Abstract
To better describe the community structure of sulfate-reducing bacteria in environmental systems, we compared several dissimilatory sulfite reductase (dsr) primer sets for terminal restriction fragment length polymorphism application. A new reverse primer that increased allelic diversity estimates up to 5-fold was applied to hydrocarbon seep samples to examine the relationship between guild activity and diversity.
A major scientific challenge in ecology is to link community function with community structure. For the sulfate-reducing microorganisms (SRM) that make up a large polyphyletic guild, with species belonging to at least five bacterial phyla and two archaeal phyla (14), community diversity of SRM is often assessed by using the dissimilatory (bi)sulfite reductase (dSir [EC 1.8.99.3]) subunits encoded by dsrA and dsrB gene sequences as functional markers (1, 5, 11). While these genes exhibit high conservation, considerable polymorphisms exist at the traditionally targeted primer sites (18, 20). While a number of molecular methods have recently been employed to address the functional gene content of communities, such as functional gene arrays (19) or metatranscriptome sequencing (for example, reference 16), there is still a need for relatively inexpensive and high-throughput methods, like community fingerprinting.
Numerous dsrAB-based studies of community structure have employed primers DSR1F and DSR4R (17) as the sole PCR primer set to sample SRM diversity via clone library analysis (see, for example, references 1, 2, and 10). More recently, mixes of five or six degenerate forward and reverse primers have been used to explore dsrAB diversity in environmental samples, thus enabling the discovery of new phylotypes (4, 9, 18). These forward and reverse mix primer sets have amplified no archaeal sequences in studies in which they were employed, despite their capacity to do so at least in silico (4, 9), suggesting that sulfate-reducing archaea may be rare in cold and temperate environmental systems. Here, we focused on sulfate-reducing bacteria (SRB) and improved their target primer set for high-throughput molecular screening of sediments and soils by the community fingerprinting method terminal restriction fragment length polymorphism (T-RFLP) analysis (6). This will facilitate studies of the spatial and temporal distribution of SRB in various environmental samples, such as natural or accidental hydrocarbon-impacted sediments.
We designed a novel reverse primer (DSR1334R) taking into account the technical requirements of T-RFLP, including a high primer specificity for this PCR-based method. dsr-based clone libraries were generated using various primer sets under similar conditions as used for dsrA-based T-RFLP. The improved dsrA-based fingerprinting strategy was then applied to sediment samples from an active natural hydrocarbon seep (8) to investigate the link between community function and diversity.
Primer design and comparison.
The Probe Match tool in ARB software program (7) and plotcom of the EMBOSS package (13) were employed to identify highly conserved regions for the bacterial dsrAB sequences (20). Candidate primers (Fig. 1 and Table 1) were further tested for appropriate thermodynamic stability, GC content, melting temperature, and length with the Primer3Plus algorithm (15).
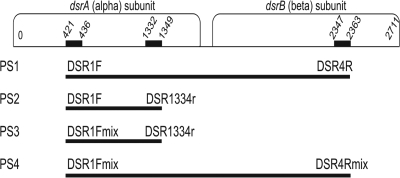
FIG. 1.
Positions of the forward and reverse primers on the dsrA and dsrB genes. Numbers indicate the target site for each primer (according to the Desulfovibrio vulgaris Hildenborough strain [GenBank accession no. U16723]). The expected amplification size (approximately 1,900 bp for primer sets PS1 and PS4 and approximately 930 bp for PS2 and PS3) is denoted for each primer set as a bar (not drawn to scale).
TABLE 1.
