (pBBR1MCS-2) were used to infect three different plants by using an established method (13, 14). Briefly, actively growing bacteria were collected by centrifugation and were resuspended in fresh LB broth to an OD600 of 1.0 (∼2 × 109 CFU/ml). To infect plants, 5 μl of bacterial suspension was added to a cut surface or a wound site that had been treated with 70% ethanol. For potato and radish tissues, the inoculated samples were incubated at 28°C for 48 h in petri dishes moisturized with wet filter papers. For experiments to determine the virulence of P. carotovorum subsp. carotovorum strains on Chinese cabbage, the inoculated plants or detached leaves were incubated at 28°C. The maceration area was recorded 48 h after inoculation, and images of the samples were taken after 5 days.
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA gene and the aidH gene from strain T63 have been deposited in the GenBank database under accession no. GQ849009 and GQ849010, respectively.
RESULTS
Isolation and identification of an Ochrobactrum isolate capable of degrading AHLs.
By using the sensitive AHL reporting system from A. tumefaciens (7), we initiated a project to identify bacterial isolates capable of inactivating AHLs from soil samples obtained from different locations in China. After screening about 2,000 independent isolates, we obtained 37 strains that exhibited different levels of AHL inactivation activity. Among these, isolate T63, which was able to completely eliminate AHL activity in the assay within 1 h, was chosen for further study. Subsequent analyses indicated that strain T63 is a Gram-negative bacterium that forms yellow colonies on an LB plate (data not shown). Further characterization revealed that the 16S rRNA gene of strain T63 is 97% identical to those of Ochrobactrum sp. strain BH3, Ochrobactrum intermedium, Ochrobactrum anthropi, Ochrobactrum sp. strain TK14, and Ochrobactrum sp. strain bmh-1 (data not shown); it was therefore designated Ochrobactrum sp. T63.
Cloning and characterization of the aidH gene from strain T63.
To identify the gene encoding the AHL inactivation activity from strain T63, we constructed a cosmid library of its genome in E. coli. The resulting cosmid clones were screened for AHL-inactivating activity, and seven clones exhibiting such activity were obtained from 670 candidates examined. Restriction mapping analyses led to the identification of a ∼6-kbp SalI-EcoRI fragment that is shared among seven clones. By subcloning analysis, we further localized the AHL-degrading activity to a 2.3-kb HindIII fragment (Fig. 1). Sequencing analyses revealed that the 6-kb DNA fragment contains five significant open reading frames (ORFs) (Fig. 1). The first complete ORF (orf1) is predicted to encode a peptidyl-tRNA hydrolase domain protein of 145 amino acids. The second complete ORF (aidH) is predicted to encode a protein of 271 amino acids related to members of the alpha/beta-hydrolase fold family. The third complete ORF (orf2) is predicted to encode a pyruvate dehydrogenase of 575 amino acids. The fourth complete ORF (orf3) is predicated to encode a pantothenate kinase of 337 amino acids. The last complete ORF (orf4) is predicted to encode a phosphoribosyl-ATP pyrophosphohydrolase of 107 amino acids. Consistent with the results from the subcloning analysis, the insertion of the ORF of the 2.3-kb HindIII fragment into the expression vector pET-22b(+) conferred AHL degradation activity in E. coli, indicating that this gene is responsible for this phenotype; it was thus referred to as AidH (autoinducers degrading hydrolase).
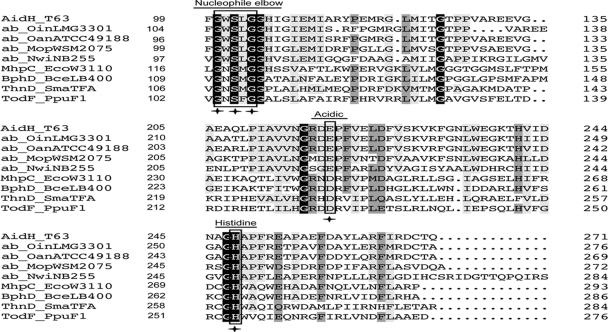
Sequence analysis revealed that AidH is a 271-amino-acid protein with a predicted molecular mass of 29.5 kDa. In agreement with this, purified His6-AidH migrated as a ∼30-kDa protein on SDS-PAGE gels (see Fig. S1 in the supplemental material). Whereas the predicted amino acid sequence of AidH has no significant similarity to any of the known AHL-inactivating enzymes, it exhibits 85% identity to members of the alpha/beta-hydrolase fold from Ochrobactrum anthropi ATCC 49188 (GenBank accession no. YP001369382) and Ochrobactrum intermedium LMG3301 (accession no. EEQ96967), 65% identity with the alpha/beta-hydrolase fold from Mesorhizobium opportunistum WSM2075 (accession no. EEW31886), and 44% identity with the alpha/beta-hydrolase fold from Nitrobacter winogradskyi Nb-255 (accession no. YP317111.1) (Fig. 2 and data not shown). Furthermore, AidH contains the “nucleophile-acid-histidine” catalytic triad that is conserved among members of alpha/beta-hydrolase family (20). The common “nucleophile elbow” element G-X-Nuc-X-G is also present in AidH (G-W-S102-Leu-G), in which the nucleophile residue is a serine, which, among Cys or Asp residues, is an essential component of the catalytic triad of alpha/beta-hydrolases (20, 39) (Fig. 2). Finally, AidH does not contain a detectable N-terminal hydrophobic signal peptide, suggesting that it is not a secreted protein. In agreement with this observation, a culture supernatant of strain T63 or E. coli expressing AidH was unable to inactivate AHLs (data not shown).
