Abstract
Bacteriophages specific to Salmonella strains were isolated from sewage effluent and characterized. A five-strain bacteriophage mixture was applied to dairy manure compost inoculated with Salmonella enterica serotype Typhimurium. Bacteriophage treatment resulted in a greater than 2-log-unit reduction of Salmonella within 4 h at all moisture levels compared to the controls.
Composting is a complex process designed to mitigate the risk of pathogen contamination while producing a nutrient-rich substrate, suitable for land application (19). When performed properly, pathogenic enteric microorganisms, such as Salmonella and Escherichia coli O157:H7, are reduced to undetectable levels in most cases (18). Some studies, however, have revealed that Salmonella strains are able to survive if composting is performed improperly (11). Furthermore, the surfaces of compost heaps have been shown to reach insufficient temperatures for the complete inactivation of pathogenic bacteria (27) and may result in pathogen regrowth (12).
The growing demand for organically grown fruits and vegetables emphasizes the need for safe soil amendments and organic fertilizers. Despite increased awareness of the potential risk of pathogen contamination of crops, multiple outbreaks of food-borne illnesses associated with fresh produce have occurred (3, 21). The persistence of human pathogens in compost has led researchers to explore different approaches for pathogen reduction, such as irradiation or ammonia supplementation (20, 24). To date, there are no reports on the potential for using bacteriophage to reduce pathogen contamination of compost. Recent bacteriophage studies have evaluated their effectiveness in live animals (2, 26, 28), on fresh produce (15, 23, 25), and on meat products (7, 30). Application of bacteriophages may therefore be a preventive step in the preharvest stages of food production.
The objectives of this study were to isolate and characterize bacteriophages specific to Salmonella serovars and to develop a bacteriophage mixture effective in reducing pathogen contamination in compost under different environmental conditions.
Isolation, characterization, and selection of Salmonella bacteriophages.
Bacteriophages specific to Salmonella strains were isolated using five sewage samples collected from treatment facilities in three cities in South Carolina (Pendleton, Anderson, and Clemson). Bacteriophage enrichments were performed by mixing 12 Salmonella host strains at 108 CFU/ml with a sewage mixture. The 12 strains belonged to Salmonella enterica serotypes Enteritidis (n = 5), Typhimurium (n = 3), Newport (n = 2), Poona (n = 1), and Heidelberg (n = 1). The bacteriophages were then purified by standard procedures (29). Briefly, the presence of phage was determined by spotting a 10-μl drop of filtered enrichment mixture onto the surface of a tryptic soy agar (TSA) plate overlaid with 0.6% agar containing specific host bacterial strains. The bacterial strains positive for plaque formation were then purified by selecting plaques with a sterile pipette tip.
Bacteriophages were successfully isolated for 10 of the 12 Salmonella strains used as hosts for enrichment (Table 1) with a total of 33 phages isolated. Bacteriophage strain Felix O1 (FO) was kindly provided by the University of Calgary. Bacterial lysis was determined by spotting 10 μl of bacteriophage suspension onto lawns of Salmonella prepared according to standard protocols (1). The host range of 34 bacteriophage isolates was determined with 53 strains of Salmonella, and the results for 10 phages were presented in Table 1. Host range comparison revealed that most bacteriophages were able to lyse Salmonella strains used during the initial bacteriophage enrichment, with 5 bacteriophages being able to infect more than 20 of the 53 strains, but none were effective against all strains (Table 1). Bacteriophage FO, which has been previously characterized (14), had a unique host range, as it was able to lyse 10 serotypes. Most Salmonella serotypes that were previously isolated from rendered animal by-products (n = 18) were not susceptible to the bacteriophages used in this study (data not shown). Most of these serotypes were from animal sources; however, they were rarely associated with food-borne illnesses (13). Due to the diversity of Salmonella serotypes and host-specific properties of bacteriophages, a mixture of multiple bacteriophages is required for Salmonella treatment.
TABLE 1.
