Abstract
Functional metagenomics was used to search for florfenicol resistance genes in libraries of cloned DNA isolated from Alaskan soil. A gene that mediated reduced susceptibility to florfenicol was identified and designated pexA. The predicted PexA protein showed a structure similar to that of efflux pumps of the major facilitator superfamily.
Antimicrobial resistance in various bacterial pathogens is an escalating global problem, with multidrug resistance in many pathogens becoming increasingly common (15, 32). To develop alternative treatments or to use existing therapies judiciously and efficaciously, it is important to understand the origins and ecological reservoirs of antimicrobial resistance genes and the underlying resistance mechanisms (4). Identifying the sources of the resistance genes and their association with mobile genetic elements will aid in efforts to predict their emergence and dissemination in clinically relevant pathogens (14).
The dynamics of emergence and persistence of antimicrobial resistance determinants are complex and still not fully understood. Spread of resistance genes can be caused by use of an antimicrobial agent, thereby selecting for clonal dissemination of a bacterium harboring the corresponding resistance gene or—if the gene is located on a mobile element—by horizontal transfer of the respective mobile genetic element among bacteria of the same or different species and genera (10, 18). However, there is also evidence that antimicrobial treatment at a specific site is not the sole risk factor for the development or dissemination of resistance (17, 26). Resistance genes have been found in remote, “pristine” environments far removed from human influence (2). The discovery of what Waksman and Woodruff termed “antagonistic” microorganisms (35) led to the early assumption that resistance genes have arisen as a self-defense mechanism against self-produced “war munitions” or attacks from other microbes trying to gain an advantage in the competitive environment that exists in microbial communities. It has also been suggested that resistance genes serve functions other than those based on anthropomorphic definitions (8, 36). If this assumption is correct, the occurrence of genes conferring resistance to both currently used therapeutics and those yet to be approved is likely underestimated. Understanding the frequency and diversity of these resistance genes in environmental reservoirs will aid in predicting the emergence and dissemination of antimicrobial resistance genes (26, 29).
Many antimicrobial agents are produced by soil bacteria (23), and soil bacteria are still believed to represent not only a source of novel antimicrobial agents but also a source of novel resistance genes. It has been hypothesized that only 0.5% of microbes residing in soil are culturable by current methods (34), and consequently, investigations into the diversity of resistance genes that exist in nature are biased if they focus solely on cultivable microorganisms. Metagenomics is a culture-independent method of examining the DNA present in a given sample (19, 28). DNA is extracted directly from the sample and cloned into commercial vectors. Classically, metagenomic analysis was based on random sequencing of inserted DNA or amplification by PCR of target genes (9, 33). Using a similar approach but searching for a specific function using heterologous expression in a surrogate host has been designated functional metagenomics and has already been used to identify antimicrobial resistance genes (1, 2). This approach allows exploration of genes whose function may not be obvious based on their sequence. Functional metagenomics could provide powerful insight into the genetic diversity of antimicrobial resistance not yet accounted for in complex microbial communities such as those that exist in soil.
Florfenicol is a synthetic fluorinated derivative of chloramphenicol. It is a broad-spectrum antimicrobial agent approved for the control of respiratory tract infections in cattle and swine, infectious pododermatitis in cattle, and furunculosis in salmon. It acts by binding to the ribosome, thus inhibiting protein synthesis in bacteria (24). Resistance to florfenicol has been observed in many diverse bacteria, and a variety of mechanisms have been described (5, 11, 12, 16, 17, 24). These have all been discovered by analysis of organisms that exhibited phenotypic resistance to florfenicol or elevated MICs in cases where no CLSI-approved interpretive criteria were available. In some instances, the genes responsible for florfenicol resistance have also been found to be physically linked to genes conferring resistance to other antimicrobial agents (5, 11, 25). This could imply that, although the drug is used strictly in animals, the use of florfenicol might select for and amplify resistances to antimicrobials that are relevant to human health (18, 22).
The present study aimed to discover florfenicol resistance genes by using metagenomic libraries constructed from DNA extracted from the soil of remote sites in Alaska. Our hypothesis is that genes that confer resistance to florfenicol exist in the environment, even in the absence of a sufficiently high selective pressure imposed by the presence of florfenicol. Identifying novel resistance genes, particularly those that might be found in noncultivable microbes, can help to predict the emergence of resistance. Identifying the genes that are linked to a resistance gene will aid in our understanding of coselection and persistence of resistance genes.