FIG. 2.
Comparison of amino acid sequences of AidH and several alpha/beta-hydrolases. The alignment was generated by DNAMAN. Sections from left to right are the protein, species, and number of amino acids for the gene before the sequences shown. Identities are highlighted in white with a black background, and similarities are shaded gray. The catalytic triad residues are boxed with rectangles. The amino acid residues essential for AHL-degrading activity are indicated by asterisks. AidH_T63, AidH of Ochrobactrum sp. T63 (GenBank accession no. GQ849010); ab_OinLMG3301, alpha/beta-hydrolase fold of O. intermedium (accession no. EEQ96967); ab_OanATCC49188, alpha/beta-hydrolase fold of O. anthropi ATCC 49188 (accession no. YP001369382); ab_MopWSM2075, alpha/beta-hydrolase fold of Mesorhizobium opportunistum WSM2075 (accession no. EEW31886); ab_NwiNB255, alpha/beta-hydrolase fold of Nitrobacter winogradskyi Nb-255 (accession no. YP317111); MhpC_EcoW3110, MhpC from Escherichia coli W3110 (accession no. D86239); BphD_BceLB400, BphD from Burkholderia cepacia LB400 (accession no. X66123); ThnD_SmaTFA, ThnD from Sphingomonas macrogoltabidus TFA (accession no. AF204963); TodF_PpuF1, TodF from Pseudomonas putida F1 (accession no. Y18245).
AidH is the sole gene in Ochrobactrum sp. T63 responsible for the AHL inactivation phenotype.
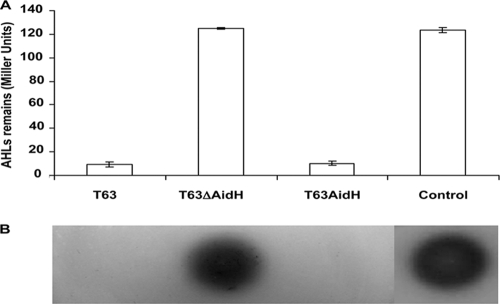
To determine whether any other gene in Ochrobactrum sp. T63 contributes to its AHL-inactivating function, we constructed an aidH gene in-frame deletion mutant. When mixed with OOHL signals, this strain, T63ΔAidH, is completely defective in the AHL-degrading activity. Moreover, the introduction of a plasmid expressing AidH (pB8C-1) restored AHL-inactivating function to the wild-type level (Fig. 3). These results indicated that under our experimental conditions, aidH is the only gene responsible for degrading AHL signals in Ochrobactrum sp. T63.
FIG. 3.
aidH is the sole Ochrobactrum sp. T63 gene involved in AHL-degrading activity. (A) N-(3-Oxooctanoyl)-l-homoserine lactone (OOHL) was incubated with the wild-type, the aidH deletion mutant, or the complementation strain. Culture supernatants were extracted, and the AHLs were detected by the biosensor A. tumefaciens NTL4(pZLR4) as described in Materials and Methods. Error bars denoting standard deviations from three experiments are shown. (B) Plate assay of samples described above (A). The extracts were spotted onto an ABM minimal medium plate seeded with an A. tumefaciens NTL4(pZLR4) culture and X-Gal (40 μg/ml) and incubated at 28°C for 16 h. The control was an OOHL standard (10 pmol).
AidH encodes an AHL-lactonase.