Host range of 10 Salmonella bacteriophage isolates determined on 35 Salmonella host strainsa
| S. enterica serotype and/or strainb | Lysis by bacteriophagec: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S5p1 | S5p2 | S6p1 | S6p2 | p1 | p2 | p4 | FO | (1)-2 | {2} | |
| 3:19 | ++ | − | ++ | ++ | − | − | − | +++ | +++ | ++ |
| Anatum N5396 | − | − | ++ | ++ | − | − | − | − | +++ | ++ |
| Anatum | − | − | − | − | − | − | − | − | +++ | +++ |
| Cornwallis | ++ | − | ++ | ++ | + | − | − | +++ | +++ | +++ |
| Dublin 23742 | ++ | ++ | − | ++ | ++ | ++ | ++ | +++ | ++ | ++ |
| Enteritidis 15060 | +++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ |
| Enteritidis 30661 | +++ | +++ | ++ | +++ | +++ | +++ | +++ | ++ | ++ | ++ |
| Enteritidis Benson | +++ | +++ | + | +++ | +++ | +++ | +++ | S | ++ | ++ |
| Enteritidis H2292 | +++ | +++ | − | ++ | ++ | +++ | +++ | − | ++ | ++ |
| Enteritidis H3353 | +++ | +++ | + | +++ | +++ | +++ | +++ | ++ | +++ | +++ |
| Enteritidis H4639 | +++ | +++ | − | +++ | +++ | +++ | +++ | +++ | ++ | ++ |
| Enteritidis H4717 | +++ | +++ | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Enteritidis ME18 | +++ | +++ | + | + | + | +++ | +++ | +++ | +++ | +++ |
| Heidelberg UNL | − | S | − | − | − | − | − | ++ | ++ | ++ |
| Heidelberg 21380 | − | S | − | ++ | ++ | S | − | +++ | +++ | +++ |
| Infantis | − | − | ++ | ++ | − | − | − | +++ | − | − |
| Kentucky | − | + | − | − | − | − | − | +++ | +++ | +++ |
| Kentucky N11150 | − | − | − | − | − | − | − | +++ | +++ | +++ |
| Montevideo | − | − | − | ++ | − | − | − | +++ | − | − |
| Newport H9113 | ++ | ++ | − | ++ | − | − | − | S | ++ | ++ |
| Newport H9116 | +++ | +++ | +++ | +++ | + | − | +++ | +++ | +++ | +++ |
| Newport M | ++ | − | ++ | ++ | − | − | − | +++ | ++ | ++ |
| Newport N635 | ++ | − | ++ | ++ | − | − | − | +++ | ++ | ++ |
| Poona H9G77 | S | S | − | S | − | − | − | − | − | − |
| Poona H9301 | ++ | − | − | ++ | − | − | − | + | +++ | +++ |
| St. Paul 22398 | ++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Tennessee | − | − | − | − | − | − | − | − | − | − |
| Tennessee K4720 | − | − | − | − | − | − | − | ++ | − | − |
| Thompson M | − | − | − | − | − | − | − | − | +++ | +++ |
| Thompson G | − | − | − | − | − | − | − | − | − | − |
| Typhimurium 8243 | ++ | +++ | +++ | +++ | +++ | +++ | − | S | ++ | +++ |
| Typhimurium ATCC 14028 | S | ++ | + | +++ | +++ | ++ | S | S | − | S |
| Typhimurium Copenhagen | ++ | ++ | − | S | ++ | ++ | ++ | ++ | − | S |
| Typhimurium DT104 | ++ | ++ | − | S | ++ | ++ | ++ | ++ | − | S |
| Typhimurium 32463 | ++ | − | ++ | +++ | +++ | ++ | ++ | +++ | +++ | +++ |
| Total no. of strains lysed | 21 | 17 | 17 | 24 | 18 | 15 | 14 | 24 | 26 | 26 |
A subset of the host range data obtained for 34 bacteriophage isolates screened against 53 Salmonella strains.
Boldface Salmonella strains were used for bacteriophage enrichment experiments.