Construction and screening of metagenomic libraries.
The metagenomic libraries screened in this study were reported previously (2). Briefly, they were constructed using soil samples collected from an island in the Tanana River in the National Science Foundation's Long-Term Ecological Research site at Bonanza Creek Experimental Forest near Fairbanks, AK. The samples were transported at 4°C, stored at −20°C, and thawed at room temperature just before use. Cells were lysed either directly in the sample or after being separated from the sample matrices. DNA from the cells was ligated into pCC1BAC (Epicentre, Madison, WI) or pCC1FOS (CopyControl fosmid library production kit; Epicentre). Escherichia coli Epi300 (Epicentre) was used as the host for these vectors. Recombinant clones were scraped from Luria-Bertani (LB) agar supplemented with chloramphenicol (12.5 μg/ml) into selective LB broth plus 20% glycerol. The libraries were stored in pools at −80°C.
The metagenomic libraries were inoculated into 3 ml LB broth plus chloramphenicol (12.5 μg/ml) and incubated for 2 to 3 h at 37°C with shaking. Cultures were plated onto LB agar with florfenicol (16 μg/ml). Although breakpoints for resistance to florfenicol exist for only a few animal pathogens (7), we previously reported that a common genetic basis for florfenicol resistance in E. coli resulted in a MIC of ≥16 μg/ml (27). Half of the plates were incubated at 37°C, and half were incubated at room temperature. Clones growing on these selective plates were evaluated by restriction endonuclease analysis and retransformation into chemically competent E. coli DH5α to confirm the phenotype (Table 1).
TABLE 1.
Bacterial cloning strains, plasmids, and metagenomic clones described in this work
| Strain, plasmid, gene, or clone | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| E. coli strains and plasmids for cloning | ||
| Epi300 | mcrA Δ(mrr-hsdRMS-mcrBC) endA1 recA1; high-transformation efficiency of large DNA | Epicentre, Madison, WI |
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF) U169 recA endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen, Carlsbad, CA |
| pCC1FOS | Chlr; fosmid cloning vector | Epicentre, Madison, WI |
| pDrive | Ampr Kanr; TA PCR cloning vector | Qiagen, Valencia, CA |
| EZ-Tn5 <KAN-2> | Kanr; Tn5 for transposon mutagenesis | Epicentre, Madison, WI |
| Metagenomic clone in pCC1FOS from Alaskan soil DNA, Ak20-3 | Fflr Chlr; contains a gene encoding a major facilitator superfamily drug exporter | This work |
| ORF of active gene from the metagenome, pexA | Confers resistance to florfenicol and chloramphenicol | This work |
Abbreviations: Amp, ampicillin; Chl, chloramphenicol; Ffl, florfenicol; Kan, kanamycin.
A total of 13,201 Mb of DNA from Alaskan soil was screened. A single fosmid clone, Ak20-3, grew on LB plates supplemented with florfenicol (16 μg/ml) when incubated at room temperature. Restriction endonuclease analysis of this clone with NotI and XhoI showed an insert of approximately 40 kb.
Identification and analysis of active gene.
The clone Ak20-3 was subjected to in vitro transposon mutagenesis using the commercially available EZ-Tn5 <KAN-2> insertion kit (Epicentre). Mutants exhibiting susceptibility to florfenicol were sequenced using the manufacturer's primers to identify the inactivated gene. Other insertion mutants were randomly chosen and used to sequence the remaining inserted DNA by using the manufacturer's primers. The sequence was assembled using Sequencher (Genecodes, Ann Arbor, MI) and SeqMan (Lasergene software; DNAStar, Madison, WI) programs. Finishing was done by primer walking. The Artemis program (21) served to identify putative open reading frames (ORFs), which were annotated using BLAST (Basic Local Alignment Search Tool) (3). Predicted ORFs within the DNA insert are listed in Table 2.
TABLE 2.
