Abstract
The effects of three temperatures (5, 15, and 25°C) on the survival of Salmonella enterica serovar Typhimurium in topsoil were investigated in small microcosms by three different techniques: plate counting, invA gene quantification, and invA mRNA quantification. Differences in survival were related to the effect of protozoan predation. Tetracycline-resistant Salmonella serovar Typhimurium was inoculated into soil and manure-amended soil at 1.5 × 108 cells g soil−1. Population densities were determined by plate counting and by molecular methods and monitored for 42 days. Simultaneous extraction of RNA and DNA, followed by quantitative PCR, was used to investigate invA gene levels and expression. Analysis by these three techniques showed that Salmonella serovar Typhimurium survived better at 5°C. Comparing DNA and CFU levels, significantly higher values were determined by DNA-based techniques. invA mRNA levels showed a fast decrease in activity, with no detectable mRNA after an incubation period of less than 4 days in any of the soil scenarios. A negative correlation was found between Salmonella serovar Typhimurium CFU levels and protozoan most probable numbers, and we propose the role of the predator-prey interaction as a factor to explain the die-off of the introduced strain by both culture- and DNA quantification-based methods. The results indicate that temperature, manure, and protozoan predation are important factors influencing the survival of Salmonella serovar Typhimurium in soil.
Salmonella bacteria excreted in the feces of asymptomatic animals may constitute an important source of freshwater and food contamination when manure is spread directly on land (43). The bacteria can be shed from manure and are reported to survive in soil for 160 to 200 days (23, 25).
Numerous methods have been developed for the detection and quantification of Salmonella in different matrices (11, 14, 35). The introduction of molecular techniques has become an especially important advance in reducing the time required for detection of Salmonella and in detecting active bacteria in environmental samples through their DNA and RNA (10, 18, 51).
Due to a lack of sensitivity and problems with PCR inhibitors in environmental samples, quantitative PCR (qPCR) of Salmonella using DNA isolated directly from soil or manure without enrichment has not yet been widely applied (34). Another problem with DNA-based detection assays is also the possible detection of DNA from inactive or nonviable bacteria, causing false-positive results (31). Quantification of mRNA, however, allows analysis of which genes are being expressed and thereby quantitative measurement of the activity levels of specific functional traits of interest. As mRNA, in general, is considered an extremely labile molecule, it is commonly accepted that analysis of mRNA is a better measure of microbial activity than is DNA analysis (46).
A sequence of the invA gene (10) has been utilized as a target for the detection of Salmonella nucleic acids in soil samples. This gene is highly conserved in almost all Salmonella serotypes (7, 17, 44), and detection of Salmonella based on the presence of this gene has previously been reported (11, 12, 24). invA mRNA has been used as a biomarker for active cells (13, 18, 26); it has been discussed, however, whether transcription of the invA gene may differ with different physiological states of the cell, which may affect assay specificity (13). Jacobsen and Holben (26) showed a detection limit of 5 × 104 seeded Salmonella serovar Typhimurium cells per g of soil. Based on these data, invA mRNA seems to be a feasible candidate for reverse transcriptase PCR assays to specifically identify living Salmonella cells.
The survival of Salmonella in environmental habitats can be influenced by different factors. In several studies, survival of Salmonella in soils has been examined by culture-dependent methods testing the influence of different factors like manure addition, temperature, and interaction with other microorganisms (23, 28, 39, 48). Salmonella spp. spread with manure have been reported to survive for up to 300 days in soils (4, 29), but the duration of their survival depends on several factors such as, for example, the incubation temperature (20, 50). Another important factor influencing Salmonella survival in soil is predation by protozoa. The role of protozoa in food-borne pathogens' survival in the environment is often a neglected factor in microbial ecology, and therefore this has only been investigated in a few studies (5). Brandl et al. (8) investigated the viable form of Salmonella enterica in vesicles of the protozoan Tetrahymena. They showed that this protozoan releases vesicles containing a high density of S. enterica, leading to an underestimation of actual population sizes of the pathogen during predation studies.
The primary objective of the present study was to evaluate the survival of tetracycline-resistant Salmonella serovar Typhimurium in soil and manure-amended soil at three different temperatures (5, 15, and 25°C) by using three different techniques: plate counting, invA DNA qPCR, and direct quantification of invA mRNA. Furthermore, the role of predation was evaluated to relate the survival of Salmonella as measured by different methods with the estimated most probable number (MPN) of protozoa present in soil and manure-amended soil.
MATERIALS AND METHODS
Soils.
Soil samples were collected from Sjællands Odde, Denmark. Topsoil was obtained from the 0- to 30-cm layer of an agricultural field containing 19% clay, 18% silt, 62% sand, and 1.2% carbon and with a pH of 7.2. Approximately 100 kg of soil was obtained in total by using a manual composite sampling technique in which subsamples were taken from scattered locations within a 10-m2 area, mixed thoroughly, and stored frozen at −20°C in aliquots of ∼2 kg. Prior to experiments, aliquots were thawed and acclimatized in the dark at 10°C for 10 days as described by Mortensen and Jacobsen (38). Three subsamples of soil were negative for Salmonella when tested by qPCR, and no CFU of tetracycline-resistant bacteria were detected according to the procedure described below (detection limit, 100 CFU g−1).