Ten Salmonella bacteriophages were tested on Salmonella host strains. The 10 bacteriophages are ME18 S5p1, H4717 S5p2, H9116 S6p1, H3353 S6p2, H3353 p1, ME18 p2, ME18 p4, Felix O1 (FO), H9301 (1)-2, and H2292 {2}; only the last portion of the bacteriophage designation is shown for 9 of the 10 bacteriophages (Felix O1 [FO] is the exception). Lysis results were recorded as follows: +++, confluent lysis; ++, semiconfluent lysis; +, individual plaques; S, shadow lysis; −, no lysis.
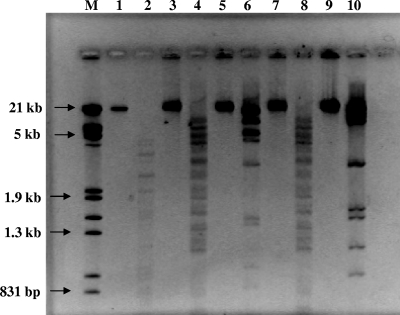
Five bacteriophages [H4717 S5p2, ME-18 p2, H9301 (1)-2, H2292 {2}, and FO] were selected for use based on host range and lytic ability in tryptic soy broth (TSB). The preparation of high concentrations of bacteriophage was performed according to the protocol of Yoichi et al. (31). DNA extraction was performed with the MoBio UltraClean microbial DNA isolation kit (MoBio, Carlsbad, CA). DNA samples were digested with the restriction endonucleases DraI and NdeI (Promega, Madison, WI) according to the supplier's recommendations. Restriction analysis by DraI of the five Salmonella bacteriophages revealed that bacteriophages H9301 (1)-2 and H2292 {2} shared the same patterns, whereas the other bacteriophages had unique patterns (Fig. 1). Further restriction analysis by digestion with NdeI confirmed that H9301 (1)-2 and H2292 {2} had indistinguishable patterns; however, host range analysis revealed differences between the two (Table 1). These two bacteriophage strains, isolated from different sources, could be closely related but divergent enough to change host range.
FIG. 1.
Digestion of bacteriophage DNA using the restriction enzyme DraI. Lane M, Lambda HindIII/EcoRI DNA marker; lane 1, undigested FO DNA; lane 2, digested FO DNA; lane 3, undigested H9301 (1)-2 DNA; lane 4, digested H9301 (1)-2 DNA; lane 5, undigested H4717 S5p2 DNA; lane 6, digested H4717 S5p2 DNA; lane 7, undigested H2292 {2} DNA; lane 8, digested H2292 {2} DNA; lane 9, ME-18 p2 DNA; lane 10, digested ME-18 p2 DNA.
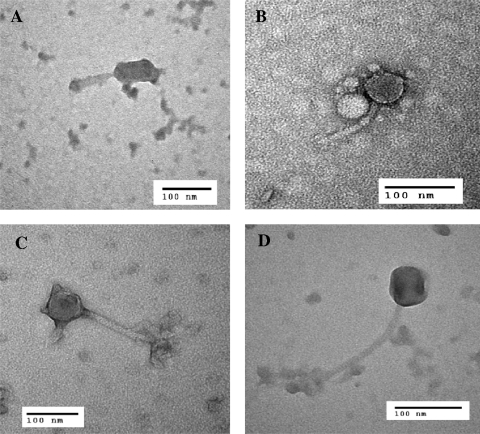
Transmission electron microscopy images of four selected Salmonella bacteriophages revealed that all had unique morphologies (Fig. 2). The presence of a contractile tail for H4717 S5p2 suggests that it belongs to the family of Myoviridae. Bacteriophages ME-18 p2, H9301 (1)-2, and H2292 {2} had flexible tails and were belong to the family Siphoviridae. Bacteriophage H2292 {2} had an unusually long tail compared to the tail of bacteriophage H9301 (1)-2.
FIG. 2.
Transmission electron micrographs of four Salmonella-specific phages. (A) H4717 S5p2 observed at a magnification of ×346,000; (B) ME-18 p2 observed at a magnification of ×333,000; (C) H9301 (1)-2 observed at a magnification of ×324,000; (D) H2292 {2} observed at a magnification of ×459,000.
