Abstract
Aggregation-promoting factors (Apf) are secreted proteins that have been associated with a diverse number of functional roles in lactobacilli, including self-aggregation, the bridging of conjugal pairs, coaggregation with other commensal or pathogenic bacteria, and maintenance of cell shape. In silico genome analysis of Lactobacillus acidophilus NCFM identified LBA0493 as a 696-bp apf gene that encodes a putative 21-kDa Apf protein. Transcriptional studies of NCFM during growth in milk showed apf to be one of the most highly upregulated genes in the genome. In the present study, reverse transcriptase-quantitative PCR (RT-QPCR) analysis revealed that the apf gene was highly induced during the stationary phase compared to that during the logarithmic phase. To investigate the functional role of Apf in NCFM, an Δapf deletion mutant was constructed. The resulting Δapf mutant, NCK2033, did not show a significant difference in cell morphology or growth compared to that of the NCFMΔupp reference strain, NCK1909. The autoaggregation phenotype of NCK2033 in planktonic culture was unaffected. Additional phenotypic assays revealed that NCK2033 was more susceptible to treatments with oxgall bile and sodium dodecyl sulfate (SDS). Survival rates of NCK2033 decreased when stationary-phase cells were exposed to simulated small-intestinal and gastric juices. Furthermore, NCK2033 in the stationary phase showed a reduction of in vitro adherence to Caco-2 intestinal epithelial cells, mucin glycoproteins, and fibronectin. The data suggest that the Apf-like proteins may contribute to the survival of L. acidophilus during transit through the digestive tract and, potentially, participate in the interactions with the host intestinal mucosa.
Lactobacillus species are phylogenetically diverse and are found naturally in milk, plants, meats, and the mucosal surfaces (oral, intestinal, and reproductive tracts) of humans and animals. They also represent one of the most prominent groups of microorganisms, having been used for centuries in the bioprocessing of foods, notably in fermented dairy products, vegetables and meats and in sourdough. Due to their explored gastrointestinal (GI) health-promoting properties, certain strains of lactobacilli, mostly of human origin, have been used as probiotic adjuncts in cultured dairy and functional food products. Lactobacillus acidophilus is among the most widely used probiotic species in yogurt and fermented milks. The probiotic functionality of L. acidophilus has been well documented both in vitro and in vivo, and it includes the alleviation of lactose intolerance, maintenance of the GI microflora balance, immunomodulation via stimulation of host cytokine expression and serum immunoglobulin A (IgA), exclusion of pathogenic microorganisms, and alleviation of cold-like symptoms in children (16, 18, 20, 27, 32, 36).
The ability of probiotic bacteria to form cellular aggregates is considered a desirable characteristic, as they can potentially inhibit adherence of pathogenic bacteria to intestinal mucosa either by forming a barrier via self-aggregation or coaggregation with commensal organisms on the intestinal mucosa or by direct coaggregation with the pathogens to facilitate clearance (12, 41). In addition, studies have suggested aggregation as an important mechanism for genetic exchange, adhesion, and colonization in the host environments, as well as immunomodulation of colonic mucosa (13, 39, 46, 48). In general, the mechanism of cellular aggregation in probiotic lactobacilli remains unclear, although it appears that the aggregation phenomenon is a result of complex interactions among components on the cell surface, such as proteins, glycoproteins, techoic or lipotechoic acids (LTA), and secreted factors. Enzymes such as glycosyltransferases and a putative DEAD-box helicase from Lactobacillus reuteri have also been implicated as mediators of cell aggregation (35, 40, 47).
Aggregation-promoting factors (Apf) and the genes encoding them have previously been characterized for several Lactobacillus species, including Lactobacillus crispatus, Lactobacillus johnsonii, Lactobacillus gasseri, Lactobacillus paracasei, and Lactobacillus coryniformis. Some of these Apf proteins serve as major factors in aggregation, whereas others do not appear to be directly involved in aggregation. L. gasseri 2459, a vaginal isolate, produced a 2-kDa pheromone-like, hydrophilic peptide that mediates autoaggregation and induces aggregation of specific strains of Lactobacillus plantarum and Enterococcus faecalis (10). A surface coaggregation-promoting factor (Cpf) was identified in a silage strain, L. coryniformis DSM 20001T, which mediates coaggregation with specific pathogenic strains of Escherichia coli and campylobacter (41). An earlier study by Reniero et al. (39) showed that a putative 32-kDa Apf protein from L. gasseri 4B2 contributes to cell aggregation and enhanced conjugation efficiency. Subsequent studies, however, failed to functionally correlate this Apf protein with aggregation phenotype (26, 45). The authors demonstrated that the Apf proteins in 4B2, encoded by apf1 and apf2, had characteristics of S-layer proteins and contributed to the maintenance of cell shape in L. gasseri. Similarly, the Apf proteins from L. crispatus M247 and L. paracasei BGSJ2-8 did not restore the aggregation phenotype of their respective nonaggregative mutants (33, 34).
