Abstract
Dynamic, multicompartment in vitro gastrointestinal simulators are often used to monitor gut microbial dynamics and activity. These reactors need to harbor a microbial community that is stable upon inoculation, colon region specific, and relevant to in vivo conditions. Together with the reproducibility of the colonization process, these criteria are often overlooked when the modulatory properties from different treatments are compared. We therefore investigated the microbial colonization process in two identical simulators of the human intestinal microbial ecosystem (SHIME), simultaneously inoculated with the same human fecal microbiota with a high-resolution phylogenetic microarray: the human intestinal tract chip (HITChip). Following inoculation of the in vitro colon compartments, microbial community composition reached steady state after 2 weeks, whereas 3 weeks were required to reach functional stability. This dynamic colonization process was reproducible in both SHIME units and resulted in highly diverse microbial communities which were colon region specific, with the proximal regions harboring saccharolytic microbes (e.g., Bacteroides spp. and Eubacterium spp.) and the distal regions harboring mucin-degrading microbes (e.g., Akkermansia spp.). Importantly, the shift from an in vivo to an in vitro environment resulted in an increased Bacteroidetes/Firmicutes ratio, whereas Clostridium cluster IX (propionate producers) was enriched compared to clusters IV and XIVa (butyrate producers). This was supported by proportionally higher in vitro propionate concentrations. In conclusion, high-resolution analysis of in vitro-cultured gut microbiota offers new insight on the microbial colonization process and indicates the importance of digestive parameters that may be crucial in the development of new in vitro models.
The human gastrointestinal tract harbors a complex microbial ecosystem with a coding capacity exceeding that of the host genome by a factor of 100 (13). These gut microbes play a determining role in host health by converting otherwise indigestible compounds (14, 19), protecting against gut epithelial cell injury (46), regulating host fat storage (49), and inducing immunity (20, 48). Modulation of the composition and metabolic activity of these microbes to improve host health attracts a lot of attention and is referred to as gastrointestinal resource management (15, 37). Such new strategies are often evaluated during human trials or in vivo studies of animals associated with conventional or human microbiota (50).
Despite the physiological relevance, in vivo experimental setups are inherently associated with some drawbacks. First, apart from fecal analyses over time, most in vivo data are derived from endpoint measurements, thereby limiting the dynamic monitoring of the gut microbiota. Second, troublesome sampling of different gut regions makes it difficult to locate the effects of a treatment. For mechanistic reasons, a third drawback of an in vivo approach is the inability to focus solely on gut microbial activity, because there is always a host involved. For these reasons, different types of in vitro systems have been developed, ranging from simple nonstirred batch cultures without pH control (44) to more complex continuous models involving pH-controlled single (55) or dynamic multicompartment (2, 29, 32, 34) culture systems. Other advantages are the lack of ethical constraints and a higher reproducibility due to strict control of environmental factors that can influence the microbiota, such as retention time, pH, temperature, and food intake. Therefore, in vitro methods are widely used to elucidate the mechanism behind the degradation of prebiotics (17, 52), bioactivation of polyphenols (10, 36, 38), adhesion of microbes to mucins (51), or bioavailability of environmental contaminants (53, 54).
Dynamic in vitro gut models need to fulfil certain criteria before they can be used to monitor the modulating potency of specific treatments toward the microbiota. To ensure that effects are due solely to the treatment and not to the adaptation of microbes to the in vitro environment, steady-state conditions in terms of microbial community composition and metabolic activity need to be established prior to the actual start of the experiment (39). Moreover, the stabilization of this in vitro microbiota needs to be reproducible, as comparison of different treatments requires identical starting communities. Former studies assumed but never fully substantiated this requirement (17, 38). Further, in vitro microbiota need to be gut region specific, be representative for the in vivo situation, and maintain a high diversity. The potency of in vitro models thus relies on a good characterization of its microbiota. Molecular techniques, such as denaturing gradient gel electrophoresis (DGGE) (17, 39, 52), fluorescent in situ hybridization (FISH) (6), and quantitative real-time PCR (Q-PCR) (17, 29), provide useful information but do not provide direct phylogenetic information or target only a limited group of previously identified organisms, therefore limiting current knowledge. Recently, high-resolution techniques, such as microarrays (41, 42) and pyrosequencing (59), have provided access to phylogenetic and metagenomic analysis of the gut microbiota in unprecedented detail.
In this study, we performed conventional metabolic analysis, applied existing molecular techniques (DGGE), and for the first time provided an in-depth phylogenetic analysis on the simulator of the human intestinal microbial ecosystem (SHIME) in vitro microbiota using the recently developed human intestinal tract chip (HITChip) microarray (41, 42). We evaluated the microbial colonization process in two parallel in vitro simulators (Twin-SHIME) simultaneously inoculated with the same human fecal microbiota. The aims of this study were (i) to determine when the microbial community composition and metabolic activity reach steady-state conditions, (ii) to assess the reproducibility of the stabilization process in two identical in vitro simulators, (iii) to obtain a high-resolution characterization of the colon region specificity of the residing communities, and (iv) to evaluate how the in vivo fecal inoculum changes to the in vitro colon microbial communities.
MATERIALS AND METHODS
Twin-SHIME experiment.
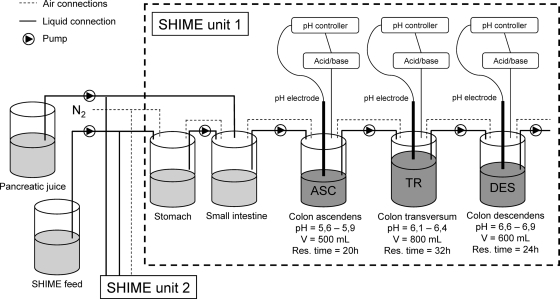
The SHIME is a dynamic in vitro model of the human gastrointestinal tract that is composed of five double-jacketed vessels, simulating the stomach, small intestine, and the three colon regions (ascending, transverse, and descending colon), with a total retention time of 72 h (Fig. 1). Three times per day, 140 ml SHIME feed and 60 ml pancreatic juice were added to the stomach and small intestine compartments, respectively. The SHIME feed contained (in grams/liter) arabinogalactan (1.0), pectin (2.0), xylan (1.0), starch (4.0), glucose (0.4), yeast extract (3.0), peptone (3.0), mucin (1.0), and cystein (0.5). The pancreatic juice contained (in grams/liter) NaHCO3 (12.5), bile salts (6.0) (Difco, Bierbeek, Belgium), and pancreatin (0.9) (Sigma, Bornem, Belgium). All vessels were kept anaerobic by flushing them with N2, and they were continuously stirred and kept at 37°C. A Twin-SHIME setup comprises two SHIME units in parallel (17, 38) in order to attain identical environmental conditions for both units. In this experiment, the three colon regions of each SHIME unit of a Twin-SHIME were inoculated with human fecal microbiota from a healthy male volunteer (22 years old) who had no history of antibiotic treatment 6 months prior to the study. More details on reactor setup and inoculum preparation were as previously described (34, 39). A specific feature of the SHIME setup is the initial 2-week stabilization period, which allows the microbiota to adapt to the imposed in vitro conditions and to evolve from a fecal microbial community to one representative of a specific colon region. During the first 26 days after inoculation, samples were collected and DGGE was applied to investigate the stabilization of the microbial communities in terms of microbiota composition. Metabolic parameters were analyzed to evaluate this stabilization also in terms of metabolic activity. Between days 19 and 26, samples were taken to characterize the metabolite production and enzyme activity of the stabilized microbial communities. Samples of the inoculum from days 19 and 26 were used for in-depth community composition analysis with the HITChip.