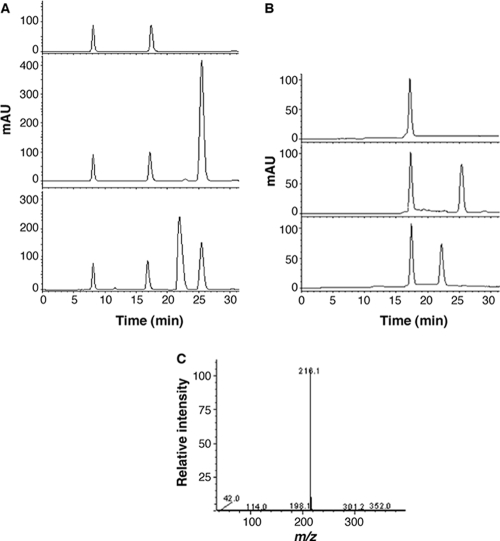
To determine the mechanism of action of AidH on AHLs, we treated N-hexanoyl-l-homoserine lactone (HHL) with purified AidH, and the resulting products were analyzed by reverse-phase HPLC and ESI-MS. The HPLC profile of HHL is characterized by a single peak with a retention time of 25.4 min, which has an (M + Na) ion at an m/z (mass-to-charge ratio) of 222.1. After being treated with AidH, a compound with a retention time of 22.1 min (Fig. 4 A) was detected. ESI-MS analysis of this product revealed an (M − H) ion at an m/z of 216.1 (Fig. 4C), indicating that the effect of AidH on HHL caused a mass increase of 18, which corresponds to a water molecule. These results suggest that the treatment of HHL with AidH leads to the cleavage of the homoserine lactone ring on the substrate, thus producing an N-hexanoyl-l-homoserine molecule. To prove this hypothesis, we produced N-hexanoyl-l-homoserine from HHL by lactonolysis and compared the resulting compound with the AidH-catalyzed product. Upon HPLC, these two samples exhibited an identical retention time of 22.1 min (Fig. 4B). Furthermore, when a mixture containing these two samples was analyzed by HPLC, a single product peak was observed, suggesting that these compounds are identical (data not shown). These results strongly suggested that aidH encodes an AHL-lactonase that hydrolyzes the ester bond of the homoserine lactone ring of AHLs.
FIG. 4.
HPLC and ESI-MS spectrometry analysis of the AidH-catalyzed HHL product. (A) HPLC elution profiles of the reaction buffer (top), reaction buffer containing N-hexanoyl-l-homoserine lactone (HHL) (middle), or AidH-digested HHL products in the reaction buffer (bottom). The HHL peak eluted at 25.4 min (middle); the product peak eluted at 22.1 min (bottom). mAU, milli-absorbance unit. (B) HPLC elution profiles of lactonolysis solution (top), reaction solution containing undigested HHL (middle), and the lactonolysis reaction mixture (bottom). Both the lactonolysis product and AidH enzymatically digested HHL appeared with a retention time of 22.1 min. (C) ESI-MS analysis of the hydrolysis product of HHL. The fraction at 22.1 min from HPLC was collected and analyzed by ESI-MS as described in Materials and Methods.
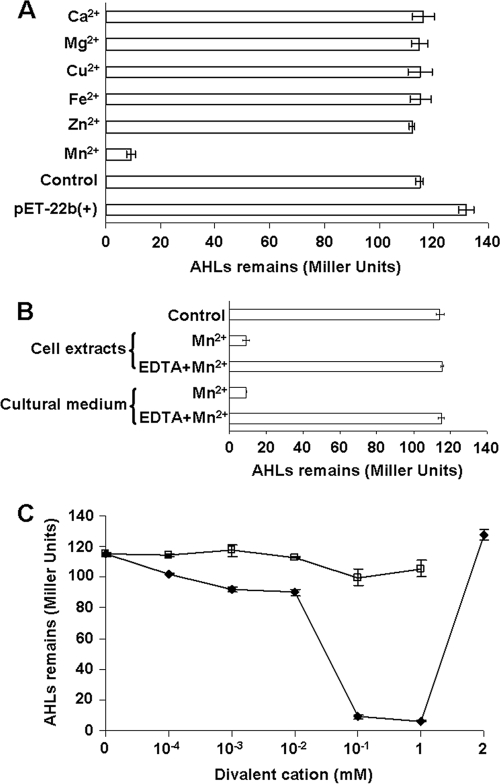
Manganese ions are important for the activity of AidH.
To determine whether AidH requires any cofactor for its enzymatic activity, we added several divalent ions to cultures expressing AidH, and proteins purified from these cultures were evaluated for their AHL-inactivating functions. These ions did not detectably influence the solubility of AidH (data not show). However, when assayed for enzymatic activity, AidH purified from bacteria grown in medium containing 1 mM Mn2+ showed the highest degrading activity, whereas no activity was observed for proteins purified from cultures incubated with several other divalent cations (Fig. 5 A). Consistently, the addition of the chelator EDTA to the cultures severely reduced AidH activity even in the presence of exogenous Mn2+ (Fig. 5B). Furthermore, 1 mM Mn2+ gave maximal enzymatic activity, indicating that the effect of Mn2+ is dose dependent (Fig. 5C).
FIG. 5.