L. acidophilus NCFM (6) exhibits a strong autoaggregation phenotype and has previously been demonstrated to efficiently coaggregate with pathogenic strains of Clostridium histolyticum and Staphylococcus aureus in vitro (15). In silico analysis of the NCFM genome sequence revealed the presence of a putative apf gene (LBA0493). Previous microarray gene expression studies have also showed that apf was consistently expressed at high levels when NCFM was grown on different carbohydrate sources, and notably during growth in milk, suggesting some physiological importance of the encoded protein (5, 8). The objective of the present study was to investigate the functional roles of the apf gene in L. acidophilus NCFM. Mutational analysis demonstrated that the apf gene potentially plays important roles in the probiotic functionality of L. acidophilus NCFM, specifically, in bile and acid tolerance and in adherence to the intestinal mucosa.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are summarized in Table 1. L. acidophilus strains were propagated in de Man-Rogosa-Sharpe (MRS) broth (Difco Laboratories, Inc., Detroit, MI) statically under aerobic conditions or on MRS agar (1.5% [wt/vol]; Difco) under anaerobic conditions at 37°C or 42°C, as indicated below. Recombinant strains were selected in the presence of 2 μg/ml of erythromycin (Em) (Sigma-Aldrich, St. Louis, MO) and/or 2 to 5 μg/ml of chloramphenicol (Cm) (Sigma) when appropriate. For upp-based counterselectable gene replacement procedures (23), plasmid-free double recombinants were selected on a semidefined agar medium containing 2% (wt/vol) glucose (GSDM) (28) and 100 μg/ml of 5-fluorouracil (5-FU) (Sigma) as described previously (23). Escherichia coli strains were grown in brain heart infusion (BHI) (Difco) medium at 37°C with aeration. E. coli EC101 (31) was propagated in the presence of 40 μg/ml of kanamycin (Kn). When necessary, Em was added at a final concentration of 150 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and PCR primers used in this study
| Strain, plasmid, or primer | Genotype or characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| L. acidophilus | ||
| NCFM | Human intestinal isolate | 6 |
| NCK1909 | NCFM carrying a 315-bp in-frame deletion within the upp gene | 23 |
| NCK1910 | NCK1909 harboring pTRK669; host for pORI-based counterselective integration vector | 23 |
| NCK2033 | NCK1909 carrying a 606-bp in-frame deletion within the apf gene | This study |
| E. coli | ||
| EC101 | RepA+ JM101; Kmr; repA from pWV01 integrated in chromosome; host for pORI-based plasmids | 31 |
| Plasmids | ||
| pTRK935 | 3.0 kb; pORI28 with a upp expression cassette and the lacZ′ from pUC19 cloned into BglII/XbaI sites; serves as counterselective integration vector | 23 |
| pTRK977 | 4.4 kb; pTRK935 with a mutated copy of apf cloned into BamHI/SacI sites | This study |
| Primers | ||
| Construction of Δapf deletion mutant | ||
| apf-1 | GTATTAGGATCCGCAGGAGCAGACGTGATTTT | This study |
| apf-2 | AGCAGCTACTGCAATTGACTT | This study |
| apf-3 | AGTCAATTGCAGTAGCTGCTTGGACTGCTGCTAAGGCATT | This study |
| apf-4 | TTATGTAGAGCTCCTTTCCCCTTTAGCAACACG | This study |
| PCR analysis and DNA sequencing of deletion targets | ||
| apf-5 | GGAAGCGGCGTAACAAATAA | This study |
| apf-6 | AAGGATGCAAACCAAGTTGC | This study |
| RT-QPCR analysis of apf gene | ||
| apf-F | TGATGCTAGCGTAGTTACAG | This study |
| apf-R | CATTGGTTCTTGCCTAAGTC | This study |
| 383-F | GCAAATCCTATCAGGTAATC | 37 |
| 383-R | ACCGTCTACTACTTTCAAC | 37 |
For primers, the 5′-to-3′ sequences are given and restriction enzyme sites are underlined.
DNA manipulation and transformation.
Genomic and plasmid DNA isolation, standard DNA techniques, and transformation were performed as described previously (23). PCR primers (Table 1) were synthesized by Integrated DNA Technologies (Coralville, IA). For cloning purposes, PCR amplicons were generated using PfuUltra II Fusion HS DNA polymerase (Stratagene Corp., La Jolla, CA) based on the manufacturer's instructions.
Sequence analysis.
Deduced protein sequences were compared against the nonredundant protein database using BlastP (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Theoretical physical and chemical properties of the deduced proteins were assessed using the ProtParam tool (http://au.expasy.org/tools/protparam.html). The signal peptide and transmembrane domain were predicted using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (9) and TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), respectively. Multiple alignments of protein sequences were carried out in ClustalX v2.0.12 with the BLOSUM series protein weight matrix, and clustering was performed by the neighbor-joining method (30). An unrooted phylogenetic tree was constructed with ClustalX v2.0.12 and visualized using the MEGA 4.0.2 software package (43).
RT-QPCR.
L. acidophilus NCFM was propagated in MRS broth using a 2% inoculum from an overnight culture. Cells were collected during the log phase (optical density at 600 nm [OD600] of 0.6 to 0.8) and stationary phase (16 h of growth) by centrifugation at 1,717 × g for 5 min at room temperature. Cell pellets were flash-frozen in an ethanol-dry ice bath and stored at −80°C. Total RNA was extracted, treated with DNase, and purified as described previously (24). Samples were collected from two independent experimental replicates (representing two biological replicates). The absence of genomic DNA in purified RNA samples was verified by PCR using NCFM gene-specific primers. Reverse transcriptase-quantitative PCR (RT-QPCR) was performed using an iScript one-step RT-PCR kit with SYBR green (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's suggestions, with each reaction scaled down to a total volume of 25 μl containing 50 ng of RNA template and a 300 nM final concentration of each primer. Primer pairs apf-F/apf-R (Table 1) and 383-F/383-R (37) were used for transcript amplification of apf and LBA0383 (DNA polymerase III, delta subunit), respectively. RT-QPCR was performed with an iCycler MyiQ single-color detection system (Bio-Rad). Data were analyzed using iCycler MyiQ software v1.0 (Bio-Rad). The number of threshold cycle per well was determined using the auto-calculated “threshold cycle calculation” and “PCR baseline subtracted curve fit” analysis mode. Transcript copy numbers of apf and LBA0383 were then quantified from the apf and LBA0383 standard curves, respectively, generated from known concentrations of PCR products. The correlation coefficients for the standard curves and PCR efficiencies for apf and LBA0383 were 0.999 and 1.000, and 82.1% and 88.0%, respectively. The transcript copy number of apf was normalized to the mRNA copy number of LBA0383 (38).
Deletion of the apf gene.