FIG. 1.
Schematic representation of a Twin-SHIME which consists of two identical SHIME units, SHIME units 1 and 2. Liquid SHIME feed and pancreatic juice enter the compartments which simulate the stomach and small intestine, respectively. After a residence time of 4 h in these sterile compartments, the suspension goes to three consecutive colon compartments, the ascending (ASC), transverse (TR), and descending (DES) colon compartments, each characterized by distinct pHs and residence times. These compartments are inoculated with human fecal microbiota. All vessels are kept anaerobic by flushing the headspace with N2, continuously stirred, and kept at 37°C.
Microbial community analysis: DGGE and HITChip.
Total DNA extractions were performed according to Boon et al. (4). Denaturing gradient gel electrophoresis (DGGE) was applied to separate PCR products of 16S rRNA genes obtained with the general bacterial primers 338F-GC and 518R. Gels had a denaturing gradient ranging from 45% to 60% (52) and were run on an Ingeny PhorU apparatus (Ingeny International, Goes, Netherlands). Normalization and further analysis of the gels were carried out using BioNumerics software, version 5.10 (Applied Maths, Sint-Martens-Latem, Belgium). Extracted data were processed according to Marzorati et al. (30) for ecological interpretation, including calculation of the range-weighted richness (RR) (approximate environmental carrying capacity for microbial diversity), the dynamics (Dy) (approximate changes within the community over a fixed time frame), and a functional organization (FO) parameter (approximate structure of the microbial community in terms of its evenness).
Microbiota composition was determined using the human intestinal tract chip (HITChip), a gastrointestinal tract-specific phylogenetic microarray, as described by Rajilic-Stojanovic et al. (41, 42). Using 4,800 oligonucleotide probes, this microarray targets 1,140 unique microbial phylotypes (with <98% identity), providing data from 131 genus-like groups. Briefly, 16S rRNA genes were amplified and subsequently in vitro transcribed, labeled with the Cy3 and Cy5 dyes, and hybridized to the array. After the arrays were washed and scanned, data were extracted using the Agilent Feature Extraction software, version 7.5 (Agilent Technologies), and normalized using a set of R-based scripts (http://www.r-project.org/). Analysis of the microarray was performed in a custom relational database which runs under the MySQL database management system (MySQL) using a series of custom R scripts, as previously described (41, 42). These robust statistical packages allow the processing of data for comparative analysis of microbiota patterns, various forms of microbiota composition visualization (e.g., microbial grouping), and semiquantitative microbiota reconstruction at different phylogenetic levels, from phylum level to bacterial group level. Quantitative analyses at the bacterial group level were validated with Q-PCR (41). To provide an ecological interpretation of these data, the Simpson reciprocal index was calculated as a measure of diversity (27). An increase in the Simpson reciprocal index reflects a diversity increase, with 1 being the lowest possible number and the number of bacterial species present in the sample being the maximal number.
Metabolic activity analysis: SCFAs, lactate, succinate, ammonium, and enzymatic assays.
Short-chain fatty acid (SCFA) and ammonium ion analyses were performed as reported by Nollet et al. (35) and De Boever et al. (10), respectively. Lactate and succinate were measured using a d-lactate/l-lactate and succinate kit (R-Biopharm, Mannheim, Germany), according to the manufacturer's protocols. β-Glucosidase, β-glucuronidase, α-l-arabinofuranosidase, β-d-xylosidase, and endo-1,4-β-xylanase activities were determined with p-nitrophenyl-β-d-glucopyranoside, p-nitrophenyl-β-d-glucuronide, p-nitrophenyl-α-l-arabinofuranoside, p-nitrophenyl-β-d-xylopyranoside (Sigma-Aldrich, Bornem, Belgium), and azo-wheat-arabinoxylan (Megazyme, Bray, Ireland), respectively (17, 52). One unit is the enzyme activity needed to release 1 μmol of p-nitrophenol from the appropriate substrate (2.5 mM) in 1 min under the assay conditions.
Enzyme assays of azoreductase and nitroreductase were optimized for measurement in SHIME suspensions based on earlier protocols (18, 57). For azoreductase activity, 0.05 ml of an amaranth solution (1.5 mM, dissolved in anaerobic phosphate buffer) was added to 1.5 ml of SHIME suspension. After anaerobic incubation for 4 h at 37°C, the mixture was centrifuged for 2 min (3,000 × g). Discoloration of the medium was monitored by measuring the absorbance of the supernatant at 520 nm with a multiwell spectrophotometer (PowerWave 340 [Bio-Tek Instruments Inc., Winooski, VT]). Nitroreductase activity was determined by adding 0.5 ml of 1.5 mM p-nitrobenzoic acid to 1 ml SHIME suspension. After anaerobic incubation for 4 h at 37°C, the amount of p-aminobenzoic acid was determined by using the method of Bratton and Marshall, as changed by Goldbarg and Rutenburg (16). This included addition of the following solutions: 1 ml NaNO2 (0.1%), 1 ml NH4 sulfamate (0.5%), and, after intensive mixing, 2 ml N-(1-naphthyl)ethylenediamine dihydrochloride (0.05%) dissolved in 100% ethanol. After 10 min of incubation at 25°C, the absorbance was measured at 550 nm. Both assays were also performed using a positive and a negative control (boiled sample), so that results are expressed as percentages of amaranth and p-nitrobenzoic acid conversion.
Statistics.
All data were analyzed using SPSS 16 software (SPSS Inc., Chicago, IL). Before investigating the probability of intergroup differences, the normality and homogeneity of variances were studied with Kolmogorov-Smirnov and Levene tests, respectively. Under normal distribution and homogeneity of variances, an analysis of variance (ANOVA) with a (post hoc) Bonferroni test was performed; otherwise, a Kruskal-Wallis with a Mann-Whitney U test was applied. To investigate differences between both SHIME units of the Twin-SHIME, a paired t test was applied when inter-SHIME differences were normally distributed; otherwise, a Wilcoxon signed-rank test was applied. Differences were considered significant at P values of <0.05.