Predicted genes encoded by the metagenomic clone Ak20-3
| ORF no. | ORF start | ORF stop | Predicted function of closest match | Accession no. of closest match (% identity) |
|---|---|---|---|---|
| 1 | 726 | 1 | Translation elongation factor Tu | ACU90852 (78) |
| 2 | 2150 | 1398 | rRNA methylase | EEUO3523 (36) |
| 3 | 3403 | 2147 | Hypothetical protein | No matches |
| 4 | 4751 | 3366 | Two-component, sigma54-specific, transcriptional regulator, Fis family | ABB31018 (52) |
| 5 | 7010 | 4764 | PAS/PAC sensor signal transduction histidine kinase | ABA89583 (37) |
| 6 | 7699 | 7007 | Hypothetical protein | No matches |
| 7 | 7821 | 8717 | Integrase/recombinase XerD/RipX family | EEI76895 (27) |
| 8 | 8701 | 9171 | Nucleoside deaminase | EAQ78406 (54) |
| 9 | 11575 | 9146 | Acyl coenzyme A dehydrogenase | ABC44659 (48) |
| 10 | 12510 | 11662 | Hypothetical protein | No matches |
| 11 | 13390 | 12572 | tRNA/rRNA methyltransferase | ABQ05252 (41) |
| 12 | 13876 | 13394 | Hypothetical protein | No matches |
| 13 | 14256 | 13873 | Hypothetical protein | No matches |
| 14 | 15545 | 14451 | Chaperone protein DnaJ | EFA67184 (50) |
| 15 | 18316 | 15575 | Excinuclease ABC, A subunit | ACU05744 (39) |
| 16 | 18381 | 18830 | Hypothetical protein | No matches |
| 17 | 18972 | 20777 | GTP-binding protein LepA | CAN93996 (62) |
| 18 | 20725 | 21231 | Methyltransferase | EDS77032 (53) |
| 19 | 21260 | 22489 | Sulfite dehydrogenase subunit SorA | EEO97828 (50) |
| 20 | 22467 | 22847 | Hypothetical protein | No matches |
| 21 | 22875 | 23780 | Universal stress protein | CAE79992 (31) |
| 22 | 25302 | 24055 | pexAa; drug resistance transporter | ACN95065 (33) |
| 23 | 25586 | 26110 | Hypothetical protein | No matches |
| 24 | 27709 | 26126 | ATP-dependent RNA helicase | EDL56381 (49) |
| 25 | 28854 | 27946 | Ribosomal protein S6 modification protein | ACA98689 (67) |
| 26 | 29351 | 28854 | Conserved hypothetical protein | EDL56110 (52) |
| 27 | 29561 | 29673 | 5S rRNA | CP001661 (85) |
| 28 | 29750 | 32672 | 23S rRNA | CP001089 (80) |
| 29 | 32838 | 34362 | 16S rRNA | CP001629 (83) |
| 30 | 34847 | 37603 | Hypothetical protein | No matches |
| 31 | 37616 | 38896 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | BAH38593 (50) |
| 32 | 40406 | 38973 | Glutamyl-tRNA synthetase | ACY17153 (53) |
| 33 | 41537 | 40455 | N-Acetylglucosaminyltransferase | BAC13049 (44) |
| 34 | 42378 | 41530 | Oligopeptide ABC transporter, ATP-binding protein | AAD35151 (56) |
pexA is the designation given in this work.
The gene encoding the decreased susceptibility to florfenicol was identified by transposon mutagenesis. A single transposon insertion at bp 24262 resulted in the clone being unable to grow on LB agar plates supplemented with florfenicol (16 μg/ml). This insertion site was located within an ORF at bp 24055 to 25302 that was tentatively designated pexA (phenicol exporter A) and coded for a protein of 415 amino acids (aa).
Amino acid alignment was done using MEGA4 (30). The new resistance protein was aligned with known phenicol exporter proteins of the major facilitator superfamily (MFS) using the ClustalW method. The resulting alignment was used to construct a phylogenetic tree using the minimum evolution method. The resulting tree was optimized using 1,000 bootstrap replicates and a random seed.
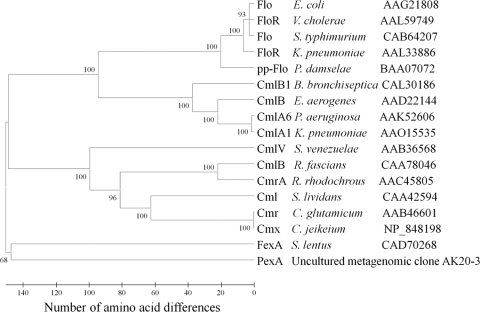
The PexA amino acid sequence showed only low similarity with other protein sequences deposited in the GenBank protein database. All similar sequences were part of the major facilitator superfamily of secondary transporters. The closest similarities, of 33% amino acid identity, were observed between PexA and drug resistance transporters from Wolbachia spp. (accession no. YP_002726856, YP_198189, and NP_966057). Phylogenetic analysis of the amino acid sequence of PexA shows very low identity with any of the known florfenicol/chloramphenicol exporters (Fig. 1). The Tmpred program (http://www.ch.embnet.org/software/TMPRED_form.html) was used to detect possible transmembrane helices in the PexA structure. The results predicted that the PexA protein has 11 transmembrane helices.