The apf gene from L. acidophilus NCFM was deleted using a upp-based counterselectable gene replacement system (23). Briefly, a 606-bp in-frame deletion within the apf gene was constructed by first amplifying a 703-bp and a 675-bp DNA segment flanking the region upstream and downstream of the deletion target, respectively, using apf-1/apf-2 and apf-3/apf-4 primer pairs (Table 1). Both purified PCR products were fused and amplified to generate copies of the Δapf allele via splicing by overlap extension PCR (SOE-PCR) (25), using 10 ng of each PCR product as amplification templates in a 50-μl PCR with the apf-1/apf-4 primer pair. All PCRs were performed with 25 to 30 amplification cycles. The purified SOE-PCR product was digested with BamHI and SacI and ligated into the pTRK935 counterselectable integration vector. The ligation mixture was transformed into E. coli EC101 with selection on BHI agar containing Kn, Em, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and isopropyl-β-d-thiogalactopyranoside (IPTG). The resulting recombinant plasmid carrying the Δapf allele, pTRK977, was electroporated into the L. acidophilus Δupp host strain harboring pTRK669, NCK1910 (Table 1). One Emr Cmr transformant carrying both plasmids was grown overnight in MRS broth containing 2 μg/ml each of Em and Cm and transferred three times (1% inoculum) in MRS broth with Em (ca. 30 generations) in a 42°C water bath. Chromosomal integrants were selected by replica plating onto MRS agar supplemented with Em or 5 μg/ml Cm. One Emr Cms pTRK977 integrant was selected and propagated in MRS broth with Em at 37°C overnight, followed by 1 transfer (1% inoculum) in MRS broth without Em (ca. 10 generations). To recover plasmid-free recombinants, the culture was plated at a 10−3 dilution on GSDM supplemented with 5-FU and incubated at 37°C anaerobically for 48 to 72 h until colonies were visible. Double recombinants with Δapf allele were screened by colony PCR using the apf-5/apf-6 primer pair (Table 1) that specifically anneals to the flanking region of the apf gene. In-frame deletion and sequence integrity were confirmed by DNA sequencing using the apf-5/apf-6 primer pair.
Stress challenge assays.
Strains were grown in MRS broth to an OD600 of 0.25 to 0.3 (early log phase) before being subjected to a stress challenge (4). The cultures were divided into equal-volume portions and harvested at room temperature. Cells were resuspended in equal volume of (i) MRS broth with 2.5% (wt/vol) oxgall bile (Difco), (ii) MRS broth with 10% (wt/vol) NaCl, or (iii) acidified MRS broth at pH 3.5 (adjusted with lactic acid) or at pH 2.0 (adjusted with HCl). Viable cell counts were determined after a 1- or 2-h incubation at 37°C by diluting and plating onto MRS agar. For sodium dodecyl sulfate (SDS) and Triton X-100 treatment assays, early-log-phase cultures were inoculated at 1% into MRS broth containing 0.01% or 0.02% (wt/vol) of SDS, or 0.25% or 0.50% (vol/vol) of Triton X-100. Growth was monitored by OD600 after a 24-h incubation at 37°C.
Survival in acidified milk.
The assay was performed essentially as described previously (5). Briefly, MRS-grown overnight cultures were pelleted, washed twice with phosphate-buffered saline (PBS; Invitrogen, Carlsbad, CA), pH 7.4, and resuspended in the same buffer. Approximately 5 × 108 to 8 × 108 CFU of each culture was inoculated into 50-ml aliquots of 11% skim milk (SM) acidified to pH 4.5 with lactic acid. The samples were stored at 4°C for 2 weeks. Viable cells were determined every 48 h during the 2-week period by diluting 1-ml aliquots and plating them on MRS agar.
Exposure to simulated gastric and small-intestinal juices.
Simulated gastric and small-intestinal juices were prepared essentially as described previously (14, 21). Briefly, overnight cultures were pelleted, washed twice, and resuspended in sterile distilled water. The cell suspension (0.2 ml) was mixed with 1 ml of freshly prepared simulated gastric juice (0.5% [wt/vol] NaCl solution containing 3 g/liter pepsin [Fisher Scientific, Pittsburg, PA], pH 2.0) or small-intestinal juice (0.5% NaCl solution containing 1 g/liter pancreatin [Sigma] and 3 g/liter oxgall [Difco], pH 8.0) and incubated at 37°C. The viable cell count was determined by plating onto MRS agar at 30-min or 1-h intervals.
Adherence assays.
Caco-2 (ATCC HTB-37) epithelial cell adherence assays were performed as described previously (23) using MRS medium-grown stationary-phase cells (16 h of growth). Washed cells were resuspended in PBS and adjusted to an OD600 equivalent to ∼1 × 108 CFU/ml prior to adding them into each well containing a Caco-2 cell monolayer. For adherence to extracellular matrix (ECM) components, collagen (type IV from human cell culture; Sigma), laminin (from Engelbreth-Holm-Swarm murine sarcoma/basement membrane; Sigma), or fibronectin (from human plasma; Sigma) was diluted in 50 mM carbonate-bicarbonate buffer, pH 9.6 (Sigma), to a final concentration of 10 μg/ml. Mucin (type III from porcine stomach; Sigma) was dissolved in PBS at 10 mg/ml. Nunc Maxisorp 96-well microplate wells were coated with the protein solutions (100 μl/well) by incubation at 4°C overnight. After two washes with PBS to remove excess protein substrates, the wells were blocked with 2% bovine serum albumin (BSA) solution (Invitrogen) for 2 h at 37°C, followed by two additional washes with PBS to remove excess BSA. To prepare bacterial cells for the adherence assay, MRS medium-grown stationary-phase cultures were pelleted, washed once with PBS (pH 5.0), and resuspended in the same PBS buffer in order to mimic the pH condition of the culture supernatant during stationary phase (pH ∼4.0 to 4.5) when cells were harvested. The cell density was adjusted to an OD600 equivalent to ∼1 × 108 CFU/ml, and 100 μl of the cell suspension was added to each protein-coated well. After 1-h incubation at 37°C, the wells were washed five times with PBS (pH 6.0; 200 μl/well). Adhered cells were recovered by treating each well with 100 μl of 0.05% Triton X-100 solution for 15 min with agitation. Cell suspensions were diluted and plated on MRS agar to enumerate adhered cells (expressed as the percent relative adherence of mutant in CFU/parent in CFU). All adherence experiments were performed with at least four independent cultures, each with quadruplicate wells containing individual substrate layers.
RESULTS
In silico analysis of apf.