The manganese(II) ion is important for the activity of AidH. (A) Effects of different divalent cations on the activity of AidH. E. coli BL21(DE3) cells carrying recombinant AidH were cultivated for 8 h after IPTG induction at 28°C. The indicated metal ions were added to the medium to 1 mM. E. coli BL21(DE3) carrying the vector pET-22b(+) or pET-AidH cultivated without exogenous metal ions was used as a control. Crude cell extracts were incubated with N-(3-oxooctanoyl)-l-homoserine lactone (OOHL) (final concentration, 100 nM) for 1 h at 37°C, extracted with ethyl acetate, and evaporated to dryness. The sample was dissolved in methanol, and the activity of AHLs was measured by using the biosensor A. tumefaciens strain NTL4(pZLR4). (B) EDTA abolishes AidH activity in the presence of Mn2+. Mn2+ and EDTA added to medium or in cell extracts were at 1 mM and 5 mM, respectively. Control indicates lysate from cells not expressing AidH. AHL activity was measured as described above (A). (C) Effects of different concentrations of Mn2+ and Zn2+ on the activity of AidH. Mn2+ (♦) or Zn2+ (□) at the indicated concentration was added to E. coli cultures expressing AidH, and the AHL-degrading activity of the cell extracts was evaluated with the A. tumefaciens biosensor. Error bars indicate standard deviations determined from three independent experiments.
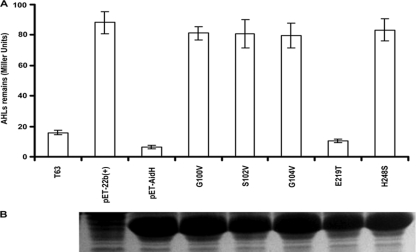
The conserved Gly-X-Ser-X-Gly motif and the histidine residue in the catalytic triad are essential for AidH activity.
The nucleophile-His-acid motif conserved among members of alpha/beta-hydrolase family constitutes the catalytic triad that acts on different substrates in various biological contexts (20). An alignment of AidH with other alpha/beta-hydrolase family proteins revealed the presence of a G100-X-S102-X-G104 motif and the Glu and His residues (E219 and H248) that are highly conserved in the “nucleophile-acid-histidine” catalytic triad (Fig. 2). We determined the importance of these motifs in AidH activity by replacing G100, S102, or G104 with a Val residue and H248 with a Ser residue. To determine whether every conserved residue is important for AidH activity, we also replaced E219 with a Thr residue. When analyzed for the AHL-inactivating function, with the exception of the E219T mutant, which is still active at wild-type levels, all other mutants had completely lost the enzymatic activity (Fig. 6 A). The loss of activity is not due to changes in other biochemical properties like the solubility of the proteins because they can be expressed and purified in a manner indistinguishable from that of wild-type AidH (Fig. 6B). These results indicate that the nucleophile and His248, but not the Glu219 residue, are required for the AHL-degrading activity of AidH.
FIG. 6.
Residues in the predicted catalytic triad are important for the enzymatic activity of AidH. (A) Mutations in G100, S102, G104, or H248 abolished AidH activity. The crude cell extracts of bacterial strains expressing the indicated AidH mutants were incubated with 100 nM N-(3-oxooctanoyl)-l-homoserine lactone (OOHL) for 1 h at 37°C and extracted with ethyl acetate, and AHL activity was detected by the A. tumefaciens biosensor. For samples, T63 indicates wild-type Ochrobactrum strain T63, pET-22b(+) indicates E. coli BL21(DE3) carrying pET-22b(+), and pET-AidH indicates E. coli BL21(pET-AidH). G100V, S102V, G104V, E219T, and H248S are substitution mutants of AidH in the indicated amino acids. The experiment was repeated three times, and data shown are means of three replicates. (B) Substitution mutants of AidH code for stable proteins. Total cellular proteins of IPTG-induced E. coli strains harboring each mutant on pET-22b(+) were analyzed by SDS-PAGE.
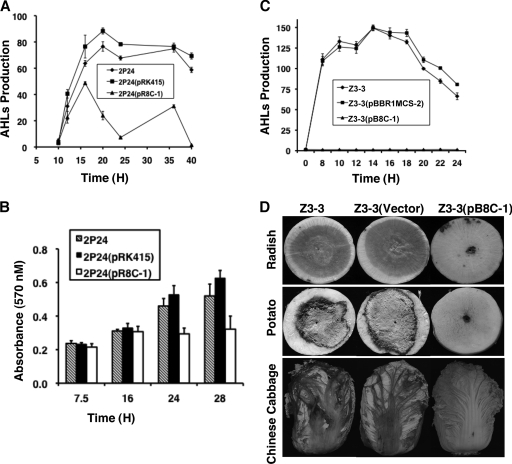
AidH interferes with QS-mediated functions in P. fluorescens 2P24 and P. carotovorum subsp. carotovorum.