FIG. 1.
Phylogenetic tree of the known chloramphenicol/florfenicol exporter proteins. For the different exporter proteins, information on bacterial hosts, database accession numbers, and gene designations (as given in the database entries) is provided. Numbers above each node show the percentage of tree configurations that occurred during the 1,000 bootstrap trials. The tree was constructed using MEGA4 (30).
Susceptibility testing.
MICs were determined by broth microdilution according to the recommendations given in the document M31-A3 of the Clinical and Laboratory Standards Institute (CLSI) (7). Florfenicol MICs were determined by using a microtiter plate according to CLSI guidelines with a range of 2 to 64 μg/ml of florfenicol. The clone was tested for susceptibility to the antibiotics amikacin, amoxicillin-clavulanic acid, ampicillin, cefoxitin, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim-sulfamethoxazole with a commercially available panel (CMV1AGNF, Sensititre Gram-negative NARMS plate; Trek Diagnostic Systems, Westlake, OH). For each strain, both tests were performed at 30°C and at 37°C and read manually after 24 h and 48 h. Based on the growth in the wells of the microtiter plates, the MICs of 17 different antimicrobial agents were determined. The commercially available E. coli ATCC 25922 served to ensure the quality of the plates, and E. coli DH5α and E. coli DH5α carrying the empty cloning vector were used for comparative reasons.
To evaluate the activity of the gene against chloramphenicol, it was necessary to move the gene into a plasmid that did not have a chloramphenicol resistance gene. PCR was conducted with primers (20) that bind 248 bp upstream and 177 bp downstream of the reading frame coding for florfenicol resistance. These primers (F, 5′-TTCACTGCAGGGATCGTGAC-3′; R, 5′-CAACTGCAGAAAAGCGAAAAG-3′) yielded a 1,701-bp PCR amplicon that contained the 1,248-bp coding sequence of interest. The amplicon was cloned into the pDrive cloning vector (Qiagen PCR cloning kit) according to kit instructions.
MICs were recorded after 48 h of incubation (Table 3). Clones with the insert had florfenicol and chloramphenicol MICs of 16 μg/ml, but only on the microtiter plates grown at 30°C. At 37°C, clones with and without the insert showed MICs of florfenicol and chloramphenicol of 2 μg/ml each. The Ak20-3 mutant with the transposon insertion within the pexA ORF had a florfenicol MIC of 2 μg/ml at 30°C and 37°C. The clones with the insert did not differ in their susceptibility to any of the antimicrobials, aside from chloramphenicol and florfenicol, from those without the insert at 30°C or 37°C (data not shown). This suggests that pexA mediates resistance to phenicols and not to any other antimicrobial agents.
TABLE 3.
MICs of control strains, fosmid clone, and subclone from the Alaskan soil metagenome for florfenicol and chloramphenicol at 30°C after 48 h of incubation
| Strain (vector) | Clone (mutation) | MIC (μg/ml)a |
|
|---|---|---|---|
| Chl | Ffl | ||
| DH5α(pCC1FOSb) | Ak20-3 | 64 | 16 |
| DH5α(pCC1FOS) | Ak20-3 (EZ-Tn5 <KAN-2>) | 64 | 2 |
| DH5α(pCC1FOS) | 64 | 2 | |
| DH5α(pDrive) | pexA | 16 | 16 |
| DH5α | 4 | 2 | |
| DH5α(pDrive) | 4 | 2 | |
Abbreviations: Chl, chloramphenicol; Ffl, florfenicol.
pCC1FOS contains a cat gene for chloramphenicol resistance.