The Pearson correlation coefficient and unweighted-pair group method using average linkages (UPGMA) clustering algorithm were used to create dendrograms of DGGE profiles from the stabilization period, taking into account both band position and band density. Cosine correlation coefficients were calculated on the basis of normalized values for metabolic parameters during the stabilization period (ammonium, acetate, propionate, butyrate, and branched SCFAs). To assess the correlation of microbial groups detected by the HITChip with specific colon regions, redundancy analysis (RDA) was used as implemented in Canoco for Windows 4.5 (22). Average signal intensities for 131 bacterial groups were used as species data, and diagrams were plotted using the CanoDraw for Windows utility. The Monte Carlo permutation procedure (21) was used to assess statistical significance of the variation in data sets in relation to sample origin, i.e., the ascending, transverse, or descending colon compartment.
RESULTS
Stabilization of microbial community composition in a Twin-SHIME.
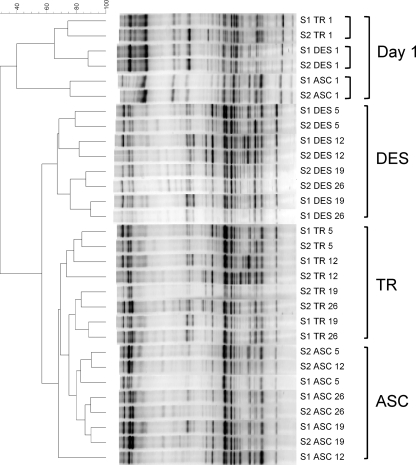
DGGE was applied to assess the microbiota composition of the stabilizing communities in both SHIME units after inoculation of the human fecal sample (Fig. 2). Colon samples from day 1 were more similar to one another than colon samples from later time points. From day 5 onwards, samples from the different colon compartments clustered separately, independently of the time point of sampling. Interestingly, the microbial communities of both SHIME units developed similarly during the stabilization period, with day 26 displaying correlations of 90%, 82%, and 76% in the ascending, transverse, and descending colon compartments, respectively.
FIG. 2.
Clustering tree based on Pearson and UPGMA correlations of the DGGE profiles in the ascending (ASC), transverse (TR), and descending (DES) colon compartments of a Twin-SHIME (S1, SHIME 1; S2, SHIME 2) at different times after inoculation (days 1, 5, 12, 19, and 26).
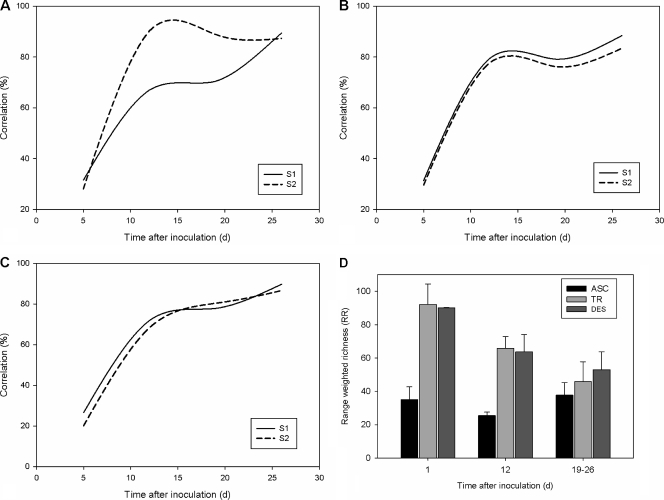
The initial weekly rates of change (100% − percent correlation) were around 70% in all colon regions of both SHIME units. After 14 days, rates of change dropped below 20% for each colon compartment, indicating steady-state microbial compositions (Fig. 3 A to C). The range-weighted richness (RR; approximate diversity) of the ascending colon was low on the first day after inoculation, while both the transverse and descending colons were characterized by initially high RR values that decreased during further stabilization (Fig. 3D). Finally, microbial communities from all colon compartments were highly structured, as the functional organization (FO) parameter (ranging between 0.40 and 0.48) was stable throughout the stabilization period; i.e., species abundances were distributed among dominant and subdominant members.
FIG. 3.
Correlation coefficients (percentages) between DGGE profiles of consecutive time points of the Twin-SHIME (SHIME 1 and 2) during the stabilization period in the ascending (A), transverse (B), and descending (C) colon compartments. (D) Average range-weighted richness (RR) of SHIME 1 and 2, based on the DGGE patterns of the ascending (ASC), transverse (TR), and descending (DES) colon at different time points after inoculation, specifically, day (d) 1 (n = 2), day 12 (n = 2), and during the control period (days 19 to 26 [n = 4]) (n indicates the number of replicate experiments).
Stabilization of microbial community activity in a Twin-SHIME.
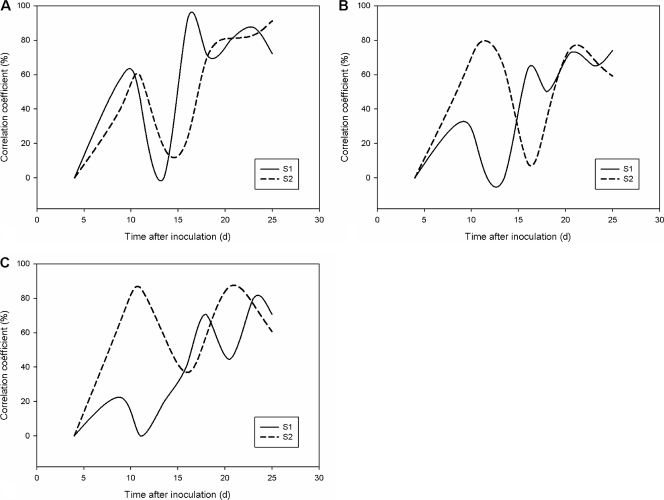
The metabolic fingerprints (ammonium, acetate, propionate, butyrate, and branched SCFAs) were variable in all colon regions of both SHIME units during the stabilization, reaching a steady-state level after approximately 17 to 20 days (Fig. 4 A to C).
FIG. 4.
Correlation coefficients (percentages) between the metabolic profiles of consecutive time points (ammonium, acetate, propionate, butyrate, and branched SCFAs) of the Twin-SHIME (SHIME 1 and 2) during the stabilization period in the ascending (A), transverse (B), and descending (C) colon compartments.
In-depth analysis of microbial community composition in a stabilized Twin-SHIME.