Analysis of the genome sequence of L. acidophilus NCFM (1) identified the presence of a gene, designated apf (LBA0493), that encodes an Apf ortholog (GenBank accession no. YP_193409) with 231 amino acids in length. The apf gene is flanked by two terminators, indicating its transcription as a monocistronic mRNA. Its proximal region shares conserved gene order with the corresponding apf loci in L. acidophilus ATCC 4796 (GenBank accession no. HMPREF0492_0847), Lactobacillus ultunensis (GenBank accession no. HMPREF0548_0528), and L. crispatus (GenBank accession no. HMPREF0506_0073). An N-terminal signal peptide cleavage site is predicted at position 34 and 35 (AQA-AT) of the putative Apf precursor. The resulted mature Apf has a predicted size of 21 kDa and a basic isoelectric point (pI ∼9) and is constituted of 46.1% hydrophobic residues. No transmembrane domain was found in the deduced Apf protein sequence.
BlastP analysis showed that the Apf protein from L. acidophilus shared a high level of sequence homology to the corresponding proteins from Lactobacillus helveticus (76% identity; GenBank accession no. ZP_05753058 and YP_001577001), L. ultunensis (75% identity; GenBank accession no. ZP_04010810), and strains of L. crispatus (62 to 70% identity) and had moderate sequence homology (40% to 55% identity) to the Apf proteins from L. gasseri (Apf1), L. johnsonii (Apf1), Lactobacillus jensenii, and Lactobacillus iners. Sequence conservation among the Apf orthologs was confined mostly to the C-terminal regions of the proteins, with a moderate degree of similarity at the N-terminal region. As reported earlier (26, 34, 44), the C-terminal region of the Apf proteins also shared significant sequence homology with the LysM domain-containing proteins from other mostly heterofermentative Lactobacillus species, as well as species of enterococci, listeria, and streptococci. An unrooted phylogenetic tree constructed with proteins exhibiting sequence similarity to the Apf of L. acidophilus shows defined grouping between proteins with and without LysM domain, with the latter group of proteins found only in Lactobacillus species (Fig. 1). Among the non-LysM domain-containing proteins, the Apf proteins formed a distinct cluster, with divergence appearing between the Apfs from group B members of the L. acidophilus complex (L. gasseri and L. johnsonii) and the Apfs from L. acidophilus, L. helveticus, L. ultunensis, and L. crispatus. It is noteworthy that a small lipoprotein (120 amino acid residues) from NCFM of unknown function, encoded by LBA1850, is the only protein encoded in the NCFM genome that shared C-terminal sequence conservation with the Apf of NCFM. Orthologs of this lipoprotein are found exclusively in the closely related species of L. helveticus, L. ultunensis, and L. crispatus and form a tight cluster. Meanwhile, the LysM domain proteins are mostly putative peptidoglycan-binding proteins (Pbp) from lactobacilli and other Gram-positive species and formed separate clusters from the Apf proteins. Subclusters within this group appeared to be genus dependent. Overall, Apf, Pbp, and the putative lipoproteins have a conserved YG motif (22) at their C-terminal region, which was proposed to be likely involved in noncovalent anchoring of these proteins onto the cell surface (44).
FIG. 1.
Unrooted phylogenetic tree of proteins with sequence similarity to the Apf of L. acidophilus NCFM. BlastP hits on L. acidophilus NCFM Apf protein sequence were aligned, and the phylogenetic tree was constructed using ClustalX v2.0.12 as described in Materials and Methods. The phylogenetic tree was presented using MEGA 4.0.2 software. (For the GenBank accession numbers of all proteins included in the analysis, see Table S1 in the supplemental material.) PG, peptidoglycan, Pbp, peptidoglycan-binding protein.
Transcription of the apf gene was induced during the stationary-growth phase.
To investigate whether the expression pattern of the apf gene in L. acidophilus NCFM is growth phase dependent, the transcriptional level of the apf gene was compared during log and stationary growth phases using RT-QPCR analysis. Results from two independent biological replicate samples showed that at the log phase, the transcript level of the apf gene was approximately 23-fold lower than that of the LBA0383 reference gene, whereas during the stationary phase, the apf transcript level was 3-fold higher than that of LBA0383. Overall, the apf gene was induced 20-fold during the stationary phase compared to the log growth phase.
Phenotype of the Δapf mutant (NCK2033).
A mutant carrying an in-frame deletion within the apf gene was constructed and isolated using the upp-based counterselective gene replacement system. One Δapf deletion mutant, designated NCK2033, was selected for further analysis.
(i) General characteristics.
When cultured in MRS medium, NCK2033 did not show a significant difference in growth or cell morphology compared to that of the NCK1909 wild-type reference strain (data not shown). The mutant cells appeared to settle to the bottom of the test tube during growth in MRS liquid medium in a manner similar to that seen with the NCK1909 culture. Thus, deletion of the apf gene did not affect the ability of the mutant to autoaggregate in planktonic culture.
(ii) Stress tolerance.
Stress challenge assays were performed with early-log-phase cultures to screen for any stress-sensitive phenotype of the Δapf mutant compared to the NCK1909 reference. No significant difference in the survival rate was observed when both strains were exposed to MRS broth containing 10% NaCl for 2 h at 37°C. In contrast, a nearly 3-log reduction of the NCK2033 cells was observed compared to a <2-log reduction of the NCK1909 after exposure to 2.5% oxgall bile (Fig. 2). When exposed to MRS broth at pH 3.5 in the presence of lactic acid, no difference in survival rates was observed between NCK2033 and the control strain, even after 2-h incubation (data not shown). However, an approximately 40% decrease in survival of NCK2033 was observed compared to NCK1909 after a 1-h exposure to MRS broth adjusted to pH 2.0 with HCl (Fig. 2), an acidic pH condition that mimics the gastric environment.
FIG. 2.
