Abstract
Some aquatic microbial oxygenic photoautotrophs (AMOPs) make hydrogen (H2), a carbon-neutral, renewable product derived from water, in low yields during autofermentation (anaerobic metabolism) of intracellular carbohydrates previously stored during aerobic photosynthesis. We have constructed a mutant (the ldhA mutant) of the cyanobacterium Synechococcus sp. strain PCC 7002 lacking the enzyme for the NADH-dependent reduction of pyruvate to d-lactate, the major fermentative reductant sink in this AMOP. Both nuclear magnetic resonance (NMR) spectroscopy and liquid chromatography-mass spectrometry (LC-MS) metabolomic methods have shown that autofermentation by the ldhA mutant resulted in no d-lactate production and higher concentrations of excreted acetate, alanine, succinate, and hydrogen (up to 5-fold) compared to that by the wild type. The measured intracellular NAD(P)(H) concentrations demonstrated that the NAD(P)H/NAD(P)+ ratio increased appreciably during autofermentation in the ldhA strain; we propose this to be the principal source of the observed increase in H2 production via an NADH-dependent, bidirectional [NiFe] hydrogenase. Despite the elevated NAD(P)H/NAD(P)+ ratio, no decrease was found in the rate of anaerobic conversion of stored carbohydrates. The measured energy conversion efficiency (ECE) from biomass (as glucose equivalents) converted to hydrogen in the ldhA mutant is 12%. Together with the unimpaired photoautotrophic growth of the ldhA mutant, these attributes reveal that metabolic engineering is an effective strategy to enhance H2 production in AMOPs without compromising viability.
Genetic engineering is currently being employed to increase the yield of biofuels from terrestrial crops (29), both by improving the total biomass yield as well as by increasing the efficiency of microbial recovery of fuels from harvested biomass (14). While much research is focused on crop-based approaches to biofuel production, aquatic microbial oxygenic photoautotrophs (AMOPs) offer several advantages over the use of terrestrial plants, both in terms of biomass accumulation as well as the direct excretion of alternative fuel precursors, such as hydrogen (10). For example, the levels of sustained fermentative hydrogen produced by AMOPs were increased through modification of environmental stressors (1, 11), and genetic manipulation of respiratory oxidases increased transient photohydrogen (11).
The maximum obtainable fermentative hydrogen energy conversion efficiency (ECE; ratio of combustion enthalpies of hydrogen to carbohydrate catabolized) via glycolysis terminating at acetyl-coenzyme A (CoA) formation is 41%, corresponding to 4 mol H2 per mol glucose equivalent. Nonphototrophs display corresponding glucose ECEs of about 20% and 30% for Enterobacter and Escherichia coli, respectively, catabolizing exogenously supplied glucose (6, 16). Metabolic engineering has been successful in enhancing fermentative hydrogen production from exogenous glucose in E. coli, in which null mutations for genes involved in the conversion of pyruvate to lactate, either alone or in combination with other pathways, resulted in incremental increases in hydrogen yields (18, 26, 33, 34). Similar studies using chemical mutagenesis have been carried out in Enterobacter sp. strains, which increased fermentative hydrogen production less than 2-fold (17, 22). Currently, the analogous mutants have not been constructed in any AMOP, and thus the feasibility of genetic alteration for enhancing fermentative hydrogen production remains unproven.
Cyanobacteria are physiologically and evolutionarily diverse prokaryotic AMOPs, which are tolerant of widely variable environmental conditions, including pH, temperature, and salinity. Because certain circumstances, such as the formation of dense mats, require these organisms to survive under dark, anoxic conditions for extended periods of time, many cyanobacteria are facultative anaerobes with the ability to carry out fermentative metabolism as a means of satisfying the need for ATP regeneration (27). Many cyanobacteria produce H2 as a by-product of the dark anoxic catabolism of photosynthetically derived, energy-storage compounds (principally glycogen), and for this reason these organisms are under investigation as a potential renewable source of hydrogen (8).