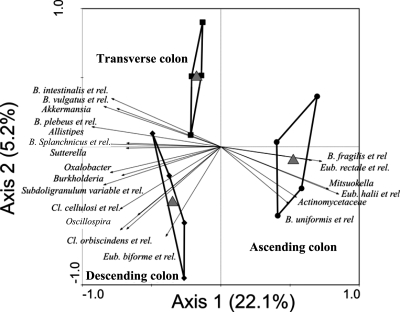
The HITChip provided detailed characterization of the stable microbial communities (samples from days 19 and 26) at the phylum and bacterial group levels in the respective colon compartments of the SHIME. Corresponding colon regions of both SHIME units showed similar microbiota abundances, so the averages from both units were calculated per colon compartment (Tables 1 and 2 and Fig. 5). There was a large similarity between the different colon parts at the phylum level (Table 1). Three phyla dominated the microbial communities, with Bacteroidetes being much more abundant than Firmicutes (Clostridium clusters IV, IX, and XIVa) and Proteobacteria. There were trends of higher proportions of Clostridium clusters IX and XIVa in the ascending colon and of Clostridium cluster IV and Proteobacteria in the descending colon. Verrucomicrobia was the only phylum for which the numbers of organisms significantly differed between the colon regions: the ascending colon was hardly colonized with Verrucomicrobia, whereas the transverse and descending colons possessed much higher numbers (P = 0.02). The Simpson reciprocal index tended to increase from the ascending (90 ± 15) to the transverse (100 ± 3) and descending (106 ± 7) colon compartments. At a lower phylogenetic level, HITChip analysis elucidated the presence of numerous bacterial groups in the in vitro microbiota (see Table S1 in the supplemental material). Redundancy analysis (RDA) at the bacterial group level revealed that the colon compartments were significantly different (P = 0.008) (Fig. 5). The RDA highlighted that 20 bacterial groups could be allocated to a specific colon compartment. The percentage of variation exhibited by these 20 bacterial groups was four times higher along the x axis of the RDA than along the y axis. Nonparametric statistical tests also indicated a significant correlation between most of these 20 groups and a colon compartment (Table 2). Bacteria that were characteristic for the ascending colon included Actinomycetaceae, Mitsuokella multacida, and relatives and groups belonging to the Bacteroidetes and Clostridium cluster XIVa. Akkermansia spp. and four Bacteroidetes groups were correlated to the transverse colon. Finally, Bacteroidetes, Clostridium clusters IV and XVI, and Proteobacteria displayed an increased abundance in the descending colon.
TABLE 1.
Abundances of higher taxonomic groupsa
| Higher taxonomic group | % abundance in: |
|||
|---|---|---|---|---|
| ASC | TR | DES | Human feces | |
| Actinobacteria | 1.11 | 0.47 | 0.71 | 0.27 |
| Bacteroidetes | 75.50 | 80.59 | 75.60 | 52.49 |
| Firmicutes | ||||
| Bacilli | 0.38 | 0.34 | 0.36 | 1.12 |
| Clostridium cluster I | 0.12 | 0.11 | 0.11 | 0.05 |
| Clostridium cluster III | 0.06 | 0.04 | 0.05 | 0.07 |
| Clostridium cluster IV | 1.54 | 1.19 | 2.41 | 26.07 |
| Clostridium cluster IX | 4.65 | 2.71 | 3.14 | 0.38 |
| Clostridium cluster XI | 0.14 | 0.12 | 0.12 | 0.20 |
| Clostridium cluster XIII | 0.03 | 0.03 | 0.03 | 0.01 |
| Clostridium cluster XIVa | 9.44 | 5.63 | 6.46 | 15.95 |
| Clostridium cluster XV | 0.04 | 0.04 | 0.08 | 0.12 |
| Clostridium cluster XVI | 0.06 | 0.06 | 0.06 | 0.14 |
| Clostridium cluster XVII | <0.01 | <0.01 | <0.01 | <0.01 |
| Clostridium cluster XVIII | 0.05 | 0.04 | 0.05 | 0.13 |
| Uncultured Clostridiales | 0.29 | 0.26 | 0.37 | 0.33 |
| Total | 16.81 | 10.56 | 13.23 | 44.57 |
| Cyanobacteria | <0.01 | <0.01 | <0.01 | <0.01 |
| Fusobacteria | 0.03 | 0.03 | 0.02 | 0.01 |
| Proteobacteria | 5.90 | 7.69 | 9.83 | 2.36 |
| Spirochaetes | 0.01 | 0.01 | 0.01 | <0.01 |
| Tenericutes | ||||
| Asteroleplasma species | <0.01 | <0.01 | <0.01 | <0.01 |
| Uncultured Mollicutes | 0.07 | 0.07 | 0.07 | 0.22 |
| Verrucomicrobia | 0.02 A | 0.17 B | 0.11 B | 0.01 |
Abundances of groups (approximate phylum level) are based on the HITChip analysis of the ascending (ASC) (n = 4), transverse (TR) (n = 4), and descending (DES) (n = 4) colon compartments of a stabilized Twin-SHIME (days 19 and 26) and the human fecal microbiota that was used as the inoculum for the in vitro model (n = 2) (n indicates the number of replicate experiments). Corresponding colon regions of both SHIME units showed similar abundances, so the averages from both units were calculated. The Simpson reciprocal index reflects the diversity within these communities, i.e., species richness and evenness. Values with different letters are significantly different between the different colon regions of the SHIME. The Simpson reciprocal indexes for ASC, TR, DES, and human feces are 90 ± 15, 100 ± 3, 106 ± 7, and 193 ± 11, respectively.
TABLE 2.
Abundances of bacterial groups belonging to higher taxonomic groupsa
| Higher taxonomic group | Bacterial group | P value (Kruskal-Wallis) | % abundance in: |
||
|---|---|---|---|---|---|
| ASC | TR | DES | |||
| Actinobacteria | Actinomycetaceae | 0.026 | 0.03 A | 0.02 B | 0.02 B |
| Bacteroidetes | Bacteroides fragilis-like | 0.015 | 8.41 A | 5.97 B | 5.42 B |
| Bacteroides uniformis-like | 0.023 | 11.35 A | 7.64 B | 8.12 B | |
| Alistipes-like | 0.039 | 2.80 A | 4.54 B | 4.79 B | |
| Bacteroides intestinalis-like | 0.023 | 4.55 A | 8.65 B | 6.74 AB | |
| Bacteroides plebeius-like | 0.024 | 1.92 A | 4.25 B | 4.02 B | |
| Bacteroides vulgatus-like | 0.059 | 4.08 A | 8.95 B | 7.18 AB | |
| Bacteroides splanchnicus-like | 0.873 | 2.74 A | 3.08 A | 2.95 A | |
| Firmicutes | |||||
| Clostridium cluster IV | Clostridium cellulosi-like | 0.016 | 0.03 A | 0.03 A | 0.05 B |
| Clostridium orbiscindens-like | 0.009 | 0.03 A | 0.03 A | 0.11 B | |
| Oscillospira guilliermondii-like | 0.016 | 0.03 A | 0.04 A | 0.16 B | |
| Subdoligranulum variabile-like | 0.012 | 0.04 A | 0.11 AB | 0.24 B | |
| Clostridium cluster IX | Mitsuokella multacida-like | 0.058 | 0.24 A | 0.13 B | 0.15 AB |
| Clostridium cluster XIVa | Clostridium colinum-like | 0.017 | 0.06 A | 0.03 B | 0.03 B |
| Eubacterium hallii-like | 0.015 | 0.14 A | 0.04 B | 0.05 B | |
| Eubacterium rectale-like | 0.035 | 0.08 A | 0.04 B | 0.04 B | |
| Clostridium cluster XVI | Eubacterium biforme-like | 0.090 | 0.03 AB | 0.02 A | 0.03 B |
| Proteobacteria | Oxalobacter formigenes-like | 0.029 | 0.09 A | 0.18 AB | 0.29 B |
| Burkholderia | 0.053 | 1.28 A | 3.21 AB | 5.88 B | |
| Sutterella wadsworthensis-like | 0.138 | 1.10 A | 2.14 A | 2.68 A | |
| Verrucomicrobia | Akkermansia | 0.019 | 0.05 A | 0.41 B | 0.27 B |
Abundance is based on the HITChip analysis for the ascending (ASC) (n = 4), transverse (TR) (n = 4), and descending (DES) (n = 4) colon compartments of a stabilized Twin-SHIME (days 19 and 26) (n indicates the number of replicate experiments). Corresponding colon regions of the SHIME units showed similar abundances, so the averages from both units were calculated. Nonparametric statistical tests were applied to determine the significance of the correlation of each bacterial group to a certain colon region. Boldface represents colon regions characterized by the highest abundance of a certain bacterial group. Values with different letters are significantly different.