Manure.
Fresh manure was obtained from dairy cows. It was stored at 5°C for 1 week before the beginning of the assay. Physical/chemical analysis of subsamples indicated a moisture content of 90.4%, an ammonia N concentration of 1.94 g kg−1, and total N, phosphorus, potassium, copper, and magnesium concentrations of 3.94, 0.77, 3.63, 0.00995, and 1.284 g kg−1, respectively (wet basis). qPCRs were negative for Salmonella, and no CFU of tetracycline-resistant bacteria were detected in three manure subsamples.
Bacteria.
Salmonella serovar Typhimurium tetracycline-resistant DSM554 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany, and grown in Müller broth (Oxoid) containing 25 μg/ml tetracycline (Sigma-Aldrich) at 37°C in the dark for 24 h at 100 rpm. Ten milliliters of the culture was placed into 100 ml Müller broth with tetracycline and incubated again at 37°C for at least 4 h until an optical density at 600 nm of ∼0.6 was attained. Subsequently, the culture was centrifuged (6,000 × g, 10 min) and resuspended in 100 mM sterile phosphate buffer (PB), pH 7, resulting in an inoculum of 2 × 109 CFU ml−1 based on direct plate counts (Müller agar plates with 25 μg/ml tetracycline, 24 h, 37°C).
Microcosm setup and Salmonella serovar Typhimurium inoculation.
Triplicate microcosms of topsoil and manure-amended topsoil were set up in 100-ml glass flasks with airtight glass stoppers (Schott Glaswerke, Mainz, Germany).
Prior to inoculation, 30 g (wet weight) of soil was placed in each flask and sterilized PB or liquid manure was added to bring the soil to 85% of the field water-holding capacity. The amount of manure in the manure-amended topsoil corresponded to a manure application rate of 3 kg m−2, which is commonly practiced on farms in the region.
At the onset of the experiment, 1 ml Salmonella serovar Typhimurium (Tet+) culture (described above) was inoculated into the microcosm soil surface without mixing, producing 1.5 × 108 CFU g soil−1 in each microcosm. In manure-amended topsoil microcosms, manure amended with the bacteria was applied to the soil surface, producing the same level of CFU g soil−1. The two different soil scenarios (with/without manure) were incubated in triplicate at three different temperatures, 5, 15, and 25°C.
Direct plate counts of Salmonella serovar Typhimurium were done on days 0, 1, 2, 4, 7, 11, 15, 19, and 24 after inoculation and once a week thereafter until the pathogen concentration dropped below the detection limit (100 CFU g soil−1). Samples of 0.5 g (wt/wt) as a composite of about five plugs were removed from the flasks and transferred to 50-ml screw-cap glass tubes, 9.5 ml sterile PB (100 mM, pH 7) was added, and the mixture was agitated on a horizontal shaker (150 rpm, 30 min). Serial 10-fold dilutions were prepared in PB, and 0.1-ml aliquots were surface plated on Müller agar containing tetracycline. Colonies were counted after incubation for 24 h at 37°C.
At the same time points as for direct plate counting, another 0.5 g of soil was taken for nucleic acid extraction. In order to ensure a snapshot freeze event, these samples were frozen immediately in liquid nitrogen and stored at −80°C.
DNA/RNA extraction and cDNA synthesis.
DNA and RNA were coextracted as described by Nicolaisen et al. (40). In short, the method involves bead beating in the presence of cetyltrimethylammonium bromide (CTAB) buffer, phenol, and chloroform, followed by phenol extraction and precipitation of nucleic acids from the aqueous phase by 30% polyethylene glycol 6000 (PEG). Phenol and CTAB buffer were added to the frozen samples, and cell lysis was perfumed for 2 × 15 s at a speed setting of 5.0 m s−1 with intermittent cooling to prevent overheating of the sample. The following modification, described by Bælum et al. (3), was applied to the protocol. One microliter of glycogen (Roche, Basel, Switzerland) was added to PEG to aid in nucleic acid precipitation. After the extraction procedure, 7-μl aliquots of each sample were used for RNase-free DNase I treatment (Promega, Madison, WI) according to the manufacturer's protocol as modified by Jacobsen and Holben (26). Reverse transcription (RT) was performed using an Omniscript RT kit from Qiagen (Crawley, United Kingdom) with 2 μl of DNase-treated extract as the template, 40 pmol of invA reverse primer (described below), and a reaction volume of 10 μl. RT reaction temperatures and time of incubation were as described previously by Bælum et al. (3).
qPCR.