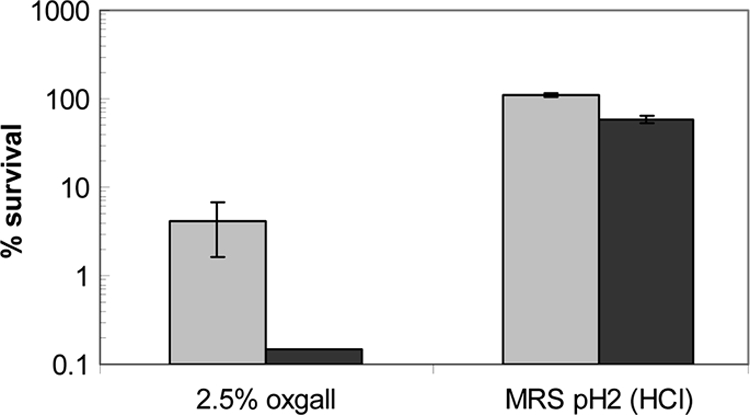
Survival of NCK2033 (Δapf mutant; dark gray bars) and NCK1909 (reference strain; light gray bars) early-log-phase cells (OD600 of 0.25 to 0.3) after exposure to MRS broth supplemented with 2.5% oxgall bile for 2 h at 37°C or exposure to MRS broth at pH 2.0 (adjusted with HCl) for 1 h at 37°C. The data are the means ± standard errors of the means for three independent replicates.
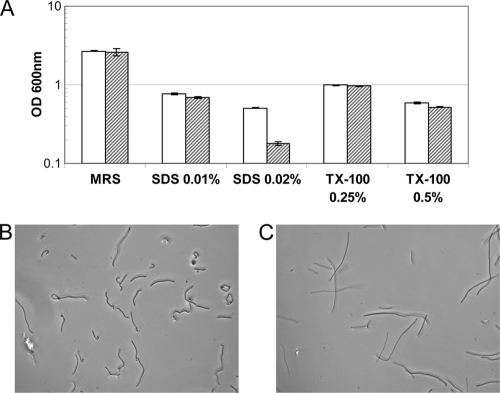
In addition, NCK2033 showed a higher susceptibility to SDS treatment at 0.02%, whereas no growth inhibition was observed in the presence of Triton X-100 (Fig. 3A). Interestingly, microscopic examination of the cellular morphology after 24 h of growth in MRS broth containing 0.02% SDS revealed that while the NCK1909 reference appeared as curly rods (Fig. 3B), the NCK2033 cells were less curly and tended to form longer rods (Fig. 3C).
FIG. 3.
Survival of NCK2033 and NCK1909 in the presence of surfactants SDS and Triton X-100. (A) Early-log-phase cultures of NCK1909 (white bars) and NCK2033 (striped bars) were inoculated (1% inoculum) into MRS broth supplemented with the indicated concentrations of SDS or Triton X-100 (TX-100), and cell density was measured after 24 h of incubation at 37°C. The data are the means ± standard errors of the means for two independent replicates. The morphologies of NCK1909 (B) and NCK2033 (C) cells were examined with phase-contrast microscopy after the 24 h of incubation in MRS broth with 0.02% SDS.
(iii) Stability in acidified 11% skim milk.
Previous gene expression studies of L. acidophilus NCFM grown in milk revealed the apf gene to be one of the most significantly upregulated genes (5). During the present study, we observed no significant difference in the growth or acidification rate of NCK2033 compared to that of the reference strain during growth in 11% SM (data not shown). To further assess whether Apf potentially contributes to survival during yogurt storage, the survival of NCK2033 was evaluated in SM acidified to pH 4.5 with lactic acid for 2 weeks at 4°C, simulating conditions during the storage of yogurt. Overall, the viability of both the Apf− and Apf+ cultures was relatively stable over the 2-week experimental period, with less than a 10% decrease in survivors at the end of the storage period (data not shown).
(iv) Survival in simulated gastric and small-intestinal juices.
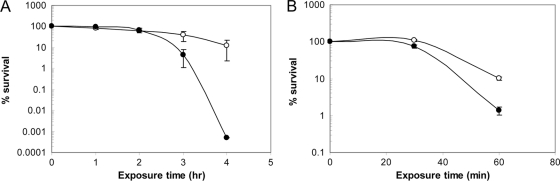
The ability of NCK2033 to survive GI transit was assessed in vitro by exposure to simulated gastric and small-intestinal juices. Remarkably, NCK2033 showed a significant decrease in survival (4-log reduction) after 3 h of exposure to the simulated small-intestinal fluid compared to that of the NCK1909 reference strain (Fig. 4A). In addition, NCK2033 exhibited a 2-log reduction after a 1-h exposure to simulated gastric juice, compared to a 1-log reduction from the reference strain (Fig. 4B). Significant cell death from both strains occurred beyond 60 min of exposure. Thus, inactivation of the apf locus rendered NCK2033 significantly more susceptible to GI environments.
FIG. 4.
Survival of stationary-phase cells of NCK2033 (dark circles) compared to NCK1909 (open circles) in simulated small-intestinal juice at pH 8.0 (A) and gastric juice at pH 2.0 (B). Percentage of survival represents viable cells (CFU/ml) after treatment at various time points versus before treatment (time zero). The data are the means ± standard errors of the means for three independent replicates.
Adherence ability of NCK2033.
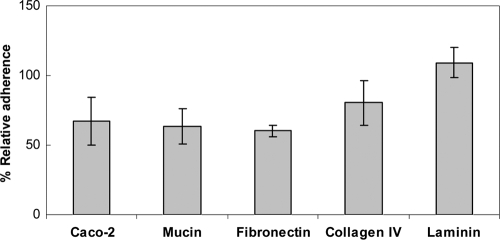
To examine the effect of apf deletion on the adherence ability to Caco-2 epithelial cells in vitro, a stationary-phase culture of NCK2033 was exposed to Caco-2 monolayers for 1 h at 37°C under ambient atmospheric conditions. An approximately 30% reduction of adherence was observed for the NCK2033 cells compared to that of the NCK1909 cells (Fig. 5). Whether the Apf proteins are involved in the adherence to mucin and major ECM components, such as collagen type IV, laminin, and fibronectin, was also determined. The NCK2033 cells showed an approximately 40% decrease in adherence to fibronectin compared to that of NCK1909. In addition, the ability of NCK2033 to adhere to mucin in vitro was reduced to an average of 63.5% of that of the parental reference strain. The adherence to both type IV collagen and laminin was unaffected.
FIG. 5.