To determine whether the activity of AidH interferes with biological processes controlled by AHL-mediated quorum sensing, we tested its effect on two independent systems. Thus, we first introduced pR8C-1 (Table 1) into biocontrol strain P. fluorescens 2P24 (56). The expression of aidH from this plasmid did not detectably affect the growth of this bacterial strain (data not shown). Importantly, compared to the strain that harbors the vector, the quorum-sensing signals in culture supernatants of recombinant strain 2P24(pR8C-1) decreased dramatically 16 h after incubation (Fig. 7 A). Consistent with this observation, biofilm formation by P. fluorescens 2P24, a phenotype positively regulated by the PcoR/PcoI QS system (56), was significantly affected 24 h after inoculation, and such a defect cannot be restored by extended incubation of the testing strains (Fig. 7B).
FIG. 7.
Effect of AidH on phenotypes of Pseudomonas fluorescens 2P24 and P. carotovorum subsp. carotovorum. Vectors or their derivatives expressing AidH were introduced into Pseudomonas fluorescens 2P24 and P. carotovorum subsp. carotovorum, and the resulting strains were tested for extracellular AHL accumulation (A and C), biofilm formation (B), and pathogenicity (D). AHLs produced by bacterial strains were evaluated for their abilities to activate traG on A. tumefaciens biosensor strain NTL4(pZLR4), measured by the expression of a TraR-dependent LacZ fusion. The activity of β-galactosidase is expressed in Miller units. The formation of biofilm in Eppendorf tubes was evaluated by crystal violet staining as described previously by Wei and Zhang (56). Error bars indicate standard deviations of data from three experiments. (D) Radish, potato, and Chinese cabbage were inoculated with 5 μl of bacterial culture (2 × 109 CFU/ml) of Z3-3, Z3-3(pBBR1MCS-2) (vector), and Z3-3(pB8C-1), respectively. The development of disease symptoms was documented by photographing the inoculated plant tissues 48 h (radish and potato) or 5 days (Chinese cabbage) after inoculation. Similar results were obtained in multiple independent experiments, and images shown are representative of one experiment.
We further examined the ability of AidH to reduce the quorum-sensing-controlled virulence of the plant bacterial pathogen P. carotovorum, which causes soft rot diseases on a variety of hosts (3, 4). We cloned the aidH gene into the broad-host-range vector pBBR1MCS-2 (27), and the resulting plasmid, pB8C-1, was introduced into P. carotovorum strain Z3-3. The expression of this enzyme did not affect the growth rate of P. carotovorum (data not shown). However, whereas the AHL production by the wild-type strain or its derivative containing the vector peaked 14 h after incubation, no AHL was detected in the culture supernatant of strain Z3-3(pB8C-1) during the entire growth period (Fig. 7C). Consistently, compared to its parental strain Z3-3 or Z3-3(pBBR1MCS-2), which caused severe tissue-macerating symptoms on detached tissues of potato, radish, or Chinese cabbage, Z3-3(pB8C-1) failed to cause soft rot disease symptoms on these hosts (Table 3 and Fig. 7D). Therefore, AidH is able to function as an AHL-degrading enzyme not only in Ochrobactrum sp. but also in species of other bacterial genera, including P. fluorescens and P. carotovorum. These results also demonstrate that AidH is capable of effectively quenching QS-dependent functions in these bacteria by degrading AHLs.
TABLE 3.
Virulence assay of P. carotovorum subsp. carotovorum strains on plant tissuesa
| Plant | Tissue | Inoculum concn (CFU/ml) | Mean maceration area (cm2) ± SD |
||
|---|---|---|---|---|---|
| Z3-3 | Z3-3 (pBBR1MCS-2) | Z3-3 (pB8C-1) | |||
| Potato | Tuber | 2 × 109 | 7.6 ± 1.6 | 13.5 ± 1.5 | 0 |
| 2 × 108 | 11.8 ± 6.6 | 9.8 ± 2.9 | 0 | ||
| Chinese cabbage | Leaf | 2 × 109 | 29.7 ± 9.6 | 23.0 ± 5.9 | 5.0 ± 2.0 |
| 2 × 108 | 19.1 ± 4.1 | 22.3 ± 8.5 | 0 | ||
| Radish | Root | 2 × 109 | 15.9 ± 1.4 | 14.4 ± 1.5 | 0 |
| 2 × 108 | 11.2 ± 4.0 | 10.4 ± 5.0 | 0 | ||
The maceration areas were measured 48 h after inoculation; data are means of data from three replicates.