qPCR was carried out in an iCycler (Bio-Rad, Hercules, CA). To include impacts of the soil matrix on extraction efficiency and coextracted PCR enzyme inhibitors, standards for quantification were prepared by individual inoculations of 10-fold dilutions (101 to 107 g soil−1) of Salmonella serovar Typhimurium to aliquots of 0.5 g soil, followed by nucleic acid extraction as described above. The primers employed were based in those described by Chiu and Ou (10) with the modifications described by Jacobsen and Holben (26). The primer sequences were as follows: invA forward, 5′-ACAGTGCTCGTTTACGACC-3′; invA reverse, 5′-ACTGGTACTGATCGATAAT-3′. All qPCRs were performed in a final volume of 20 μl containing 10 μl of premixed mastermix (Dynamo HS SYBR green qPCR kit; Finnzymes, Helsinki, Finland), 0.4 μM forward primer, 0.4 μM reverse primer, 20 μg bovine serum albumin (New England BioLabs Inc., Ipswich, MA), 7.2 μl of deionized PCR grade water, and 1 μl of template DNA (∼10 to 50 ng). DNA extractions were diluted 10-fold prior to PCR quantification to avoid interactions from coextracted enzyme inhibitors. All DNA and cDNA samples were quantified in triplicate, including negative controls (containing all of the reagents except the template DNA). Another control using DNase-treated RNA samples was performed to discard possible DNA contamination which would interfere with cDNA-based quantification of invA gene expression levels. Each qPCR consisted of the following steps (6): 15 min initial denaturation and enzyme activation at 95°C, followed by 40 cycles of 30 s denaturation at 95°C, 30 s annealing at 55°C, 30 s elongation at 72°C, and 15 s at 77°C for quantification of the invA product. The procedure ended with one cycle of 6 min at 72°C for elongation and a melting curve analysis generated by analyzing the amount of double-stranded DNA after each 0.5°C increase in temperature up to 95°C.
Counting of protozoa.
Samples for counting of protozoa were taken four times during the experiment. A 3-fold dilution series of soil and manure-amended soil samples was prepared in 96-well microtiter plates (Nunc-Thermo Scientific, Roskilde, Denmark). Dilutions were made in modified Neff's amoeba saline buffer with 0.10 g liter of tryptic soy broth−1 as described by Page (41). Well dishes were incubated at 15°C in the dark, and protozoan enumeration was done by visual inspection of single wells with an inverted microscope after 7 and 21 days of incubation. MPN calculations were performed by the computer-assisted method developed by Briones and Reichardt (9). The method uses Microsoft Excel and its associated Solver tool to generate MPNs, error estimates, and 95% confidence limits.
Statistical analysis.
All amplification and plate counting experiments were performed in triplicate, and mean values and standard deviations are given. Statistical analyses were carried out using SPSS statistics 17.0. Results were compared for statistically significant differences by using a one-way analysis of variance test. Correlation between protozoan MPN and S. Typhimurium survival levels was evaluated using Pearson's coefficient. For all tests, a P value of ≤0.05 was considered significant.
RESULTS
Quantification of Salmonella serovar Typhimurium by plate counting and DNA-based methods.
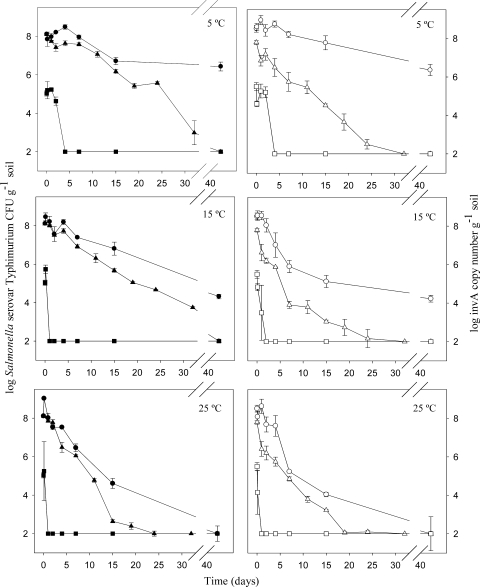
Figure 1 shows levels of Salmonella serovar Typhimurium in soil and manure-amended soil samples incubated at three temperatures: 5, 15, and 25°C. A decrease in Salmonella serovar Typhimurium levels and activity was observed with all of the three methods used throughout the experiment.
FIG. 1.
Evolution of Salmonella serovar Typhimurium (Tet+) measured by qPCR and plate counting in soil samples (closed symbols) and manure-amended soil samples (open symbols) at three different temperatures. Symbols: • and ○, log invA DNA copy number gram soil−1; ▴ and ▵, log CFU count gram soil−1; ▪ and □, log invA mRNA copy number gram soil−1. Each point represents the mean of triplicate quantifications from one sample. Detection limit = 102. Error bars represent standard errors of the mean. Please note the broken x axes.
Using plate counting, we observed a significantly (P < 0.001) lower level of survival of Salmonella serovar Typhimurium in soil samples incubated at 25°C compared to 5°C as early as 4 days. This tendency was maintained throughout the experiment. At 5°C and 15°C, the addition of manure reduced the survival of Salmonella serovar Typhimurium, while at 25°C no significant difference was observed. In manure-amended soil samples, a significant difference between temperatures was found no earlier than after 11 days, with the highest survival at 5°C.