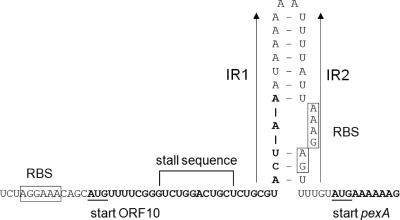
Many of the known MFS proteins from clinically relevant bacteria involved in the active efflux of antimicrobial agents are inducibly expressed. Sequence analysis of the region upstream of pexA revealed a structure similar to that of translational attenuators previously reported to be located upstream of staphylococcal cat genes for chloramphenicol resistance or the fexA gene for chloramphenicol/florfenicol resistance (11). Upstream of pexA, a small reading frame for a 10-aa peptide which contained a potential ribosome stall sequence (5′-GUCUGGACUGCU-3′) similar to previously described ones in regulatory regions of other inducibly expressed phenicol resistance genes was detected (Fig. 2). A pair of two imperfect inverted repeated sequences of 11 and 14 bp, the latter of which contained the pexA-associated ribosome binding site, was also detected in the pexA upstream region. Calculation of the stability of the mRNA secondary structure formed by these inverted repeats showed a distinctly lower stability of ΔG = −24.3 kJ/mol than that calculated for the mRNA secondary structure in the fexA upstream region (ΔG = −74.7 kJ/mol) (9).
FIG. 2.
Presentation of the predicted pexA regulatory region. The predicted regulatory region- and pexA-associated ribosome binding sites (RBS) are boxed. The start codons of the ORF for the regulatory peptide and the pexA gene are underlined, and the corresponding coding sequences are displayed in bold letters. The inverted repeated (IR) sequences IR1 and IR2 are marked by arrows, and an mRNA secondary structure formed by these IR sequences is shown. Calculation of the stability of this stem-loop structure followed the specifications given by Tinoco et al. (31).
To check whether the newly identified phenicol resistance gene was induced by low concentrations of either florfenicol or chloramphenicol, E. coli DH5α strains containing the pCC1FOS vector with and without the Ak20-3 insert were passed three times on nonsupplemented LB agar plates at 24-h intervals. Colonies from the antibiotic-free LB plate were then used to inoculate 3-ml aliquots of cation-adjusted Mueller-Hinton broth containing florfenicol or chloramphenicol (0.5 μg/ml); these aliquots were incubated with shaking at 30°C for 3 h. Cells were pelleted at 8,000 rpm for 3 min and then resuspended in sterile water, diluted in Mueller-Hinton broth, and used to inoculate the aforementioned microtiter plates. MIC determination followed CLSI standards (7). No differences in MICs of any of the antimicrobial agents tested, including chloramphenicol and florfenicol, were observed between the preincubated and nonpreincubated strains when grown at 30°C or 37°C.
The sequence upstream of pexA shows some homology to the translational attenuators upstream of other known antimicrobial resistance genes (11). Temperature could explain the inability to observe a difference in expression after preincubation with a low concentration of florfenicol or chloramphenicol. Differences in the translational components between the natural host of pexA and the surrogate E. coli host could have also played a role in the apparent noninducibility of pexA in the E. coli host despite the presence of what appears to be a translational attenuator upstream of pexA.
An rRNA operon was predicted using RNAmmer 1.2 (13) within the DNA insert that carries pexA. A BLAST (3) query to the GenBank database, excluding all uncultured bacteria, indicated that the predicted 1,525-bp 16S rRNA segment shows highest similarity to the genera Geobacter (89% query coverage, 95% identity) and Desulfomicrobium (100% query coverage, 83% identity), both members of the Deltaproteobacteria/Epsilonproteobacteria. Both of these microbes have been isolated from environmental sources.
Although the ultimate source of this gene is unknown, this finding supports the idea that resistance genes exist independently of exposure to therapeutic concentrations of antimicrobial agents and may serve unknown functions in their natural environment. Due to the proximity of pexA to an rRNA operon, it seems unlikely that it was inserted there or would be excised in a lateral transfer event because structures resembling mobile genetic elements, such as insertion sequences or transposons, have not been detected. In the present case, pexA confers an elevated MIC of chloramphenicol, which is produced by a soil bacterium. In its natural host, pexA could provide protection against chloramphenicol excreted by Streptomyces spp. However, genes which are anthropomorphically defined as “resistance genes” could also have functions other than survival in the presence of antimicrobial agents. It has been known for some time that at low concentrations antimicrobial agents have multiple effects on bacterial cells, including changes in gene expression, increased mRNA stability, increased rates of mutation, and increased genetic transfer (6, 8). Reactions due to subinhibitory concentrations of antimicrobials probably represent the true function of resistance genes in nature, and it is important to recognize that these genes are selected for under conditions other than treatment with antimicrobials. Understanding these origins could provide clues to novel interventions against resistant organisms or discovery of new antimicrobial compounds.