DISCUSSION
Strategies aimed at effective interference with the activity of autoinducers hold great potential for controlling infections and other harmful biological processes regulated by quorum sensing. Among these strategies, the enzyme-mediated degradation of autoinducers has been proven to be useful in controlling infections of plants (16). Here, we describe AidH, a novel AHL-lactonase from a strain of Ochrobactrum isolated from Yunnan Province, China, an area known for its biological diversity (58). The identification of AidH adds to a growing list of enzymes that hydrolyze the ester bond of the homoserine lactone ring of AHLs. Based on sequence similarity, these enzymes can be divided into two groups. The first group contains a sole member, the QsdA lactonase from R. erythropolis strain W2. This protein is a member of the phosphotriesterase (PTE) family of zinc-dependent metalloproteins (10, 54). The second group includes all the other known lactonases: AiiA from Bacillus sp. strain 240B1 (13), AhlD from Arthrobacter sp. strain IBN110 (40), AttM from A. tumefaciens A6 (60), and AiiB from A. tumefaciens C58 (6). Phylogenetic analyses further divided these proteins into two clusters. The AiiA-like cluster, consisting of all the AHL-lactonases from Bacillus species, shares more than 90% identity at the amino acid level, and the AttM-like cluster includes the enzymes AttM and AiiB from A. tumefaciens (6, 60) and AhlD from Arthrobacter sp. IBN110 (40). Members of this cluster share only 30 to 58% similarity in peptide sequence and less than 25% identity with the AiiA-like cluster members (16), but all four enzymes contain a highly conserved motif, HXDH-H-D, which is essential for AHL-degrading activity (13, 15, 40, 55).
Our HPLC and MS analyses demonstrated that AidH is an AHL-lactonase that hydrolyzes the lactone ring of AHLs to produce acylhomoserine (Fig. 4). However, AidH has no detectable homology with any of the known AHL-degrading proteins. Instead, this protein is highly similar to members of the alpha/beta-hydrolase fold family, particularly the alpha/beta-hydrolase fold of Ochrobactrum anthropi ATCC 49188 (Fig. 2). Alpha/beta-hydrolases are a large group of structurally related enzymes with diverse catalytic functions (20). However, these enzymes neither share sequence similarity nor act on similar substrates (20). Given the highly diverse activities of these enzymes, it is difficult to predict whether the AidH homologs from species of Mesorhizobium and Ochrobactrum are capable of inactivating AHLs. The two features shared by members of this fold family are a nucleophile-acid-histidine catalytic triad and a nucleophile elbow with a sequence of Gly-X-Nuc-X-Gly (19, 20, 37). Because of the high level of similarity between AidH and members of the alpha/beta-hydrolase fold family and the fact that mutations in the catalytic triad or the nucleophile elbow abolished its enzymatic activity (Fig. 5), AidH clearly is a member of the alpha/beta-hydrolase family. This is the first report showing that a member of the alpha/beta-hydrolase fold family is capable of cleaving the lactone bond of acylhomoserine lactone.
AidH displays strong hydrolyzing activity against all tested AHLs, including N-butanoyl-l-homoserine lactone (BHL), N-hexanoyl-l-homoserine lactone (HHL), N-decanoyl-l-homoserine lactone (DHL), N-(3-oxohexanoyl)-l-homoserine lactone (OHHL), N-(3-oxooctanoyl)-l-homoserine lactone (OOHL), and 3-hydroxy-acylhomoserine lactones produced by P. fluorescens 2-79 (26) (data not shown). These AHL signals differ in the lengths and the natures of the substitutions at the C3 position of the acyl side chain. It is consistent with a previous report by Wang et al. in which the AHL-lactonase AiiA demonstrated strong catalytic activity against all 10 AHL signal molecules (55), and AidH did not show substrate specificity on AHL-type molecules.
Although alpha/beta-hydrolases are not known to require cofactors for their activity (20, 28), our experiments revealed that the lactonase activity of AidH relies greatly on Mn2+ (Fig. 5). Although bioinformatic analyses did not reveal any potential metal ion-binding site on AidH, our recent structural study of AidH found that the shapes of AidH crystals are different under conditions with or without exogenous Mn2+ (X. X. Yan et al., unpublished results), suggesting that Mn2+ induces a conformational shift in the structure of AidH, thus influencing its enzymatic activity. These observations indicate that Mn2+ plays an important role in the activity of AidH. However, the mechanisms underlying how Mn2+ affects its activity and how Mn2+ interacts with AidH remained to be investigated.
The expression of AHL-degrading enzymes in plant pathogens whose virulence was regulated by AHL-mediated quorum sensing often leads to a significant reduction of their virulence (13, 14). In P. carotovorum subsp. carotovorum, the production and secretion of exoenzymes essential for its virulence are dependent upon an AHL-mediated quorum sensing (4, 24, 44). The introduction of aidH into Z3-3 led to the abolishment of AHL production and attenuated soft rot disease symptoms on all plants tested, including potato, Chinese cabbage, and radish (Table 3 and Fig. 7D), indicating the potential use of the aidH gene in the prevention of plant diseases caused by phytopathogens whose virulences are controlled by AHLs. Similarly, the expression of AidH significantly reduces the accumulation of QS signals in P. fluorescens strain 2P24, leading to a decrease in biofilm formation, a trait positively regulated by QS (Fig. 7A and B). Interestingly, whereas AidH abolishes AHL production in Z3-3, AHLs are still detectable in strain 2P24 expressing this enzyme from several vectors with different copy numbers and promoter activities (Fig. 7A and C). Although AidH did not display substrate specificity on AHL signals in vitro, it is possible that its AHL-degrading activity is influenced by the inner environment of different bacterial cells. Nevertheless, the identification of AidH has added another tool to control harmful processes regulated by AHL-mediated quorum sensing.