Using qPCR based on invA DNA copy numbers, we observed a 1.5- to 2-log decrease at 5°C, a 4-log decrease at 15°C, and a >6-log decrease at 25°C over a period of 42 days (Fig. 1). The generally lower invA gene levels detected in soil samples incubated at 25°C than in those incubated at 5°C corresponded well to the results obtained by the plate counting technique. Contrary to the plate counting results, however, the evolution of invA genes did not present significant differences between the soil and manure-amended soil scenarios, and only from 4 to 15 days at 15°C did we observe slightly lower levels of invA genes in soil samples with manure than in those without manure.
Quantification of Salmonella serovar Typhimurium activity levels based on invA mRNA.
At 5°C, ∼105 invA mRNA copies g soil−1 were detected until 48 h after inoculation, while in the 15 and 25°C experiments we only detected invA mRNA until 3 h after inoculation (Fig. 1). In the scenarios without manure addition, the invA mRNA levels increased until 3 h at the three temperatures assayed, while in manure-amended soil samples the amount of mRNA decreased immediately after inoculation. After 4 days of incubation, the level of invA mRNA was lower than the detection limit in all of the soil scenarios. invA mRNA could only be detected when more than 106 Salmonella serovar Typhimurium bacteria per g of soil were detected by plate counting, and the relative amount of mRNA in the samples was, in general, less than 1% of the invA DNA detected.
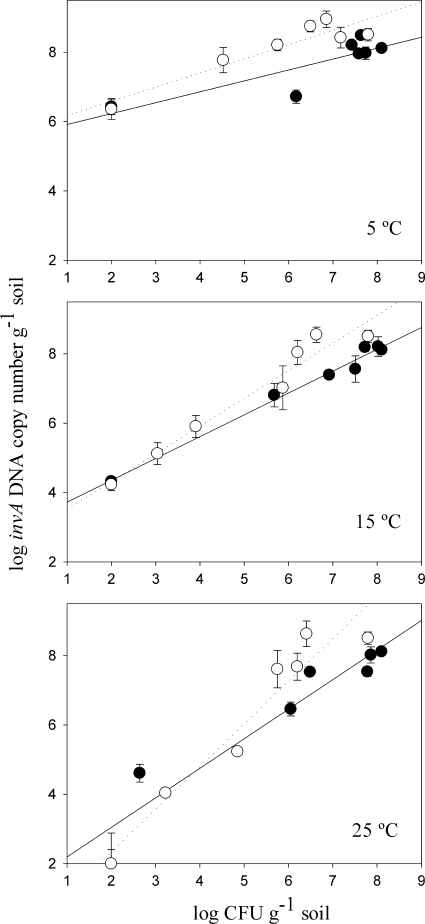
Comparison of plate counting and DNA quantification methods.
A regression of numbers of invA DNA copies compared to numbers of CFU of Salmonella serovar Typhimurium per gram of soil resulted in good correlations for the soil scenarios performed at both 15°C and at 25°C (R2, >0.9). For the soils incubated at 5°C, however, a correlation coefficient (R2) of <0.7 was observed (Fig. 2). In the samples at this low temperature, DNA quantification values were very high even when Salmonella serovar Typhimurium was not detected by plate counting. At 5°C, significant differences (P < 0.001) between DNA and CFU levels were found as early as 4 days in manure-amended soil and from 21 days in soil samples.
FIG. 2.
Linear regression of the levels of Salmonella serovar Typhimurium (Tet+) quantified by plate counting and qPCR. The logarithm of the invA DNA copy number is plotted versus the logarithm of the Salmonella serovar Typhimurium CFU count per gram of soil at three different temperatures: 5°C, 15°C, and 25°C. Symbols: •, soil; ○, manure-amended soil. Error bars represent the standard error of the mean.
Despite the good correlation between CFU values and DNA levels at 25°C, a comparison of regression slopes using the Fisher test indicated a significant difference between soils with and without manure. Also at 5°C, at high levels of CFU and invA, significant differences were found between slopes for the soils with and without manure. Addition of manure caused decreases in CFU levels especially in the beginning of the assay. This trend was not observed when quantifications were based on invA genes.
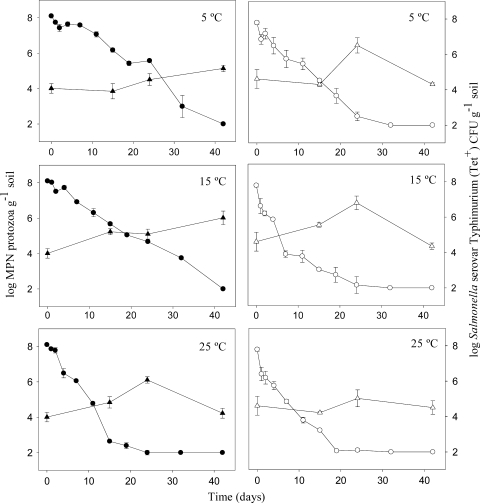
Growth of protozoa and its relation to Salmonella serovar Typhimurium survival.