If carbohydrate catabolism serves as the sole source of energy generation during anaerobic metabolism, then the carbon fluxes passing through the various pathways downstream of pyruvate, including H2 production/uptake, must balance the amount of NAD(P)H consumed with that produced upstream. Pyruvate and reductant formed by glycolysis and/or the pentose phosphate pathway in cyanobacteria can be transformed into lactate, ethanol, hydrogen, acetate, formate, and CO2, which are ultimately excreted (27). The tricarboxylic acid (TCA) pathways can additionally affect this balance. In cyanobacteria, the TCA cycle is incomplete and is thus branched into oxidative and reductive branches (27); during fermentation, the branch reducing oxaloacetate to succinate could be employed to dispose of excess NAD(P)H, although this has not been reported previously for cyanobacteria. Alanine can be produced by the reductive amination of pyruvate, though it too has not previously been reported as a product of cyanobacterial fermentation, despite its excretion during fermentation in other strains of bacteria (15, 21).
In the studies reported here, we investigated the fermentative metabolism of the unicellular, euryhaline cyanobacterium Synechococcus sp. strain PCC 7002 (here referred to as Synechococcus 7002) and an ldhA mutant strain, in which the gene encoding d-lactate dehydrogenase was inactivated. Synechococcus 7002 has a dark anaerobic metabolism that is unique among cyanobacteria thus far reported and which yields up to five fermentation products: lactate, acetate, succinate, alanine, and hydrogen (Fig. 1). Lactate was found to be the major excreted fermentation product of wild-type (WT) cells, and the enzyme producing d-lactate, which competes with hydrogenase for reducing equivalents, was eliminated genetically. Nitrate deprivation (25) and nickel supplementation (3) were previously shown to increase hydrogen yield in other strains, and therefore these treatments were examined in combination with the ldhA mutation to optimize conditions for hydrogen production. The ldhA mutant has dramatically redistributed fermentative fluxes in comparison to WT cells and a higher NAD(P)H/NAD(P)+ ratio and up to 5-fold higher hydrogen production than those of WT cells.
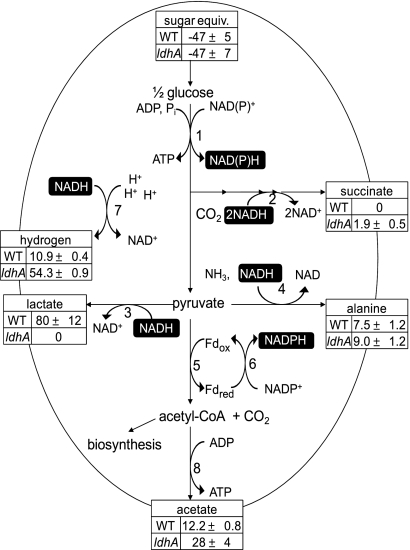
FIG. 1.
Fermentative metabolism of glucose derived from intracellular, reduced sugars in Synechococcus 7002 based on the sequenced genome and the measured metabolite concentrations for WT and ldhA mutant cultures. Tables indicate excreted metabolite concentrations in the media after 4 days (mol/1017 cells). 1, enzymes of glycolysis or pentose phosphate pathway; 2, enzymes of the TCA cycle from malate dehydrogenase to succinate dehydrogenase; 3, d-lactate dehydrogenase (insertionally inactivated in ldhA mutant); 4, alanine dehydrogenase; 5, pyruvate:ferredoxin (flavodoxin) oxidoreductase; 6, Fd:NADP+ oxidoreductase (FNR); 7, hydrogenase; 8, acetate-CoA ligase.
MATERIALS AND METHODS
Growth conditions.
Cultures of Synechococcus 7002 were grown photoautotrophically in medium A+ (28) supplemented with 2 μM NiCl2 and sparged with 2% (vol/vol) CO2 in air. Cells were grown for 4 days to early stationary phase (optical density at 550 nm [OD550] ≈ 3) at 35°C in 500-ml flasks using fluorescent lighting at a light intensity of 200 μmol photons m−2 s−1, incident on one side.
Construction of ldhA mutant.