FIG. 5.
Redundancy analysis of HITChip data at the bacterial group level of the SHIME in its three colon parts, the ascending (ASC) (n = 4), transverse (TR) (n = 4), and descending (DES) (n = 4) colon compartments (P = 0.008) (n indicates the number of replicate experiments). et rel., and related species.
To compare the data of the in vitro microbiota with the initially present microbial community, the human fecal inoculum was also analyzed with the HITChip. The microbial community of the inoculum was dominated by different Clostridium clusters (especially clusters IV and XIVa) and Bacteroidetes. Compared to the organisms in the SHIME, Firmicutes were enriched (especially Clostridium clusters IV and XIVa) in the fecal inoculum, whereas the abundance of Bacteroidetes, Proteobacteria, and Clostridium cluster IX declined (Table 1). The Simpson reciprocal index was higher for the fecal inoculum than for the SHIME compartments (193 ± 11).
Microbial community activity in a stabilized Twin-SHIME.
Corresponding colon regions of both SHIME units showed similar concentrations of metabolites and enzyme activities (P = 0.680), so the averages from both units were calculated per colon compartment (Table 3). The concentrations of these metabolic parameters were colon specific, with significant differences between the respective compartments. While succinate and lactate tended to decrease in the distal colon compartments, the other parameters increased along the different colon regions. The average acetate/propionate/butyrate ratio was 68/25/6, 65/27/8, and 66/25/9 for the ascending, transverse, and descending colon, respectively. Likewise, no significant differences in enzymatic activities were found between both SHIME units. Several enzyme activities were characteristic of specific colon regions: while glucosidase and glucuronidase activities were highest in the transverse and descending colons, nitroreductase and azoreductase activities were highest in the ascending colon. Xylosidase, arabinofuranosidase, and xylanase activities were comparable between the different regions.
TABLE 3.
Metabolite concentrations in a Twin-SHIMEa
| Metabolite or enzyme | Concn or % conversion in: |
||
|---|---|---|---|
| ASC | TR | DES | |
| Ammonium (mM) | 16.4 ± 0.9 A | 22.4 ± 1.2 B | 26.2 ± 1.4 C |
| Acetate (mM) | 31.7 ± 2.2 A | 34.5 ± 2.8 B | 38.4 ± 2.1 C |
| Propionate (mM) | 11.6 ± 1.3 A | 14.1 ± 0.6 B | 14.6 ± 1.1 B |
| Butyrate (mM) | 3.0 ± 0.8 A | 4.3 ± 0.7 B | 5.2 ± 0.7 C |
| Branched SCFA (mM) | 0.6 ± 0.1 A | 2.2 ± 0.3 B | 2.6 ± 0.1 C |
| Total SCFA (mM) | 46.7 ± 3.0 A | 55.2 ± 3.7 B | 61.3 ± 2.6 C |
| Lactate (mM) | 1.0 ± 0.3 A | 0.5 ± 0.2 B | 0.3 ± 0.3 B |
| Succinate (mM) | 3.2 ± 1.6 A | 2.7 ± 1.0 B | 2.2 ± 0.6 B |
| β-Glucosidase (U/liter) | 8.6 ± 7.3 A | 25.2 ± 3.0 B | 23.0 ± 2.1 B |
| β-Glucuronidase (U/liter) | 1.8 ± 2.1 A | 8.4 ± 3.7 B | 8.7 ± 2.0 B |
| β-d-Xylosidase (U/liter) | 64.7 ± 30.3 A | 49.4 ± 8.1 A | 36.2 ± 4.9 A |
| α-l-Arabinofuranosidase (U/liter) | 22.7 ± 21.4 A | 48.4 ± 7.2 A | 42.7 ± 7.6 A |
| Endo-1,4-xylanase (U/liter) | 2.4 ± 0.9 A | 3.0 ± 1.0 A | 3.2 ± 1.8 A |
| Nitroreductase (% conversion of p-nitro benzoic acid) | 31.6 ± 0.5 A | 9.6 ± 2.4 B | 8.4 ± 1.7 B |
| Azoreductase (% conversion of amaranth) | 94.4 ± 5.4 A | 76.9 ± 6.0 B | 76.6 ± 18.1 AB |
Concentrations are of ammonium, acetate, propionate, butyrate, branched SCFAs, total SCFA, lactate, and succinate in the ascending (ASC), transverse (TR), and descending (DES) colon compartments (10 repetitions). Average enzymatic activities are of β-glucosidase (n = 8), β-glucuronidase (n = 4), β-d-xylosidase (n = 4), α-l-arabinofuranosidase (n = 8), endo-1,4-xylanase (n = 4), nitroreductase (n = 8), and azoreductase (n = 8) in the ASC, TR, and DES colon compartments of a stabilized Twin-SHIME (days 19, 21, 23, and 26). Corresponding colon regions of the SHIME units showed similar abundances, so averages from both units were calculated. The averages ± standard deviations are shown and are within one metabolite/enzyme. Values with different letters are significantly different.
DISCUSSION
Multistage dynamic in vitro models of the human gastrointestinal tract are frequently used when evaluating strategies to steer the intestinal microbial community in a health-promoting direction (24). To compare treatments, reproducible starting points in terms of steady-state microbiota composition and metabolic activity are required. Therefore, the microbial colonization in the colon compartments of two identical multistage SHIME units was monitored for 26 days after inoculation. Analysis of the microbial community dynamics revealed an initial peak of change, due to the transit from in vivo to in vitro conditions (Fig. 3). Whereas the microbial community's composition was stable after 12 days (Fig. 3), its metabolic activity required 17 to 20 days before reaching a steady state (Fig. 4). This confirms that a minimum of 2 weeks is required for stabilization (39). Importantly, not only the final (Fig. 2 and Table 1) but also the transient microbial community compositions in the corresponding colon regions were highly similar between the two SHIME units (Fig. 2). In addition, the colon compartments of the two identical SHIME units were similar from ecological (Table 1 and Fig. 3D) and metabolic (Table 3) perspectives. We therefore conclude that multistage in vitro simulators inoculated with the same microbiota can be reproduced to contain similar microbial communities that reach compositional and functional stability after approximately 2 weeks.