The initial number of protozoa was 1.5 log higher in the manure-amended soil compared to the soil samples without manure. After 24 days, a bloom in protozoan numbers was observed in manure-amended soil at all three temperatures, with the highest levels (106) observed at 5 and 15°C (Fig. 3). In soils without manure, a slower increase in protozoan numbers was observed at 5 and 15°C, with the highest levels observed at the end of the experiment (42 days). The differences in protozoan evolution between the two soil scenarios, we believe, was a determining factor in the significant differences in Salmonella serovar Typhimurium survival. The decrease in CFU levels was faster in manure-amended soil samples at 5 and 15°C, where a faster increase in protozoan numbers was also observed. In all microcosms, prior to the bloom of protozoa, significant negative correlations were found between the abundance of protozoa and Salmonella serovar Typhimurium plate counting results (Pearson coefficient, <−0.5).
FIG. 3.
Evolution of Salmonella serovar Typhimurium CFU counts (•, ○) and protozoan levels (▵, ▴) in soil samples (closed symbols) and manure-amended soil samples (open symbols) incubated at three different temperatures. Protozoan counting was done at 15, 24, and 42 days after the beginning of the assay. Protozoan levels at time zero were determined before the addition of bacteria. Error bars represent the standard error of the mean.
DISCUSSION
Temperature has been shown to be an important factor in the survival of pathogenic bacteria in environmental samples (1, 22, 36). In our study, significant differences in Salmonella serovar Typhimurium survival at different temperatures were found by using three different detection methods. By the plate counting technique, the largest decline in bacterial levels was observed at the highest temperature (25°C), which corresponds to what was published earlier (54). Semenov et al. (47) showed that survival of Salmonella serovar Typhimurium cells in cow manure decreased faster at 23°C than at 7°C, and similar results were reported by Holley et al. (23). In several studies, enhanced survival of allochthonous bacteria in manure-amended soils has been proposed to be due to an increase in nutrient availability (16, 23, 27). In our study, however, manure addition induced a level of Salmonella serovar Typhimurium survival significantly lower than that in nonamended soil. This was proven both at 5°C and at 15°C. In our case, the high nutrient availability due to manure addition might have increased the activity of the native soil microbial community. Increasing the general competition between bacteria could lead to decreased survival of the introduced pathogen (15). The faster decrease in CFU levels in manure-amended soil can also be related to evolution in protozoan levels, since predation by protozoa is another factor affecting bacterial survival in soil. After inoculation of Salmonella serovar Typhimurium at 5 and 15°C, blooms in the numbers of protozoa occurred within 24 days, while in soils without manure the highest level of protozoa was not observed until the end of the assay. In all microcosms, before the maximum level of protozoa was reached, a significant negative correlation was found between the abundance of protozoa and Salmonella serovar Typhimurium plate counting results, corroborating other studies of the role of predation in pathogen survival (2, 45).
In all of the previous works, the influence of factors like temperature, manure addition, or protozoan predation on the survival of Salmonella in environmental samples was determined by plate counting techniques. Herein, however, we report that in addition to survival of Salmonella serovar Typhimurium based on CFU levels, these factors may influence the survival of Salmonella serovar Typhimurium based on quantification of invA DNA or mRNA as well.
Since DNA can persist in dead cells and thereby may bias the number of viable bacteria, quantification based on the extraction of DNA from environmental samples should be carefully evaluated (30, 42). Even though it has been argued that the half-life of DNA in environmental samples may be very short because of the presence of nucleases (32, 53), also very old DNA has been shown to persist in soil (33). Levels of Salmonella serovar Typhimurium based on DNA quantifications were significantly higher than those obtained by plate counting, especially in manure-amended samples at low temperatures. Such a trend has also been confirmed by others (11, 51). As previously reported by Turpin et al. (52), detection of Salmonella using a culture-based technique was not possible in any of the temperature scenarios after 42 days. Based on invA DNA levels, however, high levels of Salmonella serovar Typhimurium were still present at this time point, especially at 5°C, where a difference of 4 orders of magnitude between DNA and CFU levels was observed at the end of the assay, showing a nonculturable response higher at 5°C than at 25°C, as demonstrated by Gupte et al. (21). Due to a faster decrease in CFU levels, this difference was even more pronounced in the soil-with-manure scenario at 5°C.
The elevated invA gene levels found might be related to the presence of viable but nonculturable Salmonella serovar Typhimurium cells, as also reported by Marsh et al. (35). Especially at 5°C and 15°C, the ratio of invA gene to CFU levels was higher in soil with manure than in soil without manure. The addition of manure to soil seems to decrease the culturability of Salmonella serovar Typhimurium and support the presence of high levels of invA DNA.