Each phage was tested individually in TSB with 100 μg rifampin/ml (TSB-R) against their respective rifampin-resistant Salmonella hosts. The results of this study revealed that treatment with a single bacteriophage was not effective at completely eliminating Salmonella (data not shown). Similar studies by McLaughlin (16) and O'Flynn et al. (22) revealed that in most situations, using a single species of bacteriophage is insufficient to eliminate the growth of an entire bacterial population in broth due to the development of bacteriophage-insensitive mutants (BIMs). To determine whether BIMs developed in our study, individual isolates were collected from cultures of S. Typhimurium 8243 that remained turbid following treatment with each of the five phages. These putative mutants were then retested against the phages, revealing that there was a decrease in susceptibility to phages H9301 (1)-2 and H2292 {2} in some of the isolates, while the rest remained susceptible (data not shown). Clearly, these results highlight the importance of developing bacteriophage mixtures that can be effective for Salmonella control.
Additionally, these mutant Salmonella colonies were used to verify that the selected phages were lytic phages using mitomycin C, which induces lysogenic phages (9). Briefly, 100 μl of an overnight culture of each mutant was inoculated into two tubes of TSB and incubated at 37°C until late log phase; at this point, mitomycin C (EMD Biosciences, La Jolla, CA) was added to one tube at a concentration of 0.5 μg/ml, and the tubes were incubated overnight. The broth was tested for the presence of lysogenic phage, which was negative for all bacterial isolates tested.
In this study, five bacteriophages, H2292 {2}, H9301 (1)-2, H4717 S5p2, ME-18 p2, and FO, were combined into a mixture. All five Salmonella enterica strains (S. Enteritidis H2292, S. Poona H9301, S. Newport H9116, S. Typhimurium DT104, and S. Typhimurium 8243) were tested individually in TSB-R against the bacteriophage mixture. The strains were individually diluted to 106, 107, and 108 CFU/ml in TSB-R. Optical densities were measured every hour for 9 h, followed by a second bacteriophage treatment. Measurements were determined for up to 24 h, and visual observations were made for up to 72 h.
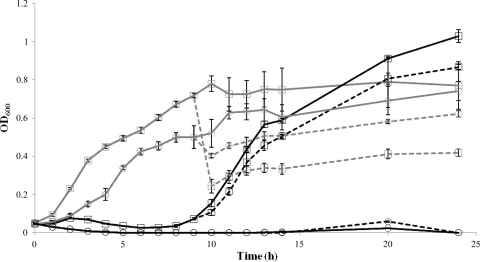
The results were nearly identical for S. Enteritidis H2292, S. Newport H9116, and S. Poona H9301. The growth of these Salmonella strains was suppressed at the same level for about 7 h relative to the controls (data not shown). Although bacterial growth did occur, it never reached the level of growth of the controls within the 24-h sampling period. The two strains of S. Typhimurium had completely opposite results, with S. Typhimurium DT104 growth being suppressed for several hours followed by regrowth above that of the controls, whereas S. Typhimurium 8243 had a brief weak peak of growth at 17 h but quickly decreased back to zero (Fig. 3). Visual observations of S. Typhimurium 8243 at 72 h confirmed that the broth remained clear, indicating that the bacteriophage mixture suppressed S. Typhimurium 8243 growth. McLaughlin and Brooks (17) also discovered that in many regrowth situations, the optical density of the bacteriophage-treated culture was greater than that of the control and concluded that this was due to the assimilation of nutrients from killed bacterial cells. O'Flynn et al. (22) developed a three-bacteriophage mixture for the elimination of Escherichia coli O157:H7 and determined that the initial inoculum of 105 CFU/ml was reduced to zero within 1 h following a bacteriophage challenge with a multiplicity of infection (MOI) between 1 and 100. However, within one additional hour, E. coli O157:H7 grew to nearly 3 log CFU/ml. In our study, subsequent bacteriophage treatments using the same bacteriophage mixture after 10 h did not reduce Salmonella numbers or growth rates in any of the experiments, suggesting that a bacteriophage-resistant population of Salmonella developed. The fact that only one strain of Salmonella was completely inhibited by the bacteriophage mixture reinforces the importance of developing bacteriophage mixtures that contain several bacteriophages specific to the strains being tested.