Primers used in this study
| Primer | Gene targeta | Sequence (5′ to 3′)b | Tmc (°C) | Reference |
|---|---|---|---|---|
| DSR1F | dsrA | ACS CAC TGG AAG CAC G | 57.0 | Wagner et al. (17) |
| DSR4R | dsrB | GTG TAG CAG TTA CCG CA | 51.1 | Wagner et al. (17) |
| DSR1334R | dsrA | TYT TCC ATC CAC CAR TCC | 57.2 | This study |
| DSR1Fmixd | ||||
| DSR1F | dsrA | ACS CAC TGG AAG CAC G | Wagner et al. (17) | |
| DSR1Fa | dsrA | ACC CAY TGG AAA CAC G | 53.0 | Loy et al. (6a) |
| DSR1Fb | dsrA | GGC CAC TGG AAG CAC G | 59.9 | Loy et al. (6a) |
| DSR1Fc | dsrA | ACC CAT TGG AAA CAT G | 49.8 | Zverlov et al. (20) |
| DSR1Fd | dsrA | ACT CAC TGG AAG CAC G | 50.4 | Zverlov et al. (20) |
| DSR4Rmixe | ||||
| DSR4R | dsrB | GTG TAG CAG TTA CCG CA | Wagner et al. (17) | |
| DSR4Ra | dsrB | GTG TAA CAG TTT CCA CA | 43.9 | Loy et al. (6a) |
| DSR4Rb | dsrB | GTG TAA CAG TTA CCG CA | 47.8 | Loy et al. (6a) |
| DSR4Rc | dsrB | GTG TAG CAG TTK CCG CA | 56.0 | Loy et al. (6a) |
| DSR4Rd | dsrB | GTG TAG CAG TTA CCA CA | 44.3 | Zverlov et al. (20) |
| DSR4Re | dsrB | GTG TAA CAG TTA CCA CA | 40.6 | Zverlov et al. (20) |
Primer annealing position according to Desulfovibrio vulgaris Hildenborough strain (GenBank accession no. U16723).
Wobble positions are shown as follows: S = G or C, Y = C or T, R = A or G, and K = G or T.
Tm, melting temperature.
The final primer mixture is an equimolar mixture of each variant (50 μM) of the following primers: DSR1F, DSR1Fa, DSR1Fb, DSR1Fc, and DSR1Fd.
The final primer mixture is an equimolar mixture of each variant (50 μM) of the following primers: DSR4R, DSR4Ra, DSR4Rb, DSR4Rc, DSR4Rd, and DSR4Re.
dsr-based T-RFLP.
Environmental genomic DNA from hydrocarbon-rich, sulfidic sediments of a Gulf of Mexico cold seep (see the supplemental material) was extracted and used in triplicate 35-μl PCRs prepared with GoTaq DNA polymerase reagents in accordance with the manufacturer's instructions. While primer set PS1 (named PS1 for primer set 1) was used to amplify the 1.9-kb dsrAB target region, PS2 and PS3 amplified a nearly 930-bp dsrA amplicon. PS4 was not used for T-RFLP given the nonspecific amplification observed during the clone library protocol (see supplemental material).
To avoid nonspecific priming, reaction mixtures were prepared on ice and were transferred directly onto a preheated (94°C) thermocycler block. PCR amplification was performed as follows: (i) 3 min at 94°C; (ii) 30 cycles, with 1 cycle consisting of 40 s at 94°C, 40 s at 54°C, and 2 min at 72°C; and (iii) a final elongation step of 8 min at 72°C. A common annealing temperature (54°C) was selected for all PCR amplifications based on gradient PCR analyses (data not shown); the aim was to allow the amplification of as many variant dsrA targets as possible but also to keep the PCR specificity high enough. Bands corresponding to the predicted dsr amplicon size were then excised and purified (QIAquick PCR purification kit; Qiagen, Hilden, Germany).
Approximately 100 ng of each elutant was digested using 5 units of NdeII and recommended reagents (Promega, Mannheim, Germany) for 2 h at 37°C, followed by 15 min of heat deactivation at 65°C. After purification, restriction products were prepared for capillary electrophoresis as described elsewhere (12). Fragment sorting and binning were done with a window size of 2 bp and shift size of 0.1 bp as described previously (12).
Primer design and in silico assessment.