The initial numbers of protozoa in our manure-amended soils were 1.5 logs higher than those in soil samples without manure, indicating that a significant number of protozoa was added along with the manure. These favorable conditions for predatory activity can be related directly to the fast decrease in Salmonella serovar Typhimurium CFU levels in the manure-amended soil. Brandl et al. (8) observed that the protozoan ciliate Tetrahymena contained intracellular feeding vesicles with high densities of ingested S. enterica. The subsequent release of these vesicles seemed to prolong bacterial survival in natural environments such as sites contaminated with manure, where this pathogen is present. The presence of Salmonella within intracellular protozoan vesicles likely underestimates the actual population of the pathogen because they cannot be detected by plate counting. Hence, if Salmonella is able to survive in protozoan vesicles, as other authors have reported (8, 19), it is in agreement with our results, where we found high numbers of invA gene levels versus low levels of CFU under conditions where predatory activity was higher. Our results also indicate that predation can be proposed to explain Salmonella dynamics in nonamended soils; the faster decline of cultivable Salmonella serovar Typhimurium at 25°C is consistent with the greater predatory levels observed under warm than under cooler soil conditions (5).
With the above-mentioned dilemma in mind, quantification of mRNA transcribed from a genus-specific gene could be the ideal biomarker for metabolically active Salmonella cells in environmental samples and it would help to determine their viability (49). In our study, we chose expression of the invA gene as such a biomarker. We observed a dramatic decrease in invA mRNA levels after only 48 h of inoculation at 5°C. At 15 and 25°C, this decrease was even faster and invA mRNA could only be detected in the 25°C microcosms until 3 h after the beginning of the experiment. Fey et al. (14) have done a study on the detection of invA mRNA in water samples, and they found a significant decrease in invA mRNA levels 3 h after inoculation, coinciding with the late logarithmic phase. In our study, however, this significant decrease in invA mRNA levels was detected 48 h after the inoculation of Salmonella serovar Typhimurium into samples incubated at 5°C.
In the present study, we were only able to detect invA mRNA in samples with more than 106 cells per g of soil (detected by plate counting), and this detection limit is higher than what has been observed in previous studies on mRNA quantification in soil samples (26, 37, 40). The fact that the number of invA transcripts in general was lower than the invA gene copy numbers may indicate that invA expression is downregulated as a response to stress due to suboptimal conditions in the environmental sample.
This study compared, for the first time, the fate of Salmonella serovar Typhimurium in soil and manure-amended soil at different temperatures by culturing and molecular methods. The Salmonella serovar Typhimurium levels detected by the three methods indicate that this pathogen survives better at 5°C. However, at this temperature, greater differences between invA DNA and CFU levels was observed. We propose the role of the predator-prey interaction as a factor to explain the differences between the culture and DNA quantification methods. The quantification of invA mRNA with our method confirms that Salmonella serovar Typhimurium shows invasive activity 48 h after inoculation into soil samples, although a high detection limit is observed. More work has to be done to improve molecular detection methods to evaluate the infectiousness of this strain in soils samples and how this ability may be affected by environmental conditions.
Acknowledgments
This study was supported by the Pathos project, which is funded by Strategic Research Council of Denmark ENV 2104-07-0015. R. García's stay at the Geological Survey of Denmark and Greenland was financed by the Spanish Institute for Agriculture, Food Research and Technology.
Footnotes
Published ahead of print on 18 June 2010.
REFERENCES
- 1.Arrus, K. M., R. A. Holley, K. H. Ominski, M. Tenuta, and G. Blank. 2006. Influence of temperature on Salmonella survival in hog manure slurry and seasonal temperature profiles in farm manure storage reservoirs. Livest. Sci. 102:226-236. [Google Scholar]
- 2.Artz, R. R. E., and K. Killham. 2002. Survival of Escherichia coli O157: H7 in private drinking water wells: influences of protozoan grazing and elevated copper concentrations. FEMS Microbiol. Lett. 216:117-122. [DOI] [PubMed] [Google Scholar]
- 3.Bælum, J., M. H. Nicolaisen, W. E. Holben, B. W. Strobel, J. Sorensen, and C. S. Jacobsen. 2008. Direct analysis of tfdA gene expression by indigenous bacteria in phenoxy acid amended agricultural soil. ISME J. 2:677-687. [DOI] [PubMed] [Google Scholar]
- 4.Baloda, S. B., L. Christensen, and S. Trajcevska. 2001. Persistence of a Salmonella enterica serovar Typhimurium DT12 clone in a piggery and in agricultural soil amended with Salmonella-contaminated slurry. Appl. Environ. Microbiol. 67:2859-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, J., T. J. Humphrey, and M. W. R. Brown. 1999. Survival of Escherichia coli O157 in a soil protozoan: implications for disease. FEMS Microbiol. Lett. 173:291-295. [DOI] [PubMed] [Google Scholar]