The results of this functional metagenomic analysis are contingent upon a given gene's ability to be expressed in E. coli. It is probable that most of the genes in these libraries are not expressed in this surrogate host, and therefore, we likely underestimate the frequency of resistance determinants in environmental samples. The gene described in this work is active at 30°C or lower temperatures, and—according to the results of broth microdilution—does not provide protection of the E. coli host against concentrations equal to or greater than 16 μg/ml of either florfenicol or chloramphenicol. This could be due to problems with heterologous expression in E. coli. Activity and stability of a given gene are likely dependent on the living conditions of the natural host. The environment from which the DNA was extracted was Alaskan soil. The microbe that originally harbored the gene was probably acclimated to life at temperatures much lower than 30°C. Perhaps pexA would have increased activity at such temperatures, but due to the limitations of using E. coli as a surrogate host, assessment of such activity is not possible.
Despite limitations, functional metagenomics has been shown to be effective in discovering diverse resistance mechanisms. It has been shown that human pathogens have likely acquired antimicrobial resistance determinants through horizontal gene transfer from other microbes within their community. Functional metagenomics could be used as a tool to screen microbial communities as a whole in order to fully assess the potential emergence and dissemination of antimicrobial resistance genes. This could be a powerful tool for the approval process of new antimicrobial compounds. Databases of existing metagenomic libraries could be constructed. Target libraries could be screened, using antimicrobial agents being considered for use, in order to evaluate whether a resistance mechanism already exists, and if so, resistance genes could be identified and attempts could be made to predict their rate of dissemination based on the genes to which the novel resistance genes are physically linked.
ID and nucleotide sequence accession numbers.
The Alaskan soil metagenome project has been registered with the NCBI (National Center for Biotechnology Information) Genome Project database (identification [ID], 28853). The GenBank accession number for metagenomic clone AK20-3 is HM537013.
Acknowledgments
We thank Jo Handelsman's laboratory for technical assistance.
This project was supported by University of Minnesota Academic Health Center Faculty Research Development grant 07-16 (R. S. Singer) and the USDA Microbial Observatory Program (J. Handelsman).
Footnotes
Published ahead of print on 11 June 2010.
REFERENCES
- 1.Allen, H. K., K. A. Cloud-Hansen, J. M. Wolinski, C. Guan, S. Greene, S. Lu, M. Boeyink, N. A. Broderick, K. F. Raffa, and J. Handelsman. 2009. Resident microbiota of the gypsy moth midgut harbors antibiotic resistance determinants. DNA Cell Biol. 28:109-117. [DOI] [PubMed] [Google Scholar]
- 2.Allen, H. K., L. A. Moe, J. Rodbumrer, A. Gaarder, and J. Handelsman. 2009. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 3:243-251. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Aminov, R. I., and R. I. Mackie. 2007. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271:147-161. [DOI] [PubMed] [Google Scholar]
- 5.Arcangioli, M. A., S. Leroy-Sétrin, J. L. Martel, and E. Chaslus-Dancla. 1999. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol. Lett. 174:327-332. [DOI] [PubMed] [Google Scholar]
- 6.Blickwede, M., R. Goethe, C. Wolz, P. Valentin-Weigand, and S. Schwarz. 2005. Molecular basis of florfenicol-induced increase in adherence of Staphylococcus aureus strain Newman. J. Antimicrob. Chemother. 56:315-323. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 3rd ed. CLSI document M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Davies, J., G. B. Spiegelman, and G. Yim. 2006. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 9:445-453. [DOI] [PubMed] [Google Scholar]