Supplementary Material
Acknowledgments
This work was funded by the National Programs for High Technology Research and Development of China (grant 2006AA10A211), the National Natural Science Foundation of China (grant 30871666), and the Chinese MOST-DEST Cooperation Project (grant 2007DFA31570).
Footnotes
Published ahead of print on 4 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andrews, H. L., J. P. Vogel, and R. R. Isberg. 1998. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bainton, N. J., P. Stead, S. R. Chhabra, B. W. Bycroft, G. P. C. Salmond, G. S. A. B. Stewart, and P. Williams. 1992. N-(3-Oxohexanoyl)-L-homoserine lactone regulates carbapenem antibiotic production in Erwinia carotovora. Biochem. J. 288:977-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard, A. M. L., S. D. Bowden, T. Burr, S. J. Coulthurst, R. E. Monson, and G. P. C. Salmond. 2007. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Philos. Trans. R. Soc. B 362:1165-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burr, T., A. M. L. Barnard, M. J. Corbett, C. L. Pemberton, N. J. L. Simpson, and G. P. C. Salmond. 2006. Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol. Microbiol. 59:113-125. [DOI] [PubMed] [Google Scholar]
- 5.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 311:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlier, A., S. Uroz, B. Smadja, R. Fray, X. Latour, Y. Dessaux, and D. Faure. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactone activity. Appl. Environ. Microbiol. 69:4989-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by Gram-negative plant-associated bacteria. Mol. Plant Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, A., Y. Cui, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl. Environ. Microbiol. 61:1959-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chilton, M. D., T. C. Currier, S. K. Farrand, A. J. Bendich, M. P. Gordon, and E. W. Nester. 1974. Agrobacterium tumefaciens and PS8 bacteriophage DNA not found in crown gall tumors. Proc. Natl. Acad. Sci. U. S. A. 71:3672-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czajkowski, R., and S. Jafra. 2009. Quenching of acyl homoserine lactone-dependent quorum sensing by enzymatic disruption of signal molecules. Acta Biochim. Pol. 56:1-16. [PubMed] [Google Scholar]
- 11.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 12.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. U. S. A. 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong, Y. H., L. H. Wang, J. L. Xu, H. B. Zhang, X. F. Zhang, and L. H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 15.Dong, Y. H., A. R. Guste, Q. Zhang, J. L. Xu, and L. H. Zhang. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68:1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong, Y. H., and L. H. Zhang. 2005. Quorum sensing and quorum-quenching enzymes. Microbiology 43:101-109. [PubMed] [Google Scholar]
- 17.Eberl, L., M. K. Winson, C. Sternberg, G. S. A. B. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-L-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Heikinheimo, P., A. Glodman, C. Jeffries, and D. L. Ollis. 1999. Of barn owls and bankers: a lush variety of α/β hydrolases. Structure 7:R141-R146. [DOI] [PubMed] [Google Scholar]
- 20.Holmquist, M. 2000. Alpha/beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr. Protein Pept. Sci. 1:209-235. [DOI] [PubMed] [Google Scholar]
- 21.Huang, J. J., J. I. Han, L. H. Zhang, and J. R. Leadbetter. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 69:5941-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishida, T., T. Ikeda, N. Takiguchi, A. Kuroda, H. Ohtake, and J. Kato. 2007. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl. Environ. Microbiol. 73:3183-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jafra, S., J. Przysowa, R. Czajkowski, A. Michta, P. Garbeva, and J. M. Van der Wolf. 2006. Detection and characterization of bacteria from the potato rhizosphere degrading N-acyl-homoserine lactone. Can. J. Microbiol. 52:1006-1015. [DOI] [PubMed] [Google Scholar]
- 24.Jones, S., B. Yu, N. J. Bainton, M. Birdsall, B. W. Bycroft, S. R. Chhabra, A. J. Cox, P. Golby, P. J. Reeves, S. Stephens, et al. 1993. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 12:2477-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 26.Khan, S. R., D. V. Mavrodi, G. J. Jog, H. Suga, L. S. Thomashow, and S. K. Farrand. 2005. Activation of the phz operon of Pseudomonas fluorescens 2-79 requires the LuxR homolog PhzR, N-(3-OH-hexanoyl)-l-homoserine lactone produced by the LuxI homolog PhzI, and a cis-acting phz box. J. Bacteriol. 187:6517-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 28.Lam, W. W. Y., and T. D. H. Bugg. 1997. Purification, characterization, and stereochemical analysis of a C-C hydrolases: 2-hydroxy-6-keto-nona-2,4-diene-1,9-dioic acid 5,6-hydrolase. Biochemistry 36:12242-12251. [DOI] [PubMed] [Google Scholar]