In order to inactivate the ldhA gene insertionally, a partial ldhA gene fragment (nucleotides +209 to +1653 relative to the ATG start codon) was amplified by PCR using the primers of Ldha1F (5′ AATACATTGCCCTACGCTGTGC 3′) and Ldha1R (5′ GGTCAACTTTTGCTTCCTTTCGG 3′). The aacC1 cassette, originally from plasmid pMS266 (2), which confers gentamicin resistance, was introduced into the AccI restriction site at +776 of the ldhA gene fragment. The AccI site was digested followed by subsequent sequential ligation of three fragments: ldhA fragment number 1 (nucleotides +209 to +776), the aacC1 cassette, and ldhA fragment number 2 (+776 to + 1653). Ligation products were directly used to transform a fresh overnight culture of wild-type Synechococcus 7002 (1 ml; OD730 of 1.5). Transformants were selected on plates containing medium A (28) supplemented with 50 mg liter−1 gentamicin. Single transformant colonies were continuously restreaked on plates containing 300 mg liter−1 gentamicin. Transformants were assayed by analytical PCR with primers Ldha1F and Ldha1R until full segregation of the ldhA and ldhA::aacC1 alleles was observed by failure to amplify the wild-type ldhA gene, as shown in Fig. S1 in the supplemental material.
Fermentative conditions.
The cells were concentrated by centrifugation and washed twice with 50 ml medium A (lacking sodium nitrate); the cells were resuspended in 130 ml of nitrate-free medium A. Final cell densities were measured by optical density at 550 nm. The final OD550 values were 3.5 for WT cells and 3.9 for ldhA mutant cells. All data for the WT and ldhA mutant were normalized to number of cells, with an OD550 of 1 containing 1.0 ± 0.2 × 108 cells per ml (24). Anoxic conditions were created to induce autofermentation; aliquots of cells (7 ml) were placed in 10-ml vials, which were then sealed with rubber septa and wrapped in foil to create dark conditions. The headspace was purged for 15 min with argon gas in order to remove most of the oxygen, and any remaining dissolved oxygen was consumed by respiration. Triplicate aliquots were prepared for both WT and ldhA mutant cells for each time point. At each time point, the headspace gas was sampled using a gas-tight syringe for gas chromatography (GC) measurements, then the vials were opened, and cells were separated from the media for analysis of the cell-free medium by NMR in ambient air conditions.
Analytical assays.
Hydrogen in the headspace of assay vials was sampled (200 μl) with a gas-tight syringe and measured by gas chromatography using a Gow Mac series 300 instrument, with a thermal conductivity detector and argon as the carrier gas. The metabolite concentrations in the extracellular medium of fermenting cells were quantified using cryoprobe-assisted proton nuclear magnetic resonance (1H NMR) spectroscopy on a Varian INOVA-600 instrument at 600 MHz at 22°C (4). The medium was spiked with 10% D2O containing 30 μg ml−1 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid, sodium salt (TSP). Excitation sculpting was utilized for water suppression (13). The peak(s) corresponding to each metabolite was integrated and then normalized to an internal standard (TSP). Quantification of each metabolite was based on a linear calibration curve, ranging from 10 μm to 1 mM, for each unique metabolite, as previously described (4). Hydrogenase activity was measured via the methyl viologen (MV) assay as described in reference 3, except cells were lysed anaerobically with BugBuster (Novagen) added to the described MV buffer solution. Respiration (oxygen uptake) and chlorophyll concentration were measured as described previously (20). Total sugars were assayed using the anthrone reagent (12). Briefly, cells were boiled in sulfuric acid for an hour, at which time the anthrone reagent was added and the cells were boiled an additional 10 min. The absorbance was measured at 620 nm for the biological samples and standards. Protein was quantified by the bicinchoninic acid assay (Sigma-Aldrich) with bovine serum albumin as the standard. Pyruvate was measured via enzymatic assay (BioVision) using sodium pyruvate as a standard. Aliquots (3 ml) of sample culture from fermentation vials were extracted via syringe under argon and injected into a vacuum-filtration apparatus (Fisher Scientific) loaded with a 25-mm nylon filter (Millipore). Following removal of the culture media, the filter was submerged in 1.8 ml of 80:20 MeOH-H2O quench-extract solvent at −20°C for 15 min. Filters were then scraped and rinsed with the extraction solvent to remove cell material. A 5-ml aliquot of the extract was dried and resuspended in the pyruvate assay buffer. Lipids were analyzed by fatty acid methyl ester analysis in a method adapted from Rodriguez-Ruiz et al. (23). Briefly, cell pellets equal to 1 ml of culture were boiled with 2 ml of 20:1 MeOH-acetyl chloride and 1 ml hexane. After boiling, the organic layer was analyzed, and fatty acid methyl esters were quantified by gas chromatography on a Shimadzu model GC-2010 instrument with flame-ionization detection and by using heptadecanoic acid (17:0) as an internal standard (M. Frada, E. Burrows, K. Wyman, G. C. Dismukes, and P. Falkowski, unpublished data).