FIG. 3.
Effect of bacteriophage cocktail applied to S. Typhimurium 8243 and S. Typhimurium DT104 in TSB-R at initial concentrations of 108 CFU/ml. S. Typhimurium 8243 (○) and S. Typhimurium DT104 (□) were treated with bacteriophage at time zero (solid black lines), followed by phage treatment at 10 h (broken black lines); control bacteria (not treated with bacteriophage) were treated with SM buffer at time zero (solid gray lines), followed by a second SM buffer treatment at 10 h (broken gray lines). Optical density measurements of control and bacteriophage treatment for the single application of bacteriophage mixture were measured every hour for 9 h. At 10 h, a second round of SM buffer or bacteriophage was added to duplicate samples of controls and treatments, respectively. OD600, optical density at 600 nm.
Bacteriophage application to compost.
Finished dairy manure compost (>60 days after composting) (pH 8.5) was obtained from a field study. The finished dairy manure compost was comprised of a mixture of manure and sawdust (specifically, a mixture of cow manure, sawdust, and calf feces), wasted feed, old hay, and vegetable wastes (squash and plant residues) at a ratio of 10:2:2:2, respectively. The compost was prepared by spreading 2,000 g of compost on trays (50.8 by 38.1 by 2.54 cm) followed by drying at room temperature for 1 day inside a biological hood to obtain moisture levels below 30%. Sterile tap water was used to prepare the compost at the desired moisture level. The moisture level was determined with an IR-35 moisture analyzer (Denver Instruments, Denver, CO). Water activity was measured with an AquaLab water activity instrument (Decagon Devices, Pullman, WA).
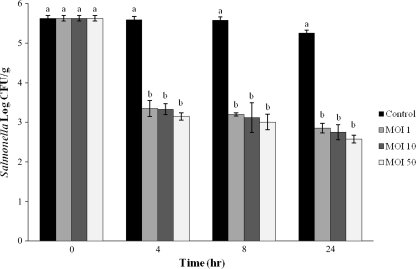
Three milliliters of 107 CFU S. Typhimurium 8243/ml was sprayed over the surface of 600 g of finished dairy compost with a final moisture content of 50%, mixed thoroughly, and stored at room temperature overnight for culture acclimation. The samples were then divided into four 150-g portions, sprayed with 3 ml of either SM buffer (50 mM Tris-Cl [pH 7.5], 0.1 M NaCl, 8 mM MgSO4·7H2O) or bacteriophage mixture at MOIs of 1, 10, and 50, and subsequently mixed by hand. Salmonella counts in compost were determined by adding 90 ml of 0.85% saline to 10-g samples of compost, shaking vigorously, and spiral plating onto XLT-4 agar supplemented with rifampin (100 μg/ml). Preliminary experiments performed to separate bacteria from phage during the sampling by centrifugation and resuspension of the bacterial pellet revealed no differences in the number of bacteria determined by plate counts. Therefore, this phage separation step was not utilized for these experiments. The use of bacteriophage in the compost resulted in significant reductions at all MOIs (Fig. 4 ). However, statistical analysis of the number of bacteria from plate counts revealed no difference (P > 0.05) for values at MOIs of 1, 10, and 50, which resulted in reductions of 2.34, 2.41, and 2.56 log CFU Salmonella/g, respectively. This suggests that at 50% moisture content under room temperature conditions (25°C), there is enough movement of bacteriophage in compost to cause significant reductions even at lower MOIs, most likely due to the increase in bacteriophage numbers upon infection of Salmonella cells.
FIG. 4.