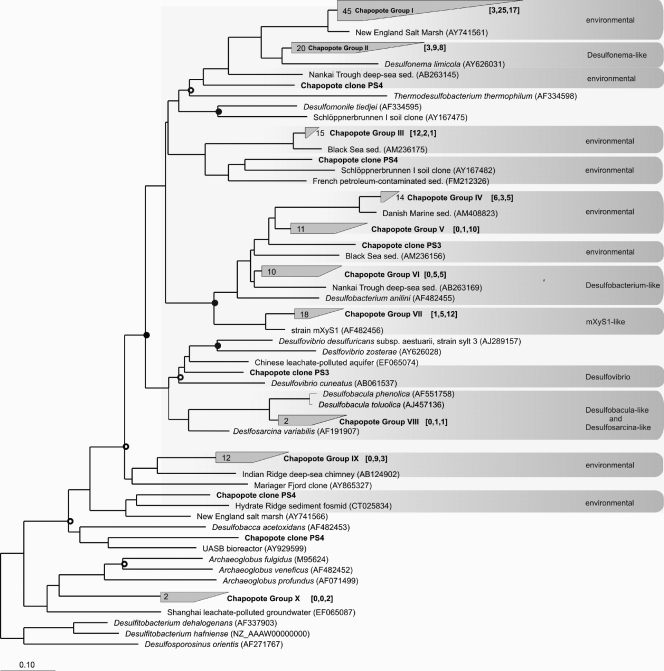
A novel reverse primer, DSR1334R (Fig. 1 and Table 1), targeting the 3′ end of the dsrA gene was designed based on alignments of nearly 100 cultured archaeal and bacterial sulfate reducers in a dsrAB gene ARB database (20). Selection of primer DSR1334R was guided by the fact that (i) short amplicons (950 bp with primer sets PS2 and PS3 versus 1,900 bp with primer sets PS1 and PS4) are generally favored under PCR conditions, given that divergent and lower abundant taxa can be more readily detected with smaller amplicon target size (3), and (ii) subsequently digested amplicons by the NdeII 4-mer restriction enzyme would most likely cut before 900 bp, according to in silico tests (11; our unpublished data). A description of the coverage of primer DSR1334R, along with its amplification efficiency in comparison to other primers, is included in the supplemental material (see Fig. S1 in the supplemental material). Furthermore, clone libraries produced with PS2, PS3, and PS4 on sample A3 revealed that dsr-possessing bacteria dominate the sample libraries (Fig. 2) (see supplemental material for protocol).
FIG. 2.
Phylogenetic tree constructed from analysis of dsrA nucleic acid sequences showing the inferred phylogenetic positions of clones from the three libraries generated using primer sets PS2, PS3, and PS4 (shown in boldface type) on sample A3. The analysis was based on approximately 600 aligned nucleic acid sequences and was calculated via distance matrix-based (Jukes-Cantor correction) analyses. GenBank accession numbers are shown in parentheses. The numbers in the brackets show the number of clones from each library, in the following order [PS2, PS3, PS4]. Gray boxes designate sequence clusters with less than 30% sequence divergence. Nodes receiving ≥50% bootstrap support are marked by open circles, while nodes receiving ≥70% bootstrap support are marked by closed circles. The bar shows 10% estimated sequence divergence. sed., sediment.
Evaluation of dsr primers for T-RFLP applications on environmental samples.
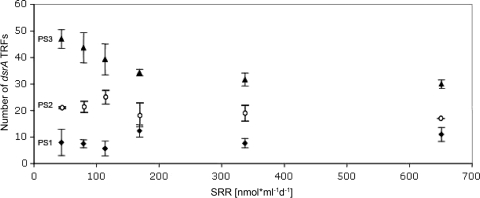
When primer DSR1334R was compared against, and used in combination with, previously published dsrAB primers, the total numbers of terminal restriction fragments (TRFs) differed substantially as a function of the primer sets used, with primer set PS3-based T-RFLP consistently identifying two to four times more TRFs than PS2 and PS1, respectively (Fig. 3). Noticeably, the greatest difference in TRF number between primer sets was observed at low sulfate reduction (SR) rates, when 47 and 9 TRFs were observed for PS3 and PS1, respectively.
FIG. 3.
Number of TRFs and the corresponding sulfate reduction rates. Three different primer sets were applied to sediment samples from the Chapopote hydrocarbon seep, and the number of binned TRFs of each sample was plotted along an axis of sulfate reduction activity. The sulfate reduction rate (SRR) is shown in nanomoles milliliter−1 day−1. Values represent the means ± standard errors of the means (error bars) (three replicates per point).
Interestingly, when the number of dsrA TRFs detected with the high-resolution primer set PS3 was plotted against the range of sulfate reduction rates sampled at the hydrocarbon seep, a negative relationship was observed in the low to high range (50 to 200 nmol ml−1 day−1) of SR (Fig. 3). This could be explained by an ecological selection for particular ecotypes of dsrA in subsurface hot spots of the hydrocarbon seep associated with higher activity, a hypothesis that would need to be tested in future studies. Such a relationship was not observed with PS2 or PS1 and the sulfate reduction rate (SRR), most likely due to the limited subsampling of SRB guild diversity.
Overall, primer DSR1334R, in combination with primer DSR1Fmix (i.e., PS3), provides reliable dsr-based T-RFLP profiles and yields significantly higher TRF number estimates (2.5- to 5-fold with our test samples) compared with previously employed dsr-based T-RFLP primers. Hence, this improved methodology may enable the characterization of a larger fraction (greater than approximately 80% of those found in current dsrA databases) of the sulfate-reducing bacterial guild and will facilitate the high-throughput study of their spatial and temporal dynamics in the environment.