Adherence levels of NCK2033 relative to that of NCK1909 on Caco-2 epithelial cells, mucin, and major ECM components in vitro. Adherence assays were performed with stationary-phase cultures grown in MRS broth at 37°C in ambient atmospheric condition. Bacterial cells were exposed to Caco-2 monolayers, or microwells coated with mucin, fibronectin, type IV collagen, or laminin for 1 h at 37°C, followed by plating on MRS agar medium for enumeration of adhered bacterial cells (see Materials and Methods). The data are the means ± standard errors of the means for four independent replicates.
DISCUSSION
The completed genome of L. acidophilus NCFM (1) has facilitated the identification and characterization of several key genetic determinants for successful delivery, persistence, and activities of L. acidophilus in the GI tract. These include loci that encode acid and bile tolerance, metabolism of prebiotic oligosaccharides, bacteriocin production, oxalate degradation, two-component regulatory systems for bile and acid responses, and adherence factors that potentially mediate attachment to gut mucosa and modulate the immune system (2-4, 7, 11, 17, 37). The present study describes the identification and functional analysis of a putative Apf in L. acidophilus that likely contributes to multiple probiotic functional roles, notably bile and acid resistance, as well as adherence to specific host cell components.
Based on sequence similarity searches in the current database, orthologs of Apf appear to be present only in the L. acidophilus group of lactobacilli. The phylogenetic relationship of the Apf proteins from this group (Fig. 1) also correlates with the phylogenetic position with respect to their 16S rRNA gene sequences (19). The divergence between the group A and B L. acidophilus (29) Apf proteins and their moderate sequence homology suggested that they are only weakly related, which is also evidenced by the lack of gene order conservation surrounding the apf loci between the group A and B L. acidophilus species. For example, both the Apf1 and Apf2 proteins from L. gasseri and L. johnsonii exhibit only moderate sequence homology with the Apf from L. acidophilus (Fig. 1). This is in contrast to the ApfA from L. acidophilus TMW1.988, which clustered with L. johnsonii Apfs and is highly similar to the Apf2 of L. johnsonii (≥87% sequence identity). Thus, it appears that at least two Apf-like proteins are present in L. acidophilus. Overall, the extensive sequence variation at the central region of the Apf proteins from these species, mostly due to insertions and deletions (indels) and residue substitutions, resulted in subclustering of the proteins, and it likely contributes to functionality differences. As previously mentioned, the Apf proteins of L. gasseri 4B2 are involved in the maintenance of cell shape, as overproduction or downregulation of the proteins resulted in morphological changes (26). Meanwhile, deletion of the apf gene in L. acidophilus did not cause any change in the morphotype of NCK2033, indicating that unlike the Apf in L. gasseri, the Apf of L. acidophilus does not play a key role in the regulation of cell shape under the growth conditions used in this study.
In order to examine whether the apf gene was associated with the aggregation phenotype of L. acidophilus, an apf deletion mutant was constructed in the NCK1909 background host by using a upp-based counterselective allelic replacement system (23). The resultant NCK2033 apf mutant did not exhibit significant defects in growth or changes in cell morphology or colony morphotype when cultured in MRS medium. Interestingly, at a given cell density as measured by optical density, the cell number of NCK2033 was approximately 1.5 times the number of the NCK1909 cells, although no significant differences in cell morphology or degree of aggregation formation could be observed from microscopic analysis. The apf mutant also showed a growth rate comparable to that of the NCK1909 reference strain in milk, despite the fact that apf is one of the most upregulated genes in NCFM during growth in 11% skim milk (5).
Consistent with the absence of direct evidence correlating apf genes with the aggregation phenotype observed for L. crispatus, L. gasseri, and L. johnsonii, inactivation of the apf gene in L. acidophilus did not affect the aggregation ability of NCK2033. Clearly, any potential aggregation phenotype involving Apf may be highly dependent on the environmental conditions and likely in vivo factors. It has been proposed that the Apf protein may not be a unique component required for aggregation, which potentially could involve a more complex interaction among proteins or major cell structural components (i.e., LTA, S-layer proteins) that influence the electrostatic property of the cell surface (41, 45). In fact, it was recently observed that an NCFM mutant carrying a deletion in one of the lipotechoic acid biosynthetic pathway genes did not aggregate when grown in MRS liquid medium (E. A. Pfeiler and T. R. Klaenhammer, unpublished data), indicating a potential role of LTA in the aggregation phenotype of L. acidophilus. A recent proteomic study by Siciliano et al. (42) also proposed elongation factor Tu (EF-Tu) as a potential major contributor to cell aggregation in L. crispatus.
To further explore the phenotype of apf deletion, NCK2033 was subjected to various stress challenges. Treatment with 10% NaCl or low pH conditions (pH 3.5) in the presence of lactic acid did not affect the survival of the mutant. Likewise, NCK2033 remained viable and at stable population levels comparable to those of the NCK1909 reference strain during refrigerated storage in acidified milk for 2 weeks, parameters that simulate typical yogurt storage conditions. Nevertheless, successful dairy delivery of probiotic bacteria also requires the microorganisms to resist the extreme low pH and bile present in the host's gastric and small-intestinal environments, respectively. Inactivation of apf significantly reduced the mutant's ability to survive in simulated gastric and small-intestinal juices, consistent with its increased sensitivity to extreme low pH condition (pH 2; in the presence of HCl) and bile, respectively, in separate survival assays. It is also plausible that by an unknown mechanism, the absence of Apf in NCK2033 may increase the mutant's susceptibility to native proteolytic activities (from pepsin and trypsin) present in the simulated gastric or small-intestinal juice preparations. Apf proteins are basic and may contribute to the overall positive charge on the cell surface at physiological pH values. In the absence of Apf, the decreased tolerance of NCK2033 against SDS, an anionic surfactant, and the associated morphological change, could possibly be attributed to changes in the cell surface electrostatic charge that increase the accessibility of SDS to cell surface molecules. Apf may also directly bind SDS and alter its denaturation effect on the surface proteins.
The ability of probiotic microorganisms to adhere to host cell components is considered an important trait, as host interactions mediated by adherence factors may promote mucosal integrity, pathogen exclusion, and host immunomodulation via host receptor recognition. In the present study, the stationary-phase NCK2033 cells exhibited reduced in vitro adherence to Caco-2 epithelial cells, mucin, and one of the major ECM components, fibronectin, but not type IV collagen or laminin. Hence, the putative Apf protein could potentially serve as an adhesion factor in vivo that participates in the interaction with the hosts' mucus layer and intestinal epithelial cells, as well as the fibronectin component of the ECM.