LC-MS.
Internal metabolite extracts for liquid chromatography-mass spectrometry (LC-MS) analysis were prepared as described above for the pyruvate assay, except 4 ml of sample culture from fermentation vials and MeOH-ACN-H2O (40:40:20) as the quench-extract solvent were used. Filters were additionally washed with 0.5 ml of fresh extraction solvent, which was added to the initial extract and centrifuged at 14,000 × g for 5 min. The supernatant was then transferred to a clean vial, and the pellet resuspended in 100 ml of fresh extraction solvent; the same extraction procedure was repeated, and the supernatant from the second extraction was combined with the initial extraction and stored at −80°C until analysis.
LC-MS metabolite quantification was conducted using an Agilent 6410 QQQ mass spectrometer coupled to a 1200-series chromatography system. A Phenomenex Synergy C18 Hydro-RP column at 40°C was employed, with 10 mM tributylamine and 10 mM acetic acid in H2O as mobile phase A and methanol as mobile phase B. The elution time table was as follows: 0 min (0% B), 8 min (35% B), 10 min (35% B), 12.5 min (90% B), 18 min (90% B), and 18.1 min (0% B). Individual metabolites were quantified by compound-specific fragmentation pattern and elution time, and intracellular concentration was quantified by calibration via addition of known concentrations of a standard mixture of target metabolites to analyzed extracts.
RESULTS
Energy metabolism in Synechococcus 7002.
The complete genome of Synechococcus 7002, accession number NC_010475 in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html), was queried by using the BLASTP algorithm for the occurrence of genes with significant sequence similarity (E value < 1 × 10−10) to other cyanobacterial genes encoding enzymes of catabolic and fermentative carbon metabolism. The predicted anoxic catabolic reactions starting from glycogen metabolism are presented in the schematic in Fig. 1. Genes in the genome which encode the enzymes that convert pyruvate to the following products were identified: lactate (d-lactate dehydrogenase, which oxidizes NADH), acetate (pyruvate:ferredoxin oxidoreductase and acetate-CoA ligase), and alanine (alanine dehydrogenase, which oxidizes NADH). The enzymes for converting pyruvate to formate (pyruvate formate lyase) or ethanol (aldehyde-alcohol dehydrogenase), as well as a common, alternate acetate pathway (acetate kinase), were absent in this strain. Only one set of genes for hydrogen metabolism was found: those for the assembly and formation of the bidirectional [NiFe] hydrogenase (hox). No genes encoding an uptake hydrogenase or nitrogenase were identified in the genome. According to genomic analyses, the reductive branch of the tricarboxylic acid (TCA) cycle is intact, and thus cells should be capable of NADH oxidation to produce succinate as a terminal product. The enzymes in this series of reactions are PEP carboxylase (unidirectional), malate dehydrogenase, fumarase, and succinate dehydrogenase (the last three catalyze reversible reactions). The reductive arm of the TCA cycle is responsible for succinate production by E. coli cells fermenting glucose under anoxic conditions (5, 7).
The fermentative pathways predicted by genome analysis were assayed experimentally by identifying and quantifying excreted metabolites. These metabolites are derived from the autofermentation of photosynthetically stored carbon, predominantly in the form of glycogen, which decreased during fermentation. Cells growing photoautotrophically were switched to dark, anoxic conditions to induce fermentative metabolism. Additionally, cells were resuspended in nitrate-free media upon induction of autofermentation to eliminate the effect of nitrate assimilation, which can act as a major competing sink for intracellular reductant (11). 1H-NMR spectra of the cell-free medium were acquired at 24-hour time intervals after the onset of fermentation, and the data were analyzed to identify possible metabolites, including lactate, formate, ethanol, acetate, succinate, and alanine.