Supplementary Material
Acknowledgments
We thank the chief scientist, crew, and scientific party of the Meteor 67/2 expedition for support with work at sea. We also thank Katrin Knittel for sharing insights regarding hydrocarbon seep systems, Gunter Wegener for processing the sulfate reduction rates, and Mirko Basen for providing positive-control strains.
Research support was provided by the Max Planck Society and by the MARUM Center for Marine Environmental Sciences.
Footnotes
Published ahead of print on 11 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansel, C. M., S. Fendorf, P. M. Jardine, and C. A. Francis. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 74:1620-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huber, J. A., H. G. Morrison, S. M. Huse, P. R. Neal, M. L. Sogin, and D. B. M. Welch. 2009. Effect of PCR amplicon size on assessments of clone library microbial diversity and community structure. Environ. Microbiol. 11:1292-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kjeldsen, K. U., A. Loy, T. R. Thomsen, T. F. Jakobsen, M. Wagner, and K. Ingvorsen. 2007. Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment in Great Salt Lake (Utah, USA). FEMS Microbiol. Ecol. 60:287-298. [DOI] [PubMed] [Google Scholar]
- 5.Kjeldsen, K. U., L. Tang, M. G. Jørgensen, and K. Ingvorsen. 2009. Enumeration and identification of dominant types of sulfate-reducing bacteria in pulp from a paper-recycling plant: a multiphasic approach. FEMS Microbiol. Ecol. 69:481-494. [DOI] [PubMed] [Google Scholar]
- 6.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Loy, A., K. Küsel, A. Lehner, H. L. Drake, and M. Wagner. 2004. Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl. Environ. Microbiol. 70:6998-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacDonald, I. R., G. Bohrmann, E. Escobar, F. Abegg, P. Blanchon, V. Blinova, W. Bruckmann, M. Drews, A. Eisenhauer, X. Han, K. Heeschen, F. Meier, C. Mortera, T. Naehr, B. N. Orcutt, B. Bernard, J. M. Brooks, and M. de Farago. 2004. Asphalt volcanism and chemosynthetic life in the Campeche Knolls, Gulf of Mexico. Science 304:999-1102. [DOI] [PubMed] [Google Scholar]
- 9.Miletto, M., A. Loy, A. M. Antheunisse, R. Loeb, P. L. E. Bodelier, and H. J. Laanbroek. 2008. Biogeography of sulfate-reducing prokaryotes in river floodplains. FEMS Microbiol. Ecol. 64:395-406. [DOI] [PubMed] [Google Scholar]
- 10.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pérez-Jiménez, J. R., and L. J. Kerkhof. 2005. Phylogeography of sulfate-reducing bacteria among disturbed sediments, disclosed by analysis of dissimilatory sulfite reductase genes (dsrAB). Appl. Environ. Microbiol. 71:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramette, A. 2009. Quantitative community fingerprinting methods for estimating the abundance of operational taxonomic units in natural microbial communities. Appl. Environ. Microbiol. 75:2495-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 14.Stahl, D. A., A. Loy, and M. Wagner. 2007. Molecular strategies for studies of natural populations of sulphate-reducing microorganisms, p. 39-117. In L. L. Barton and W. A. Hamilton (ed.), Sulfate-reducing bacteria: environmental and engineered systems. Cambridge University Press, Cambridge, United Kingdom.
- 15.Untergasser, A., H. Nijveen, X. Rao, T. Bisseling, R. Geurts, and J. A. Leunissen. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71-W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urich, T., A. Lanzén, J. Qi, D. H. Huson, C. Schleper, and S. C. Schuster. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner, M., A. Loy, M. Klein, N. Lee, N. B. Ramsing, D. A. Stahl, and M. W. Friedrich. 2005. Functional marker genes for identification of sulphate-reducing prokaryotes. Methods Enzymol. 397:469-489. [DOI] [PubMed] [Google Scholar]
- 19.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zverlov, V., M. Klein, S. Lücker, M. W. Friedrich, J. Kellermann, D. A. Stahl, A. Loy, and M. Wagner. 2005. Lateral gene transfer of dissimilatory (bi)sulfite reductase revisited. J. Bacteriol. 187:2203-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.