Previous transcriptional studies showed that the apf gene was induced during log phase when NCFM was grown on different carbohydrate sources (glucose, fructose, sucrose, fructooligosaccharides, raffinose, lactose, galactose, and trehalose) (8). In the present study, when NCFM was grown in MRS-glucose medium, RT-QPCR analysis revealed that expression of apf is growth-phase dependent, where the apf gene was significantly upregulated during the stationary phase compared to that during the log phase. Thus, Apf protein likely contributes to more significant physiological roles when cells enter the stationary phase. This is in agreement with the observation that the reduction in adherence to Caco-2 epithelial cells and mucin in vitro occurred with NCK2033 stationary-phase culture but not with log-phase culture (data not shown). In comparison, transcription of the apf1 and apf2 genes in L. gasseri and L. johnsonii also varies by growth phase, but with maximum gene expression observed during log growth (45), presumably when regulation of cell shape is critical due to rapid turnover of the cell wall.
Overall, the mechanisms involved in the complexity of cell aggregation remain largely hypothetical and likely species specific and environment dependent. Nevertheless, this study revealed that Apf potentially confers properties that promote bile tolerance and interaction with the host epithelium and, hence, may contribute to the fitness and adaptation of L. acidophilus in the small-intestinal ecosystem.
Supplementary Material
Acknowledgments
This work was supported in part by Dairy Management, Inc., the Southeast Dairy Foods Research Center, the North Carolina Dairy Foundation, and Danisco USA, Inc. (Madison, WI).
We are grateful to R. Sanozky-Dawes for technical advice and for preparing Caco-2 epithelial cells for in vitro adherence studies. We also thank S. O'Flaherty, E. Durmaz, E. Pfeiler, J. Schroeter, and G. Douglas for comments and insightful discussions.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U. S. A. 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azcarate-Peril, M. A., E. Altermann, R. L. Hoover-Fitzula, R. J. Cano, and T. R. Klaenhammer. 2004. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 70:5315-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azcarate-Peril, M. A., J. M. Bruno-Barcena, H. M. Hassan, and T. R. Klaenhammer. 2006. Transcriptional and functional analysis of oxalyl-coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes from Lactobacillus acidophilus. Appl. Environ. Microbiol. 72:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azcarate-Peril, M. A., O. McAuliffe, E. Altermann, S. Lick, W. M. Russell, and T. R. Klaenhammer. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 71:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azcarate-Peril, M. A., R. Tallon, and T. R. Klaenhammer. 2009. Temporal gene expression and probiotic attributes of Lactobacillus acidophilus during growth in milk. J. Dairy Sci. 92:870-886. [DOI] [PubMed] [Google Scholar]
- 6.Barefoot, S. F., and T. R. Klaenhammer. 1983. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl. Environ. Microbiol. 45:1808-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. U. S. A. 100:8957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrangou, R., M. A. Azcarate-Peril, T. Duong, S. B. Conners, R. M. Kelly, and T. R. Klaenhammer. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. U. S. A. 103:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 10.Boris, S., J. E. Suarez, and C. Barbes. 1997. Characterization of the aggregation promoting factor from Lactobacillus gasseri, a vaginal isolate. J. Appl. Microbiol. 83:413-420. [DOI] [PubMed] [Google Scholar]
- 11.Buck, B. L., E. Altermann, T. Svingerud, and T. R. Klaenhammer. 2005. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:8344-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bujnakova, D., and V. Kmet. 2002. Aggregation of animal lactobacilli with O157 enterohemorrhagic Escherichia coli. J. Vet. Med. B Infect. Dis. Vet. Public Health 49:152-154. [DOI] [PubMed] [Google Scholar]
- 13.Cesena, C., L. Morelli, M. Alander, T. Siljander, E. Tuomola, S. Salminen, T. Mattila-Sandholm, T. Vilpponen-Salmela, and A. von Wright. 2001. Lactobacillus crispatus and its nonaggregating mutant in human colonization trials. J. Dairy Sci. 84:1001-1010. [DOI] [PubMed] [Google Scholar]
- 14.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 1998. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 84:759-768. [DOI] [PubMed] [Google Scholar]
- 15.Collado, M. C., J. Meriluoto, and S. Salminen. 2008. Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 226:1065-1073. [Google Scholar]
- 16.Collado, M. C., J. Meriluoto, and S. Salminen. 2007. Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett. Appl. Microbiol. 45:454-460. [DOI] [PubMed] [Google Scholar]
- 17.Dobson, A. E., R. B. Sanozky-Dawes, and T. R. Klaenhammer. 2007. Identification of an operon and inducing peptide involved in the production of lactacin B by Lactobacillus acidophilus. J. Appl. Microbiol. 103:1766-1778. [DOI] [PubMed] [Google Scholar]
- 18.Engelbrektson, A. L., J. R. Korzenik, M. E. Sanders, B. G. Clement, G. Leyer, T. R. Klaenhammer, and C. L. Kitts. 2006. Analysis of treatment effects on the microbial ecology of the human intestine. FEMS Microbiol. Ecol. 57:239-250. [DOI] [PubMed] [Google Scholar]
- 19.Felis, G. E., F. Dellaglio, and S. Torriani. 2009. Taxonomy of probiotic microorganisms, p. 591-637. In D. Charalampopoulos and R. A. Rastall (ed.), Prebiotics and probiotics science and technology, vol. 1. Springer, New York, NY.