- 29.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, Y. H., J. L. Xu, L. H. Wang, S. L. Ong, J. R. Leadbetter, and L. H. Zhang. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47:849-860. [DOI] [PubMed] [Google Scholar]
- 31.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manefield, M., R. de Nys, N. Kumar, R. Read, M. Givskov, P. Steinberg, and S. Kjelleberg. 1999. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145:283-291. [DOI] [PubMed] [Google Scholar]
- 33.Manefield, M., T. B. Rasmussen, M. Henzter, J. B. Andersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2002. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148:1119-1127. [DOI] [PubMed] [Google Scholar]
- 34.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Morohoshi, T., T. Shiono, K. Takidouchi, M. Kato, N. Kato, J. Kato, and T. Ikeda. 2007. Inhibition of quorum sensing in Serratia marcescens AS-1 by the synthetic analogs of N-acylhomoserine lactone. Appl. Environ. Microbiol. 73:6339-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nardini, M., and B. W. Dijkstra. 1999. α/β Hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9:732-737. [DOI] [PubMed] [Google Scholar]
- 38.Nishiyama, M., Y. Watanabe, and T. Marumoto. 1998. Bacterial 16S rDNA sequences in immature volcanic ash soil on volcanoes Mt. Sakurajima and Mt. Fugen in Japan determined by PCR amplification. Soil Sci. Plant Nutr. 44:711-715. [Google Scholar]
- 39.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, J. Schrag, J. L. Sussman, K. H. G. Verschueren, and A. Goldman. 1992. The α/β hydrolase fold. Protein Eng. 5:197-211. [DOI] [PubMed] [Google Scholar]
- 40.Park, S. Y., S. J. Lee, T. K. Oh, J. W. Oh, B. T. Koo, D. Y. Yum, and J. K. Lee. 2003. AhlD, an N-acylhomoserine lactone in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149:1541-1550. [DOI] [PubMed] [Google Scholar]
- 41.Park, S. Y., H. O. Kang, H. S. Jang, J. K. Lee, B. T. Koo, and D. Y. Yum. 2005. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 71:2632-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U. S. A. 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piper, K. R., S. B. V. Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 44.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasmussen, T. B., T. Bjarnsholt, M. E. Skindersoe, M. Hentzer, P. Kristoffersen, M. Kote, J. Nielsen, L. Eberl, and M. Givskov. 2005. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187:1799-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmussen, T. B., M. E. Skindersoe, T. Bjarnsholt, R. K. Phipps, K. B. Christensen, P. O. Jensen, J. B. Andersen, B. Koch, T. O. Larsen, M. Hentzer, L. Eberl, N. Hoiby, and M. Givskov. 2005. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151:1325-1340. [DOI] [PubMed] [Google Scholar]
- 47.Rasmussen, T. B., M. Manefield, J. B. Andersen, L. Eberl, U. Anthoni, C. Christophersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2000. How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 146:3237-3244. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 49.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 50.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. U. S. A. 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sitnikov, D. M., J. B. Schineller, and T. O. Baldwin. 1995. Transcriptional regulation of bioluminescence genes from Vibrio fischeri. Mol. Microbiol. 17:801-812. [DOI] [PubMed] [Google Scholar]
- 52.Taga, M. E., and B. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. U. S. A. 100:14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uroz, S., P. Oger, S. R. Chhabra, M. Camara, P. Williams, and Y. Dessaux. 2007. N-Acyl homoserine lactones are degraded via an amidolytic activity in Comamonas sp. strain D1. Arch. Microbiol. 187:249-256. [DOI] [PubMed] [Google Scholar]
- 54.Uroz, S., P. M. Oger, E. Chapelle, M. T. Adeline, D. Faure, and Y. Dessaux. 2008. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl. Environ. Microbiol. 74:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, L. H., L. X. Weng, Y. H. Dong, and L. H. Zhang. 2004. Specificity and enzyme kinetics of the quorum-quenching N-acyl homoserine lactone lactonase (AHL-lactonase). J. Biol. Chem. 279:13645-13651. [DOI] [PubMed] [Google Scholar]
- 56.Wei, H. L., and L. Q. Zhang. 2006. Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek 89:267-280. [DOI] [PubMed] [Google Scholar]
- 57.Winans, S. C., and B. L. Bassler. 2002. Mob psychology. J. Bacteriol. 184:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, Y. M., K. Tian, J. M. Hao, S. J. Pei, and Y. X. Yang. 2004. Biodiversity and biodiversity conservation in Yunnan, China. Biodivers. Conserv. 13:813-826. [Google Scholar]
- 59.Zhang, L. H. 2003. Quorum quenching and proactive host defense. Trends Plant Sci. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, H. B., L. H. Wang, and L. H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 99:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.