Ecological analysis of the colon microbial community composition showed strong differences between the ascending colon on the one hand and the transverse/descending colon on the other hand. From the first day after inoculation, the ascending colon compartments deviated from the transverse and descending colon compartments through an immediate loss of species richness (Fig. 3). Moreover, the Simpson reciprocal index indicated that the species richness was consistently lower throughout the stabilization period (Table 1). This can be explained by the fact that the imposed conditions in the ascending colon (a lower pH and a higher bile salt concentration) strongly differ from fecal conditions. In contrast with the species richness and clustering of DGGE profiles, the functional organization of the microbial community was independent of time and colon regions. The values were indicative of well-adapted communities with an adequate species distribution, enabling the counteraction of stress and preservation of metabolic functionality (58). Despite the lack of direct phylogenetic information from DGGE profiles, the derived ecological descriptors provided a useful tool to deliver additional information on community dynamics, structure, and functioning. These items tend to be overlooked with conventional culture-based detection methods (2, 25, 34).
In vivo validation of in vitro microbial community development has been a common objective of many research studies (25, 32, 33), yet the applied molecular tools have often had insufficient resolving power. High-resolution HITChip analysis showed that in vitro microbial populations are very diverse (Table 1) but still less diverse than the inoculum, indicating a certain degree of specialization under in vitro conditions. First, an in vitro enrichment of Clostridium cluster IX (propionate producers) (56) but not of clusters IV and XIVa (butyrate producers) (23) was observed. This finding was supported by in vitro molar SCFA ratios, which were similar to in vivo ratios (57/21/22, acetate-propionate-butyrate) (9) but displayed higher propionate than butyrate levels. Such mild changes in SCFA profile have been observed previously (2). Second, the in vitro microbiota was enriched in Proteobacteria and Bacteroidetes but not in Firmicutes. This corresponds to earlier findings with the HITChip, with which a loss of Firmicutes as opposed to Bacteroides groups was observed during the initial in vivo-in vitro shift in the dynamic TNO intestinal model (TIM) (41). Moreover, more conventional molecular tools, such as culture-based techniques (2, 25), Q-PCR (17, 29), and DGGE (39), already determined the in vitro dominance of Bacteroidetes. There are several reasons for the discrepancy in Firmicutes/Bacteroidetes ratios between the in vitro gut model and the inoculum. First, in vitro systems may contain higher micromolar levels of oxygen, which could potentially favor aerotolerant microbes, such as Bacteroides species (5, 43) or Proteobacteria. Second, the high carbohydrate content of the in vitro SHIME nutritional medium may promote the proliferation of Bacteroidetes, which possess a larger glycobiome than Firmicutes (28). This opens perspectives to restore the Bacteroidetes/Firmicutes ratio by alteration of the SHIME feed. Third, in vitro simulators often lack the simulation of mucosal adhesion sites that are considered important for prolonged colonization. Recently, it has been proposed that the mucous layer is a protected microenvironment, containing a backup of microbes that can colonize the lumen and restore associations after luminal perturbations (P. Van den Abbeele, S. Possemiers, W. Verstraete, and T. Van de Wiele, submitted for publication). In contrast to Firmicutes, Bacteroides spp. are less capable of mucosal adhesion (31). The absence of adhesion as a selective advantage may therefore result in their higher abundance in vitro than in an in vivo gut environment. In vitro models could therefore be improved by including mucous layer simulations, such as mucin-agar tubes (26) or mucin beads (40). Finally, the selective pressure of the host on the microbiota is only partly simulated in vitro. Both retention time (6, 25) and pH (56), in addition to a variety of more specific defense mechanisms, can significantly change microbial communities, with the innate and adaptive immunity being most important (1, 7).
Application of the HITChip provided detailed characterization of the microbiota in the respective colon compartments of the SHIME. Overall, no significant differences at the phylum level were noted between colon regions (Table 1). Yet, RDA at the bacterial group level revealed that microbiota in the proximal (ascending) and distal (transverse/descending) colon compartments cluster separately (Fig. 5). From the 131 bacterial groups detected, 20 groups explained around 20% of the differences between colon regions, which is a higher percentage than expected from a flowthrough setup (Table 2). Interestingly, the colon specificity of some bacterial groups could be related to the metabolic activities of the bacteria. Although the lack of an absorption module in the SHIME gut model leads to accumulation of SCFAs in the distal colon compartment, calculation of the net SCFA production resulted in the highest values in the ascending colon (Fig. 5). This indicates it to be the main site of carbohydrate fermentation. The bacterial groups that correlated to the ascending colon should thus typically be involved in carbohydrate fermentation (e.g., Bacteroides spp. and Eubacterium spp.). Rapid fermentation by these groups could deplete essential growth factors and thus negatively affect microbes that grow on more difficult substrates, for instance, mucins. Akkermansia muciniphila, a recently isolated mucin degrader (12), is indeed virtually absent in the ascending colon yet abundantly present in the transverse and descending colon compartments (Table 2), where its presence could relate to the distally increased glucuronidase activity (11). In addition, four Bacteroides groups specifically correlated with the transverse colon. As Bacteroides species are also potent mucin degraders (8, 45, 47), one may therefore assume that mucin degradation may be a more important process in the transverse colon than in the ascending colon. Alternatively, distally occurring microbiota could participate in the distal fermentation of arabinose- and/or xylose-containing polysaccharides (xylan or arabinogalactan), as arabinofuranosidase and xylanase also increased in the distal regions (3, 17). These data illustrate the potency of HITChip analysis in revealing relationships between microbial community phylogenetics and specific metabolic activities.
In conclusion, our study shows that well-controlled multicompartment gastrointestinal in vitro models display a reproducible microbial colonization and community development upon inoculation. Moreover, phylogenetic microarray analysis evidenced the colon region specificity of the colonization process, with the proximal regions harboring saccharolytic microbes and the distal regions harboring mucin-degrading microbes (e.g., Akkermansia spp.). As a consequence, once the respective microbial populations have reached functional steady state, such gut models will thus enable placebo-controlled in vitro studies or direct comparison of the modulatory effects of two different treatments for the gut microbiota. Importantly, the shift from an in vivo to an in vitro environment resulted in an increased Bacteroidetes/Firmicutes ratio, while Clostridium cluster IX (propionate producers) was enriched compared to clusters IV and XIVa (butyrate producers), supported by proportionally higher in vitro propionate concentrations. These findings illustrate the potency of high-resolution microbial analytical techniques in revealing the importance of hitherto less explored gut parameters, such as the presence of a mucosal surface, that may need to be incorporated when new or modified dynamic gastrointestinal simulators are developed.