- 20.Foligne, B., S. Nutten, C. Grangette, V. Dennin, D. Goudercourt, S. Poiret, J. Dewulf, D. Brassart, A. Mercenier, and B. Pot. 2007. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J. Gastroenterol. 13:236-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frece, J., B. Kos, I. K. Svetec, Z. Zgaga, V. Mrsa, and J. Suskovic. 2005. Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J. Appl. Microbiol. 98:285-292. [DOI] [PubMed] [Google Scholar]
- 22.Giffard, P. M., and N. A. Jacques. 1994. Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J. Dent. Res. 73:1133-1141. [DOI] [PubMed] [Google Scholar]
- 23.Goh, Y. J., M. A. Azcarate-Peril, S. O'Flaherty, E. Durmaz, F. Valence, J. Jardin, S. Lortal, and T. R. Klaenhammer. 2009. Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 75:3093-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goh, Y. J., C. Zhang, A. K. Benson, V. Schlegel, J. H. Lee, and R. W. Hutkins. 2006. Identification of a putative operon involved in fructooligosaccharide utilization by Lactobacillus paracasei. Appl. Environ. Microbiol. 72:7518-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 26.Jankovic, I., M. Ventura, V. Meylan, M. Rouvet, M. Elli, and R. Zink. 2003. Contribution of aggregation-promoting factor to maintenance of cell shape in Lactobacillus gasseri 4B2. J. Bacteriol. 185:3288-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, H. S., and S. E. Gilliland. 1983. Lactobacillus acidophilus as a dietary adjunct for milk to aid lactose digestion in humans. J. Dairy Sci. 66:959-966. [DOI] [PubMed] [Google Scholar]
- 28.Kimmel, S. A., and R. F. Roberts. 1998. Development of a growth medium suitable for exopolysaccharide production by Lactobacillus delbrueckii ssp. bulgaricus RR. Int. J. Food Microbiol. 40:87-92. [DOI] [PubMed] [Google Scholar]
- 29.Klaenhammer, T. R., and W. M. Russell. 2000. Species of the Lactobacillus acidophilus complex, p. 1151-1157. In R. K. Robinson, C. Batt, and P. D. Patel (ed.), Encyclopedia of food microbiology, vol. 2. Academic Press, San Diego, CA.
- 30.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 31.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leyer, G. J., S. Li, M. E. Mubasher, C. Reifer, and A. C. Ouwehand. 2009. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics 124:e172-e179. [DOI] [PubMed] [Google Scholar]
- 33.Lozo, J., B. Jovcic, M. Kojic, M. Dalgalarrondo, J. M. Chobert, T. Haertle, and L. Topisirovic. 2007. Molecular characterization of a novel bacteriocin and an unusually large aggregation factor of Lactobacillus paracasei subsp. paracasei BGSJ2-8, a natural isolate from homemade cheese. Curr. Microbiol. 55:266-271. [DOI] [PubMed] [Google Scholar]
- 34.Marcotte, H., S. Ferrari, C. Cesena, L. Hammarstrom, L. Morelli, G. Pozzi, and M. R. Oggioni. 2004. The aggregation-promoting factor of Lactobacillus crispatus M247 and its genetic locus. J. Appl. Microbiol. 97:749-756. [DOI] [PubMed] [Google Scholar]
- 35.Narimatsu, M., Y. Noiri, S. Itoh, N. Noguchi, T. Kawahara, and S. Ebisu. 2004. Essential role for the gtfA gene encoding a putative glycosyltransferase in the adherence of Porphyromonas gingivalis. Infect. Immun. 72:2698-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paineau, D., D. Carcano, G. Leyer, S. Darquy, M. A. Alyanakian, G. Simoneau, J. F. Bergmann, D. Brassart, F. Bornet, and A. C. Ouwehand. 2008. Effects of seven potential probiotic strains on specific immune responses in healthy adults: a double-blind, randomized, controlled trial. FEMS Immunol. Med. Microbiol. 53:107-113. [DOI] [PubMed] [Google Scholar]
- 37.Pfeiler, E. A., M. A. Azcarate-Peril, and T. R. Klaenhammer. 2007. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 189:4624-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeiler, E. A., and T. R. Klaenhammer. 2009. Role of transporter proteins in bile tolerance of Lactobacillus acidophilus. Appl. Environ. Microbiol. 75:6013-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reniero, R., P. Cocconcelli, V. Bottazzi, and L. Morelli. 1992. High frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J. Gen. Microbiol. 138:763-768. [Google Scholar]
- 40.Roos, S., S. Lindgren, and H. Jonsson. 1999. Autoaggregation of Lactobacillus reuteri is mediated by a putative DEAD-box helicase. Mol. Microbiol. 32:427-436. [DOI] [PubMed] [Google Scholar]
- 41.Schachtsiek, M., W. P. Hammes, and C. Hertel. 2004. Characterization of Lactobacillus coryniformis DSM 20001T surface protein Cpf mediating coaggregation with and aggregation among pathogens. Appl. Environ. Microbiol. 70:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siciliano, R. A., G. Cacace, M. F. Mazzeo, L. Morelli, M. Elli, M. Rossi, and A. Malorni. 2008. Proteomic investigation of the aggregation phenomenon in Lactobacillus crispatus. Biochim. Biophys. Acta 1784:335-342. [DOI] [PubMed] [Google Scholar]
- 43.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 44.Turner, M. S., L. M. Hafner, T. Walsh, and P. M. Giffard. 2004. Identification and characterization of the novel LysM domain-containing surface protein Sep from Lactobacillus fermentum BR11 and its use as a peptide fusion partner in Lactobacillus and Lactococcus. Appl. Environ. Microbiol. 70:3673-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ventura, M., I. Jankovic, D. C. Walker, R. D. Pridmore, and R. Zink. 2002. Identification and characterization of novel surface proteins in Lactobacillus johnsonii and Lactobacillus gasseri. Appl. Environ. Microbiol. 68:6172-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voltan, S., I. Castagliuolo, M. Elli, S. Longo, P. Brun, R. D'Inca, A. Porzionato, V. Macchi, G. Palu, G. C. Sturniolo, L. Morelli, and D. Martines. 2007. Aggregating phenotype in Lactobacillus crispatus determines intestinal colonization and TLR2 and TLR4 modulation in murine colonic mucosa. Clin. Vaccine Immunol. 14:1138-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter, J., C. Schwab, D. M. Loach, M. G. Ganzle, and G. W. Tannock. 2008. Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiology 154:72-80. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi, T., K. Kasamo, M. Chuman, M. Machigashira, M. Inoue, and T. Sueda. 1998. Preparation and characterization of an Actinomyces naeslundii aggregation factor that mediates coaggregation with Porphyromonas gingivalis. J. Periodontal Res. 33:460-468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.