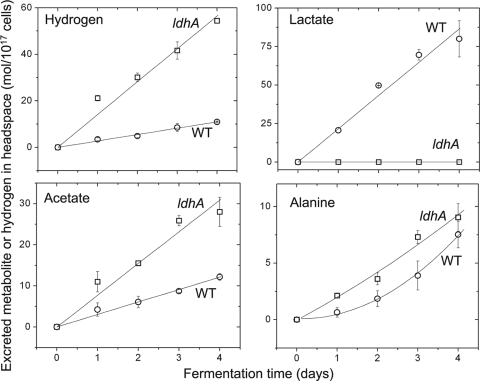
Quantitative comparisons showed that d-lactate (δ = 1.3 ppm) was the dominant fermentation product of WT cells, with a constant, linear rate of production equal to 21.6 mol per day per 1017 cells through at least 4 days (Fig. 2). The rate of d-lactate production was about 7-fold greater than the linear rate of acetate (δ = 1.9 ppm) production, 3.0 mol per day per 1017 cells. The alanine (δ = 1.4 ppm) production rate increased from an initial rate of 0.65 mol per day per 1017 cells at day 1 to 3.6 mol per day per 1017 cells from day 3 to day 4. The sum of these carbon metabolite production rates gives a lower limit to the estimated flux of glucose equivalents through glycolysis of 14 mol per day per 1017 cells, which excludes carbon flux through the pentose phosphate pathway. Hydrogen accumulated in the headspace at a rate of 2.7 mol per day per 1017 cells. As expected, due to the absence of the requisite genes for their production, ethanol (δ = 1.2 ppm) and formate (δ = 8.4 ppm) were not observed as excreted metabolites. Succinate (δ = 2.4 ppm) was also not observed as an excreted metabolite in the extracellular medium of fermenting WT cells (10 μM was less than the limit of quantification).
FIG. 2.
The kinetics of autofermentative metabolite excretion from WT (○) and ldhA mutant (□) cells of Synechococcus 7002. Lactate, alanine, and acetate were determined by 1H-NMR. Hydrogen in the headspace was measured by gas chromatography. The ordinate gives the concentration per 1017 cells. Error bars represent plus and minus one standard deviation (SD) from biological triplicates.
In Synechococcus 7002, most of the reduced pool of NADH that was generated in glycolysis was found to be reoxidized via the d-lactate pathway, with much smaller fluxes consumed through alanine and hydrogen excretion, as quantified in the rates above. To test the possibility of redirecting the flux of NADH from pyruvate to proton reduction, an ldhA mutant lacking d-lactate dehydrogenase was constructed (Fig. 1).
Comparison of wild-type and ldhA mutant cells.
Wild-type and ldhA mutant cells were grown in batch cultures to early stationary phase. The rates of photoautotrophic growth, shown in Fig. S2 in the supplemental material, and aerobic respiration of the WT and mutant cells (80 ± 10 versus 80 ± 2 μmol O2 consumed per mg Chl per h, respectively) were not significantly different. As measured by the methyl viologen assay, the enzymatic activity of hydrogenase was not significantly different for WT and ldhA mutant cells (0.49 ± 0.12 versus 0.53 ± 0.15 μmol H2 per liter culture per minute, respectively). Rates of formation of fermentative products were compared as a function of time following transfer of cells to fresh, nitrate-free media and induction of anaerobiosis (day 0), for which cells were at the early stationary phase of growth (OD550 ≈ 3.0). As observed by NMR and GC analysis, ldhA mutant cells excreted metabolite levels dramatically different than those of WT cells during fermentation. Figure 2 shows the accumulation of hydrogen in the headspace of sealed 10-ml bioreactors with 7 ml of culture, as well as the accumulation of excreted metabolites in the medium over 4 days of fermentation. d-Lactate was undetectable in the extracellular medium of the ldhA mutant, which produced hydrogen at an increased rate of 14.1 mol per day per 1017 cells, acetate at an increased rate of 7.7 mol per day per 1017 cells, and alanine at an average rate of 2.2 mol per day per 1017 cells. However, the lower limit of the flux of glucose equivalents through glycolysis, as estimated by the sum of excreted carbon products, decreased to about 5 mol per day per 1017 cells in the ldhA mutant cells.