Supplementary Material
Acknowledgments
We thank Ellen Van Gysegem and Diederik Van Driessche for technical assistance, Thomas Van Leeuwen and Thomas Soin of the Department of Crop Protection for the disposal of the spectrophotometer, and Hans Heilig, Wilma Akkermans, Ineke Heikamp, and Sebastian Tims for technical assistance with the HITChip.
Tom Van de Wiele and Sam Possemiers are postdoctoral fellows, and Pieter Van den Abbeele is a Ph.D. student from FWO—Vlaanderen (Research Foundation of Flanders, Belgium). Massimo Marzorati is a postdoctoral fellow, and Charlotte Grootaert is a Ph.D. student from the IWT (Agency for Innovation by Science and Technology in Flanders, Belgium).
This work was financially supported by a Tournesol project (T2007.11) from the Flemish Government and a GOA project (BOF07/GOA/002) from Ghent University.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 2.Allison, C., C. McFarlan, and G. T. Macfarlane. 1989. Studies on mixed populations of human intestinal bacteria grown in single-stage and multistage continuous culture systems. Appl. Environ. Microbiol. 55:672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Tamimi, M. A. H. M., R. J. Palframan, J. M. Cooper, G. R. Gibson, and R. A. Rastall. 2006. In vitro fermentation of sugar beet arabinan and arabino-oligosaccharides by the human gut microflora. J. Appl. Microbiol. 100:407-414. [DOI] [PubMed] [Google Scholar]
- 4.Boon, N., E. M. Top, W. Verstraete, and S. D. Siciliano. 2003. Bioaugmentation as a tool to protect the structure and function of an activated-sludge microbial community against a 3-chloroaniline shock load. Appl. Environ. Microbiol. 69:1511-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brioukhanov, A. L., and A. I. Netrusov. 2007. Aerotolerance of strictly anaerobic microorganisms and factors of defense against oxidative stress: a review. Appl. Biochem. Microbiol. 43:567-582. [PubMed] [Google Scholar]
- 6.Child, M. W., A. Kennedy, A. W. Walker, B. Bahrami, S. Macfarlane, and G. T. Macfarlane. 2006. Studies on the effect of system retention time on bacterial populations colonizing a three-stage continuous culture model of the human large gut using FISH techniques. FEMS Microbiol. Ecol. 55:299-310. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, M. D., and M. N. Alder. 2006. The evolution of adaptive immune systems. Cell 124:815-822. [DOI] [PubMed] [Google Scholar]
- 8.Corfield, A. P., S. A. Wagner, J. R. Clamp, M. S. Kriaris, and L. C. Hoskins. 1992. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 60:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cummings, J. H., E. W. Pomare, W. J. Branch, C. P. Naylor, and G. T. Macfarlane. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Boever, P., B. Deplancke, and W. Verstraete. 2000. Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem is improved by supplementing a soygerm powder. J. Nutr. 130:2599-2606. [DOI] [PubMed] [Google Scholar]
- 11.Derrien, M. 2007. Ph.D. thesis. Wageningen University, Wageningen, Netherlands.
- 12.Derrien, M., E. E. Vaughan, C. M. Plugge, and W. M. de Vos. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54:1469-1476. [DOI] [PubMed] [Google Scholar]
- 13.Egert, M., A. A. de Graaf, H. Smidt, W. M. de Vos, and K. Venema. 2006. Beyond diversity: functional microbiomics of the human colon. Trends Microbiol. 14:86-91. [DOI] [PubMed] [Google Scholar]
- 14.Flint, H. J., E. A. Bayer, M. T. Rincon, R. Lamed, and B. A. White. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121-131. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, G. R., H. M. Probert, J. Van Loo, R. A. Rastall, and M. B. Roberfroid. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 17:259-275. [DOI] [PubMed] [Google Scholar]
- 16.Goldbarg, J. A., and A. M. Rutenburg. 1958. The colorimetric determination of leucine aminopeptidase in urine and serum of normal subjects and patients with cancer and other diseases. Cancer 11:283-291. [DOI] [PubMed] [Google Scholar]
- 17.Grootaert, C., P. Van den Abbeele, M. Marzorati, W. F. Broekaert, C. M. Courtin, J. A. Delcour, W. Verstraete, and T. Van de Wiele. 2009. Comparison of prebiotic effects of arabinoxylan oligosaccharides and inulin in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 69:231-242. [DOI] [PubMed] [Google Scholar]
- 18.Gudiel-Urbano, M., and I. Goni. 2002. Effect of short-chain fructooligosaccharides and cellulose on cecal enzyme activities in rats. Ann. Nutr. Metab. 46:254-258. [DOI] [PubMed] [Google Scholar]
- 19.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 20.Hooper, L. V., T. S. Stappenbeck, C. V. Hong, and J. I. Gordon. 2003. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4:269-273. [DOI] [PubMed] [Google Scholar]
- 21.Hope, A. C. A. 1968. A simplified Monte Carlo significance test procedure. J. R. Stat. Soc. Series B Stat. Methodol. 30:582-598. [Google Scholar]
- 22.Lepš, J., and P. Šmilauer. 2003. Multivariate analysis of ecological data using CANOCO. Cambridge University Press, Cambridge, MA.
- 23.Louis, P., and H. J. Flint. 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294:1-8. [DOI] [PubMed] [Google Scholar]
- 24.Macfarlane, G. T., and S. Macfarlane. 2007. Models for intestinal fermentation: association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr. Opin. Biotechnol. 18:156-162. [DOI] [PubMed] [Google Scholar]
- 25.Macfarlane, G. T., S. Macfarlane, and G. R. Gibson. 1998. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 35:180-187. [DOI] [PubMed] [Google Scholar]
- 26.Macfarlane, S., E. J. Woodmansey, and G. T. Macfarlane. 2005. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl. Environ. Microbiol. 71:7483-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magurran, E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ.