- 6.Bech, T. B., K. Johnsen, A. Dalsgaard, M. Laegdsmand, O. H. Jacobsen, and C. S. Jacobsen. 2010. Transport and distribution of Salmonella enterica serovar Typhimurium in loamy and sandy soil monoliths with applied liquid manure. Appl. Environ. Microbiol. 76:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, E. F., F. S. Wang, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandl, M. T., B. M. Rosenthal, A. F. Haxo, and S. G. Berk. 2005. Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Appl. Environ. Microbiol. 71:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briones, A. M., and W. Reichardt. 1999. Estimating microbial population counts by ‘most probable number’ using Microsoft Excel. J. Microbiol. Methods 35:157-161. [DOI] [PubMed] [Google Scholar]
- 10.Chiu, C. H., and J. T. Ou. 1996. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 34:2619-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen, N. D., L. J. Martin, R. B. Simpson, D. E. Wallis, and H. L. Neibergs. 1996. Comparison of polymerase chain reaction and microbiological culture for detection of salmonellae in equine feces and environmental samples. Am. J. Vet. Res. 57:780-786. [PubMed] [Google Scholar]
- 12.Cortez, A. L. L., A. Carvalho, A. A. Ikuno, K. P. Burger, and A. M. C. Vidal-Martins. 2006. Identification of Salmonella isolates from chicken abattoirs by multiplex-PCR. Res. Vet. Sci. 81:340-344. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza, D. H., F. J. Critzer, and D. A. Golden. 2009. Real-time reverse-transcriptase polymerase chain reaction for the rapid detection of Salmonella using invA primers. Foodborne Pathog. Dis. 6:1097-1106. [DOI] [PubMed] [Google Scholar]
- 14.Fey, A., S. Eichler, S. Flavier, R. Christen, M. G. Hofle, and C. A. Guzman. 2004. Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl. Environ. Microbiol. 70:3618-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franz, E., A. D. van Diepeningen, O. J. de Vos, and A. H. C. van Bruggen. 2005. Effects of cattle feeding regimen and soil management type on the fate of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in manure, manure-amended soil, and lettuce. Appl. Environ. Microbiol. 71:6165-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagliardi, J. V., and J. S. Karns. 2000. Leaching of Escherichia coli O157: H7 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galán, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional-characterization of the salmonella invasion gene invA: homology of invA to members of a new-protein family. J. Bacteriol. 174:4338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González-Escalona, N., T. S. Hammack, M. Russell, A. P. Jacobson, A. J. De Jesus, E. W. Brown, and K. A. Lampel. 2009. Detection of live Salmonella cells in produce by a TaqMan-based quantitative reverse transcriptase real-time PCR targeting invA mRNA. Appl. Environ. Microbiol. 75:3714-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gourabathini, P., M. T. Brandl, K. S. Redding, J. H. Gunderson, and S. G. Berk. 2008. Interactions between food-borne pathogens and protozoa isolated from lettuce and spinach. Appl. Environ. Microbiol. 74:2518-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan, T. Y., and R. A. Holley. 2003. Pathogen survival in swine manure environments and transmission of human enteric illness: a review. J. Environ. Qual. 32:383-392. [DOI] [PubMed] [Google Scholar]
- 21.Gupte, A. R., C. L. E. de Rezende, and S. W. Joseph. 2003. Induction and resuscitation of viable but nonculturable Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 69:6669-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himathongkham, S., S. Bahari, H. Riemann, and D. Cliver. 1999. Survival of Escherichia coli O157:H7 and Salmonella typhimurium in cow manure and cow manure slurry. FEMS Microbiol. Lett. 178:251-257. [DOI] [PubMed] [Google Scholar]
- 23.Holley, R. A., K. M. Arrus, K. H. Ominski, M. Tenuta, and G. Blank. 2006. Salmonella survival in manure-treated soils during simulated seasonal temperature exposure. J. Environ. Qual. 35:1170-1180. [DOI] [PubMed] [Google Scholar]
- 24.Hoorfar, J., P. Ahrens, and P. Radstrom. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Islam, M., J. Morgan, M. P. Doyle, S. C. Phatak, P. Millner, and X. P. Jiang. 2004. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl. Environ. Microbiol. 70:2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobsen, C. S., and W. E. Holben. 2007. Quantification of mRNA in Salmonella sp. seeded soil and chicken manure using magnetic capture hybridization RT-PCR. J. Microbiol. Methods 69:315-321. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, X. P., J. Morgan, and M. P. Doyle. 2002. Fate of Escherichia coli O157:H7 in manure-amended soil. Appl. Environ. Microbiol. 68:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, P. W. 1976. Effect of temperature, solids content and pH on survival of Salmonellas in cattle slurry. Br. Vet. J. 132:284-293. [DOI] [PubMed] [Google Scholar]
- 29.Jones, P. W. (ed.). 1986. Sewage sludge as a vector of salmonellosis, p. 21-33. In J. C. Block, A. H. Haielaar, and P. L'Hermite (ed.), Epidemiological studies of risks associated with the agricultural use of sewage sludge. Elsevier, London, England.