- 9.de la Torre, J. R., L. M. Christianson, O. Béjà, M. T. Suzuki, D. M. Karl, J. Heidelberg, and E. F. DeLong. 2003. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc. Natl. Acad. Sci. U. S. A. 100:12830-12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 11.Kehrenberg, C., and S. Schwarz. 2004. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob. Agents Chemother. 48:615-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kehrenberg, C., S. Schwarz, L. Jacobsen, L. H. Hansen, and B. Vester. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57:1064-1073. [DOI] [PubMed] [Google Scholar]
- 13.Lagesen, K., P. Hallin, E. A. Rodland, H. Staerfeldt, T. Rognes, and D. W. Ussery. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin, B. R., R. Antia, E. Berliner, P. Bloland, S. Bonhoeffer, M. Cohen, T. DeRouin, P. I. Fields, H. Jafari, D. Jernigan, M. Lipsitch, J. E. McGowan, P. Mead, M. Nowak, T. Porco, P. Sykora, L. Simonsen, J. Spitznagel, R. Tauxe, and F. Tenover. 1998. Resistance to antimicrobial chemotherapy: a prescription for research and action. Am. J. Med. Sci. 315:87-94. [DOI] [PubMed] [Google Scholar]
- 15.Levy, S. B., and T. F. O'Brien. 2005. Global antimicrobial resistance alerts and implications. Clin. Infect. Dis. 41(Suppl. 4):S219-S220. [DOI] [PubMed] [Google Scholar]
- 16.Long, K. S., J. Poehlsgaard, C. Kehrenberg, S. Schwarz, and B. Vester. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miranda, C. D., and R. Rojas. 2007. Occurrence of florfenicol resistance in bacteria associated with two Chilean salmon farms with different history of antibacterial usage. Aquaculture 266:39-46. [Google Scholar]
- 18.O'Brien, T. F. 2002. Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 34:S78-S84. [DOI] [PubMed] [Google Scholar]
- 19.Riesenfeld, C. S., P. D. Schloss, and J. Handelsman. 2004. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38:525-552. [DOI] [PubMed] [Google Scholar]
- 20.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 21.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 22.Salyers, A. A., and C. F. Amábile-Cuevas. 1997. Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41:2321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz, S., A. Cloeckaert, and M. C. Roberts. 2006. Mechanisms and spread of bacterial resistance to antimicrobial agents, p. 73-98. In F. M. Aarestrup (ed.), Antimicrobial resistance in bacteria of animal origin. ASM Press, Washington, DC.
- 24.Schwarz, S., C. Kehrenberg, B. Doublet, and A. Cloeckaert. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28:519-542. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz, S., C. Werckenthin, and C. Kehrenberg. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singer, R. S., M. P. Ward, and G. Maldonado. 2006. Can landscape ecology untangle the complexity of antibiotic resistance? Nat. Rev. Microbiol. 4:943-952. [DOI] [PubMed] [Google Scholar]
- 27.Singer, R. S., S. K. Patterson, A. E. Meier, J. K. Gibson, H. L. Lee, and C. W. Maddox. 2004. Relationship between phenotypic and genotypic florfenicol resistance in Escherichia coli. Antimicrob. Agents Chemother. 48:4047-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein, J. L., T. L. Marsh, K. Y. Wu, H. Shizuya, and E. F. DeLong. 1996. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summers, A. O. 2002. Generally overlooked fundamentals of bacterial genetics and ecology. Clin. Infect. Dis. 34(Suppl. 3):S85-S92. [DOI] [PubMed] [Google Scholar]
- 30.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 31.Tinoco, I., P. N. Borer, B. Dengler, M. D. Levin, O. C. Uhlenbeck, D. M. Crothers, and J. Bralla. 1973. Improved estimation of secondary structure in ribonucleic acids. Nat. New Biol. 246:40-41. [DOI] [PubMed] [Google Scholar]
- 32.Tomasz, A. 1994. Multiple-antibiotic-resistant pathogenic bacteria. A report on the Rockefeller University Workshop. N. Engl. J. Med. 330:1247-1251. [DOI] [PubMed] [Google Scholar]
- 33.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 34.Vogel, T. M., P. Simonet, J. K. Jansson, P. R. Hirsch, J. M. Tiedje, J. D. van Elsas, M. J. Bailey, R. Nalin, and L. Philippot. 2009. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat. Rev. Microbiol. 7:252. [Google Scholar]
- 35.Waksman, S. A., and H. B. Woodruff. 1940. The soil as a source of microorganisms antagonistic to disease-producing bacteria. J. Bacteriol. 40:581-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yim, G., H. H. Wang, and J. Davies. 2006. The truth about antibiotics. Int. J. Med. Microbiol. 296:163-170. [DOI] [PubMed] [Google Scholar]