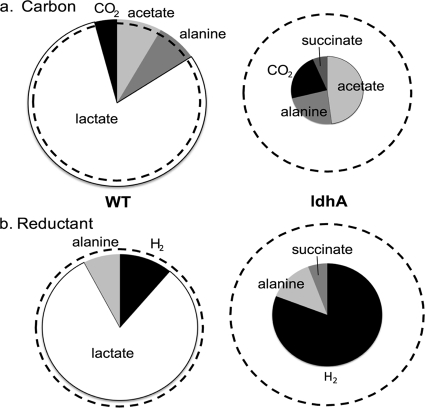
A summary of the integrated yields of excreted carbon metabolites after 4 days of fermentation is shown in the pie charts given in Fig. 3 a. The ldhA mutant cells did not excrete detectable lactate. Instead, they excreted 1.4-fold more alanine and 2.7-fold more acetate than WT cells on average over 4 days. The ldhA mutant cells also excreted a small amount of succinate during this period. As expressed by the relative areas of the pie charts in Fig. 3a, the total flux of excreted carbon metabolites decreased by 60% in the ldhA mutant. The total reduced carbohydrate catabolized by the cells during fermentation was measured using an anthrone assay (Fig. 3a, dashed circles), which indicated that the amounts of polysaccharides degraded were not significantly different in WT and ldhA mutant cells, at 47 ± 5 and 47 ± 7 mol per 1017 cells degraded, respectively. Total protein in Synechococcus 7002 was determined and found not to degrade during 4 days of fermentation (see Fig. S3 in the supplemental material). Additionally, fatty acids, which comprise a comparatively small fraction of the cell composition, were determined and found not to change appreciably in quantity during fermentation, in WT and ldhA mutant cultures. WT cells contained 5 ± 1 mg/1011 cells at the onset of fermentation and after 3 days of fermentation contained 5 ± 1 mg/1011 cells. The ldhA mutant cells contained 6 ± 1 mg/1011 cells at the onset of fermentation and after 3 days of fermentation contained 5 ± 1 mg/1011 cells.
FIG. 3.
Comparison of excreted product yields from autofermentation of wild-type versus ldhA mutant cells of Synechococcus 7002. Relative areas of the dashed circles indicate the amount of carbon (a) or reductant (b) originating from endogenous, reduced sugars catabolized over 4 days of autofermentation. Relative areas of the pie graphs represent integrated yields from all excreted metabolites produced after 4 days of autofermentation normalized to amount of carbon (a) or reductant (b) equivalents needed to produce the metabolite. CO2 is assumed to be equivalent to acetate in concentration, as described in the text. WT cultures (left) excrete the same amount of product as is catabolized, while almost 60% of the balance of catabolized carbon and reductant is not accounted for by excreted products in the ldhA mutant cultures (right).
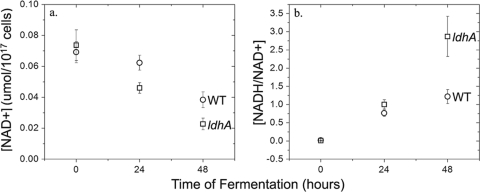
The intracellular concentrations of NAD+, NADP+, NADH, and NADPH were measured using LC-MS at the onset of fermentation (time zero) and at 24- and 48-h time points. During fermentation, the concentration of NAD+ decreased continually through 48 h following anaerobic induction (Fig. 4 a). At 96 h, oxidized pyridine nucleotides were below the detection limit. As expected at the onset of anoxic conditions, there was no NADH detectable in WT and ldhA mutant cells. The concentration of NADH increased over 48 h to 0.05 and 0.07 mol per 1017 cells for the WT and the ldhA mutant, respectively. At 24 h of fermentation, the ratio of NADH to NAD+ was 1.3-fold higher for the ldhA mutant (Fig. 4b), and at 48 h, this ratio increased to 2.3-fold. The concentration of NADPH was below the detection limit throughout fermentation. The concentration of NADP+ was 10-fold lower than that of NAD+ and decreased similarly to that of NAD+ during 96 h of fermentation.
FIG. 4.
Intracellular NAD+ concentration (a) and ratio of NADH/NAD+ (b) for WT (○) and ldhA mutant (□) cells of Synechococcus 7002. Time zero was measured at the onset of anoxic conditions. Concentrations of NAD+, NADP+, NADH, and NADPH were measured with LC-MS. The concentration of the total NAD(H) pool at 96 h was below the detection limit of our instrument. The concentration of NADP+ was over an order of magnitude lower than the concentration of NAD+ and decreased to below the detection limit by 96 h. The concentration of NADPH remained below the detection limit throughout the experiment. Error bars represent plus and minus one SD from biological triplicates.