Effects of treatment with three MOIs of a bacteriophage mixture in finished dairy compost contaminated with Salmonella Typhimurium 8243. Means with the same letter at each sampling time are not significantly different (P > 0.05).
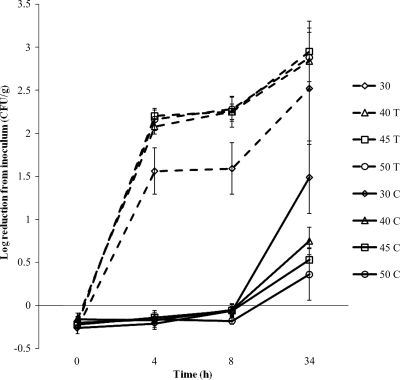
To determine the impact of compost moisture level on the effectiveness of bacteriophage treatment, three milliliters of 107 CFU S. Typhimurium 8243/ml was sprayed over the surface of 300 g of nonautoclaved finished dairy compost at each moisture content (30, 40, 45, and 50%), which were aseptically mixed, divided into two 150-g samples for treatment and control, and acclimated at room temperature. The following day, compost samples were sprayed with 3 ml of either bacteriophage mixture at an MOI of 50 or SM buffer (control). The numbers of Salmonella bacteria were reduced (P < 0.05) in dairy compost at all moisture levels (Fig. 5). The bacteriophage mixture reduced S. Typhimurium at moisture contents of 40, 45, and 50% by more than 2 log CFU within 4 h and ca. 3 log CFU after 34 h. However, at the 30% moisture content level, Salmonella reduction was about 0.5 log CFU less (P < 0.05%) at each sampling time than at 40, 45, and 50% moisture content. After 1 day, the number of Salmonella in the 30% moisture content control declined by more than 1 log CFU as a result of moisture loss, whereas less Salmonella reductions occurred in the compost of 40, 45, and 50% moisture content controls. A similar study was performed using dairy compost that had been autoclaved. Bacterial reductions in autoclaved compost were less than those observed in nonautoclaved compost (data not shown). Due to the reduction of competitive microorganisms in the autoclaved compost, the population of Salmonella was increased for several log units during overnight acclimation prior to phage application, resulting in a much lower MOI. While reduction levels were still about 2 log Salmonella CFU/g when phage was used, there was little to no difference from the initial inoculum level.
FIG. 5.
Reduction of Salmonella Typhimurium 8243 in dairy compost by bacteriophage treatment. Controls (C) are shown as solid lines, and treatments (T) are shown as broken lines. Moisture levels are represented by symbols as follows: ⋄, 30%; ▵, 40%; □, 45%; and ○, 50%.
The fact that a smaller reduction in the number of salmonellae occurred in compost with 30% moisture content suggests that fluid movement within the sample is important and that there is a minimum moisture content in compost at which bacteriophage is effective. Preliminary studies revealed that the minimum moisture content required for bacteriophage to be effective is between 23 and 30% (data not shown). It is fortunate that the best conditions for bacteriophage treatment are within the optimal conditions (ca. 55 to 60% moisture content) for proper composting in the early thermophilic phase.
Numerous review articles have been published highlighting the benefits and disadvantages of applying bacteriophages in the food and agricultural industries (4, 5, 6, 8, 10). There are limitations in preparing effective bacteriophage mixtures that will prevent the selection of resistant bacteria yet retain a species-wide host range. This study has determined that bacteriophages can reduce but not completely eliminate Salmonella from compost. Currently, a study to examine the effectiveness of bacteriophage applied to the surfaces of fresh compost mixture is being conducted. We anticipate that bacteriophages would inactivate pathogens on the surfaces in a manner similar to what was seen in the present study. However, environmental factors, such as UV radiation, fluctuating temperatures, and precipitation, could influence those results. Although there are still many hurdles to address, our research has shown that naturally occurring bacteriophages can be isolated from the environment and applied to compost surfaces contaminated with Salmonella, resulting in significant reductions.
Acknowledgments
This work was funded by a grant from the USDA-NIFSI.