- 28.Mahowald, M. A., F. E. Rey, H. Seedorf, P. J. Turnbaugh, R. S. Fulton, A. Wollam, N. Shah, C. Y. Wang, V. Magrini, R. K. Wilson, B. L. Cantarel, P. M. Coutinho, B. Henrissat, L. W. Crock, A. Russell, N. C. Verberkmoes, R. L. Hettich, and J. I. Gordon. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U. S. A. 106:5859-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mäkeläinen, H., O. Hasselwander, N. Rautonen, and A. C. Ouwehand. 2009. Panose, a new prebiotic candidate. Lett. Appl. Microbiol. 49:666-672. [DOI] [PubMed] [Google Scholar]
- 30.Marzorati, M., L. Wittebolle, N. Boon, D. Daffonchio, and W. Verstraete. 2008. How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ. Microbiol. 10:1571-1581. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto, M., H. Tani, H. Ono, H. Ohishi, and Y. Benno. 2002. Adhesive property of Bifidobacterium lactis LKM512 and predominant bacteria of intestinal microflora to human intestinal mucin. Curr. Microbiol. 44:212-215. [DOI] [PubMed] [Google Scholar]
- 32.Minekus, M., M. Smeets-Peeters, A. Bernalier, S. Marol-Bonnin, R. Havenaar, P. Marteau, M. Alric, G. Fonty, and J. Veld. 1999. A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl. Microbiol. Biotechnol. 53:108-114. [DOI] [PubMed] [Google Scholar]
- 33.Molly, K., M. Vandewoestyne, I. Desmet, and W. Verstraete. 1994. Validation of the simulator of the human intestinal microbial ecosystem (SHIME) reactor using microorganism-associated activities. Microb. Ecol. Health Dis. 7:191-200. [Google Scholar]
- 34.Molly, K., M. V. Woestyne, and W. Verstraete. 1993. Development of a 5-step multichamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 39:254-258. [DOI] [PubMed] [Google Scholar]
- 35.Nollet, L., I. VandeVelde, and W. Verstraete. 1997. Effect of the addition of Peptostreptococcus productus ATCC35244 on the gastro-intestinal microbiota and its activity, as simulated in an in vitro simulator of the human gastro-intestinal tract. Appl. Microbiol. Biotechnol. 48:99-104. [DOI] [PubMed] [Google Scholar]
- 36.Possemiers, S., S. Bolca, C. Grootaert, A. Heyerick, K. Decroos, W. Dhooge, D. De Keukeleire, S. Rabot, W. Verstraete, and T. Van de Wiele. 2006. The prenylflavonoid isoxanthohumol from hops (Humulus lupulus L.) is activated into the potent phytoestrogen 8-prenylnaringenin in vitro and in the human intestine. J. Nutr. 136:1862-1867. [DOI] [PubMed] [Google Scholar]
- 37.Possemiers, S., C. Grootaert, J. Vermeiren, G. Gross, M. Marzorati, W. Verstraete, and T. Van de Wiele. 2009. The intestinal environment in health and disease—recent insights on the potential of intestinal bacteria to influence human health. Curr. Pharm. Des. 15:2051-2065. [DOI] [PubMed] [Google Scholar]
- 38.Possemiers, S., S. Rabot, J. C. Espin, A. Bruneau, C. Philippe, A. Gonzalez-Sarrias, A. Heyerick, F. A. Tomas-Barberan, D. De Keukeleire, and W. Verstraete. 2008. Eubacterium limosum activates isoxanthohumol from hops (Humulus lupulus L.) into the potent phytoestrogen 8-prenylnaringenin in vitro and in rat intestine. J. Nutr. 138:1310-1316. [DOI] [PubMed] [Google Scholar]
- 39.Possemiers, S., K. Verthe, S. Uyttendaele, and W. Verstraete. 2004. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 49:495-507. [DOI] [PubMed] [Google Scholar]
- 40.Probert, H. M., and G. R. Gibson. 2004. Development of a fermentation system to model sessile bacterial populations in the human colon. Biofilms 1:13-19. [Google Scholar]
- 41.Rajilic-Stojanovic, M. 2007. Ph.D. thesis. Wageningen University, Wageningen, Netherlands.
- 42.Rajilic-Stojanovic, M., H. Heilig, D. Molenaar, K. Kajander, A. Surakka, H. Smidt, and W. M. de Vos. 2009. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ. Microbiol. 11:1736-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rolfe, R. D., D. J. Hentges, B. J. Campbell, and J. T. Barrett. 1978. Factors related to oxygen tolerance of anaerobic bacteria. Appl. Environ. Microbiol. 36:306-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rycroft, C. E., M. R. Jones, G. R. Gibson, and R. A. Rastall. 2001. A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 91:878-887. [DOI] [PubMed] [Google Scholar]
- 45.Salyers, A. A., J. R. Vercellotti, S. E. West, and T. D. Wilkins. 1977. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 33:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seth, A., F. Yan, D. B. Polk, and R. K. Rao. 2008. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 294:G1060-G1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanley, R. A., S. P. Ram, R. K. Wilkinson, and A. M. Roberton. 1986. Degradation of pig gastric and colonic mucins by bacteria isolated from the pig colon. Appl. Environ. Microbiol. 51:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stappenbeck, T. S., L. V. Hooper, and J. I. Gordon. 2002. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U. S. A. 99:15451-15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turnbaugh, P. J., R. E. Ley, M. A. Mahowald, V. Magrini, E. R. Mardis, and J. I. Gordon. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027-1131. [DOI] [PubMed] [Google Scholar]
- 50.Turnbaugh, P. J., V. K. Ridaura, J. J. Faith, F. E. Rey, R. Knight, and J. I. Gordon. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1:6ra14. [DOI] [PMC free article] [PubMed]
- 51.Van den Abbeele, P., C. Grootaert, S. Possemiers, W. Verstraete, K. Verbeken, and T. Van de Wiele. 2009. In vitro model to study the modulation of the mucin-adhered bacterial community. Appl. Microbiol. Biotechnol. 83:349-359. [DOI] [PubMed] [Google Scholar]
- 52.Van de Wiele, T., N. Boon, S. Possemiers, H. Jacobs, and W. Verstraete. 2004. Prebiotic effects of chicory inulin in the simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 51:143-153. [DOI] [PubMed] [Google Scholar]
- 53.Van de Wiele, T., L. Vanhaecke, C. Boeckaert, K. Peru, J. Headley, W. Verstraete, and S. Siciliano. 2005. Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environ. Health Perspect. 113:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van de Wiele, T. R., W. Verstraete, and S. D. Siciliano. 2004. Polycyclic aromatic hydrocarbon release from a soil matrix in the in vitro gastrointestinal tract. J. Environ. Qual. 33:1343-1353. [DOI] [PubMed] [Google Scholar]
- 55.Vulevic, J., R. A. Rastall, and G. R. Gibson. 2004. Developing a quantitative approach for determining the in vitro prebiotic potential of dietary oligosaccharides. FEMS Microbiol. Lett. 236:153-159. [DOI] [PubMed] [Google Scholar]
- 56.Walker, A. W., S. H. Duncan, E. C. M. Leitch, M. W. Child, and H. J. Flint. 2005. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl. Environ. Microbiol. 71:3692-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wise, A., A. K. Mallett, and I. R. Rowland. 1982. Dietary fiber, bacterial metabolism and toxicity of nitrate in the rat. Xenobiotica 12:111-118. [DOI] [PubMed] [Google Scholar]
- 58.Wittebolle, L., M. Marzorati, L. Clement, A. Balloi, D. Daffonchio, K. Heylen, P. De Vos, W. Verstraete, and N. Boon. 2009. Initial community evenness favours functionality under selective stress. Nature 458:623-626. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, H. S., J. K. DiBaise, A. Zuccolo, D. Kudrna, M. Braidotti, Y. S. Yu, P. Parameswaran, M. D. Crowell, R. Wing, B. E. Rittmann, and R. Krajmalnik-Brown. 2009. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106:2365-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.