- 30.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keer, J. T., and L. Birch. 2003. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 53:175-183. [DOI] [PubMed] [Google Scholar]
- 32.Lleo, M. M., B. Bonato, M. C. Tafi, C. Signoretto, C. Pruzzo, and P. Canepari. 2005. Molecular vs culture methods for the detection of bacterial faecal indicators in groundwater for human use. Lett. Appl. Microbiol. 40:289-294. [DOI] [PubMed] [Google Scholar]
- 33.Lorenz, M. G., and W. Wackernagel. 1991. High frequency of natural genetic transformation of Pseudomonas stutzeri in soil extract supplemented with a carbon/energy and phosphorus source. Appl. Environ. Microbiol. 57:1246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malorny, B., and J. Hoorfar. 2005. Toward standardization of diagnostic PCR testing of fecal samples: lessons from the detection of salmonellae in pigs. J. Clin. Microbiol. 43:3033-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsh, P., N. Z. Morris, and E. M. H. Wellington. 1998. Quantitative molecular detection of Salmonella typhimurium in soil and demonstration of persistence of an active but non-culturable population. FEMS Microbiol. Ecol. 27:351-363. [Google Scholar]
- 36.Mawdsley, J. L., R. D. Bardgett, R. J. Merry, B. F. Pain, and M. K. Theodorou. 1995. Pathogens in livestock waste, their potential for movement through soil and environmental pollution. Appl. Soil Ecol. 2:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meckenstock, R., P. Steinle, J. R. van der Meer, and M. Snozzi. 1998. Quantification of bacterial mRNA involved in degradation of 1,2,4-trichlorobenzene by Pseudomonas sp. strain P51 from liquid culture and from river sediment by reverse transcriptase PCR (RT/PCR). FEMS Microbiol. Lett. 167:123-129. [DOI] [PubMed] [Google Scholar]
- 38.Mortensen, S. K., and C. S. Jacobsen. 2004. Influence of frozen storage on herbicide degradation capacity in surface and subsurface sandy soils. Environ. Sci. Technol. 38:6625-6632. [DOI] [PubMed] [Google Scholar]
- 39.Natvig, E. E., S. C. Ingham, B. H. Ingham, L. R. Cooperband, and T. R. Roper. 2002. Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Appl. Environ. Microbiol. 68:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicolaisen, M. H., J. Bælum, C. S. Jacobsen, and J. Sorensen. 2008. Transcription dynamics of the functional tfdA gene during MCPA herbicide degradation by Cupriavidus necator AEO106 (pRO101) in agricultural soil. Environ. Microbiol. 10:571-579. [DOI] [PubMed] [Google Scholar]
- 41.Page, F. C. 1988. A new key to freshwater and soil gymnamoebae with instructions for culture. Culture collection of algae and protozoa at Freshwater Biological Association, Scottish Marine Biological Association. Natural Environment Research Council, Ambleside, United Kingdom.
- 42.Pedersen, J. C., and C. S. Jacobsen. 1993. Fate of Enterobacter cloacae JP120 and Alcaligenes eutrophus AEO106(pRO101) in soil during water-stress: effects on culturability and viability. Appl. Environ. Microbiol. 59:1560-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pell, A. N. 1997. Manure and microbes: public and animal health problem? J. Dairy Sci. 80:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahn, K., S. A. Degrandis, R. C. Clarke, S. A. McEwen, J. E. Galán, C. Ginocchio, R. Curtiss, and C. L. Gyles. 1992. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6:271-279. [DOI] [PubMed] [Google Scholar]
- 45.Recorbet, G., C. Steinberg, and G. Faurie. 1992. Survival in soil of genetically engineered Escherichia coli as related to inoculum density, predation and competition. FEMS Microbiol. Ecol. 101:251-260. [Google Scholar]
- 46.Selinger, D. W., R. M. Saxena, K. J. Cheung, G. M. Church, and C. Rosenow. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semenov, A. V., A. H. C. van Bruggen, L. van Overbeek, A. J. Termorshuizen, and A. M. Semenov. 2007. Influence of temperature fluctuations on Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in cow manure. FEMS Microbiol. Ecol. 60:419-428. [DOI] [PubMed] [Google Scholar]
- 48.Semenov, A. V., L. van Overbeek, and A. H. C. van Bruggen. 2009. Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Appl. Environ. Microbiol. 75:3206-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheridan, G. E. C., C. I. Masters, J. A. Shallcross, and B. M. Mackey. 1998. Detection of mRNA by reverse transcription PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sorber, C. A., and B. E. Moore. 1987. Survival and transport of pathogens in sludge-amended soil: a critical literature review. U.S. Environmental Protection Agency, Washington, DC.
- 51.Touron, A., T. Berthe, B. Pawlak, and F. Petit. 2005. Detection of Salmonella in environmental water and sediment by a nested-multiplex polymerase chain reaction assay. Res. Microbiol. 156:541-553. [DOI] [PubMed] [Google Scholar]
- 52.Turpin, P. E., K. A. Maycroft, C. L. Rowlands, and E. M. H. Wellington. 1993. Viable but non-culturable Salmonellas in soil. J. Appl. Bacteriol. 74:421-427. [DOI] [PubMed] [Google Scholar]
- 53.Wery, N., A. M. Pourcher, V. Stan, J. P. Delgenes, F. Picard-Bonnaud, and J. J. Godon. 2006. Survival of Listeria monocytogenes and Enterococcus faecium in sludge evaluated by real-time PCR and culture methods. Lett. Appl. Microbiol. 43:131-136. [DOI] [PubMed] [Google Scholar]
- 54.Zibilske, L. M., and R. W. Weaver. 1978. Effect of environmental factors on survival of Salmonella typhimurium in soil. J. Environ. Qual. 7:593-597. [Google Scholar]