We thank Darryl Krueger and JoAn Hudson at Clemson University Electron Microscope Facility for guidance with electron microscopy.
Footnotes
Published ahead of print on 4 June 2010.
REFERENCES
- 1.Adams, M. H. 1959. Bacteriophages, p. 450-454. Interscience, New York, NY.
- 2.Atterbury, R. J., M. A. P. Van Bergen, F. Ortiz, M. A. Lovell, J. A. Harris, A. De Boer, J. A. Wagenaar, V. M. Allen, and P. A. Barrow. 2007. Bacteriophage therapy to reduce Salmonella colonization of broiler chickens. Appl. Environ. Microbiol. 73:4543-4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Food and Drug Administration. 2006. FDA statement on foodborne Escherichia coli O157:H7 outbreak in spinach. Food and Drug Administration, U.S. Department of Health and Human Services, Silver Spring, MD. http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2006/ucm108761.htm. Accessed 27 October 2006.
- 4.Garcia, P., B. Martinez, J. M. Obeso, and A. Rodriguez. 2008. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47:479-485. [DOI] [PubMed] [Google Scholar]
- 5.Goodridge, L., and T. Abedon. 2003. Bacteriophage biocontrol and bioprocessing: application of phage therapy to industry. SIM News 53:254-262. [Google Scholar]
- 6.Greer, G. G. 2005. Bacteriophage control of foodborne bacteria. J. Food Prot. 68:1102-1111. [DOI] [PubMed] [Google Scholar]
- 7.Greer, G. G. 1988. Effects of phage concentration, bacterial density, and temperature on phage control of beef spoilage. J. Food Sci. 53:1226-1227. [Google Scholar]
- 8.Hagens, S., and M. Loessner. 2007. Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 76:513-519. [DOI] [PubMed] [Google Scholar]
- 9.Hoshiba, H., J. Uchiyama, S. Kato, T. Ujihara, A. Muraoka, M. Daibata, H. Wakiguchi, and S. Matsuzaki. 2010. Isolation and characterization of a novel Staphylococcus aureus bacteriophage, phi MR25, and its therapeutic potential. Arch. Virol. 155:545-552. doi: 10.1007/s00705-010-0623-2. [DOI] [PubMed] [Google Scholar]
- 10.Hudson, J. A., C. Billington, G. Carey-Smith, and G. Greening. 2005. Bacteriophages as biocontrol agents in food. J. Food Prot. 68:426-437. [DOI] [PubMed] [Google Scholar]
- 11.Jones, P. W. 1982. Waste and animal health. Public Health Eng. 10:35-39. [Google Scholar]
- 12.Kim, J., M. W. Shepherd, Jr., and X. Jiang. 2009. Evaluating the effect of environmental factors on pathogen regrowth in compost extract. Microb. Ecol. 58:498-508. doi: 10.1007/s00248-009-9524-x. [DOI] [PubMed] [Google Scholar]
- 13.Kinley, B., J. Rieck, P. Dawson, and X. Jiang. 2010. Analysis of Salmonella and enterococci isolated from rendered animal products. Can. J. Microbiol. 56:65-73. [DOI] [PubMed]
- 14.Kuhn, J., M. Suissa, D. Chiswell, A. Azriel, B. Berman, D. Shahar, S. Reznick, R. Sharf, J. Wyse, T. Bar-On, I. Cohen, R. Giles, I. Weiser, S. Lubinsky-Mink, and S. Ulitzur. 2002. A bacteriophage reagent for Salmonella: molecular studies on Felix 01. Int. J. Food Microbiol. 74:217-227. [DOI] [PubMed] [Google Scholar]
- 15.Leverentz, B., W. S. Conway, M. J. Camp, W. J. Janisiewicz, T. Abuladze, M. Yang, R. Saftner, and A. Sulakvelidze. 2003. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl. Environ. Microbiol. 69:4519-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLaughlin, M. R. 2007. Simple colorimetric microplate test of phage lysis in Salmonella enterica. J. Microbiol. Methods 69:394-398. [DOI] [PubMed] [Google Scholar]
- 17.McLaughlin, M. R., and J. P. Brooks. 2008. EPA worst case water microcosms for testing phage biocontrol of Salmonella. J. Environ. Qual. 37:266-271. [DOI] [PubMed] [Google Scholar]
- 18.Misra, R. V., R. N. Roy, and H. Hiraoka. 2003. On-farm composting methods, p. 1-33. Land and water discussion paper 2. Food and Agricultural Organization, United Nations, Rome, Italy. http://www.fao.org/docrep/007/y5104e/y5104e00.HTM.
- 19.Molbak, K. 2005. Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin. Infect. Dis. 41:1613-1620. [DOI] [PubMed] [Google Scholar]
- 20.Monteith, H. D., E. E. Shannon, and J. B. Derbyshire. 1986. The inactivation of a bovine enterovirus and a bovine parvovirus in cattle manure by anaerobic digestion, heat treatment, gamma irradiation, ensilage and composting. J. Hyg. (Lond.) 97:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nygard, K., J. Lassen, L. Vold, Y. Andersson, I. Fisher, S. Loefdahl, J. Threlfall, I. Luzzi, T. Peters, M. Hampton, M. Torpdahl, G. Kapperud, and P. Aavitsland. 2008. Outbreak of Salmonella Thompson infections linked to imported rucola lettuce. Foodborne Pathogen. Dis. 5:165-173. [DOI] [PubMed] [Google Scholar]
- 22.O'Flynn, G., R. P. Ross, G. F. Fitzgerald, and A. Coffey. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:3417-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pao, S., S. P. Randolph, E. W. Westbrook, and H. Shen. 2004. Use of bacteriophages to control Salmonella in experimentally contaminated sprout seeds. J. Food Sci. 69:M127-M130. [Google Scholar]
- 24.Park, G. W., and F. Diez-Gonzalez. 2003. Utilization of carbonate and ammonia-based treatments to eliminate Escherichia coli O157:H7 and Salmonella Typhimurium DT104 from cattle manure. J. Appl. Microbiol. 94:675-685. [DOI] [PubMed] [Google Scholar]
- 25.Sharma, M., J. R. Patel, W. S. Conway, S. Ferguson, and A. Sulakvelidze. 2009. Effectiveness of bacteriophages in reducing Escherichia coli O157:H7 on fresh-cut cantaloupes and lettuce. J. Food Prot. 72:1481-1485. [DOI] [PubMed] [Google Scholar]
- 26.Sheng, H. Q., H. J. Knecht, I. T. Kudva, and C. J. Hovde. 2006. Application of bacteriophages to control intestinal Escherichia coli O157:H7 levels in ruminants. Appl. Environ. Microbiol. 72:5359-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepherd, M. W., P. Liang, X. Jiang, and M. P. Doyle. 2007. Fate of Escherichia coli O157:H7 during on-farm dairy manure-based composting. J. Food Prot. 70:2708-2716. [DOI] [PubMed] [Google Scholar]
- 28.Smith, H., and M. Huggins. 1982. Successful treatment of experimental Escherichia coli infections in mice using phage: its general superiority over antibiotics. J. Gen. Microbiol. 128:307-318. [DOI] [PubMed] [Google Scholar]
- 29.Van Twest, R., and A. Kropinski. 2008. Bacteriophage enrichment from water and soil, p. 15-22. In M. R. J. Clokie and A. M. Kropinski (ed.), Bacteriophages: methods and protocols, vol. 1. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 30.Whichard, J. M., S. Namalwar, and P. William. 2003. Suppression of Salmonella growth by wild-type and large-plaque variants of bacteriophage Felix 01 in liquid culture and on beef frankfurters. J. Food Prot. 66:220-225. [DOI] [PubMed] [Google Scholar]
- 31.Yoichi, M., M. Morita, K. Mizoguchi, C. R. Fischer, H. Unno, and Y. Tanji. 2004. The criterion for selecting effective phage for Escherichia coli O157:H7 control. Biochem. Eng. J. 19:221-227. [Google Scholar]