Table 1 lists both excreted and catabolized carbon equivalents and reductant mobilized during fermentation.
TABLE 1.
Comparison between intracellular reduced sugars catabolized and excreted metabolite concentrationsa
| Parameter (row no.)d | Intracellular RSC | Hydrogen excreted | Acetate excreted | Lactate excreted | Alanine excreted | Succinate excreted | CO2 equivalentf | Excess equivalent excreted (%)e |
|---|---|---|---|---|---|---|---|---|
| WT measured (1)b | 47 ± 5 | 10.9 ± 0.4 | 12.2 ± 0.8 | 80 ± 12 | 7.5 ± 1.2 | 0 | 12 | |
| ldhA mutant measured (2)b | 47 ± 7 | 54.3 ± 0.9 | 28 ± 4 | 0 | 9.0 ± 1.2 | 1.9 ± 0.5 | 28 | |
| Carbon equivalent per molecule (3)c | −6 | 0 | 2 | 3 | 3 | 4 | 1 | |
| NADH equivalent per molecule (4)c | −2 | 1 | −1 | 1 | 1 | 2 | 0 | |
| WT carbon equivalent (5) | −282 | 0 | 24 | 240 | 23 | 0 | 12 | 17 (6) |
| ldhA mutant carbon equivalent (6) | −282 | 0 | 56 | 0 | 27 | 8 | 26 | −165 (59) |
| WT NADH equivalent (7) | −94 | 11 | −12 | 80 | 8 | 0 | 0 | −7 (7) |
| ldhA mutant NADH equivalent (8) | −94 | 54 | −28 | 0 | 9 | 4 | 0 | −55 (59) |
Values are given as units of mol per 1017 cell for intracellular reduced sugars catabolized (RSC), measured by anthrone, for excreted metabolite concentrations, measured by GC or NMR, and for equivalents. Negative numbers represent fluxes into the available carbon pools (or NAD+ reduction), while positive numbers represent formation of excreted carbon (or NADH oxidation).
Data measured after 4 days of autofermentation.
Theoretical equivalents of carbon (row 3) or reductant (as NADH) (row 4) expected per glucose equivalent of carbohydrate catabolized during fermentation, assuming it is formed solely by the reactions of glycolysis.
The experimental carbon equivalents are obtained in rows 5 and 6 as the products of rows 1 × 3 and 2 × 3, respectively. Likewise, the experimental NADH equivalents are obtained in rows 7 and 8 as the products of rows 1 × 4 and 2 × 4, respectively.
Excess carbon or reductant equivalent that is excreted by the cell and the percentage of the catabolized intracellular RSC that this equivalent represents. The propagated systematic error for column 8 is ±15 for carbon equivalents (rows 5 and 6) and ± 5 for NADH equivalents (rows 7 and 8) based on the systematic error for each detection method.
CO2 was not directly measured and is set equal to the acetate concentration since it forms concomitantly with acetyl-CoA via the pyruvate:ferredoxin oxidoreductase reaction (Fig. 1). Additionally, the reductant formed in this reaction is accounted for in the acetate column only, to avoid double counting.
DISCUSSION
In this study we set out to quantitatively account for the sources and sinks of carbon and reductant during autofermentation and originating from photosynthetically produced, endogenous carbohydrate. The majority of the carbon and reductant produced by autofermentation was predicted to be excreted as hydrogen, lactate, acetate, alanine, succinate, and carbon dioxide. Unlike E. coli, which is a closed metabolic system, in which quantifiable exogenous sugar is the only source of electrons and carbon for the cells, cyanobacteria must be thought of as open systems. Photosynthetically generated reductants are ultimately stored upon combination with CO2 as carbohydrates and other biological macromolecules, but the extent to which CO2 continues to be incorporated into cyanobacterial cells during dark metabolism is unknown. For example, CO2 (or HCO3−) has been shown to compete for reductant with H2 formation during autofermentation in the cyanobacterium Arthrospira maxima (1), as well as the purple nonsulfur bacterium Rhodobacter sphaeroides (31).
In certain cases, the process of in vivo hydrogen production in cyanobacteria has been shown to be limited by the concentration of substrate, NAD(P)H and/or H+, as opposed to enzyme activity/availability (1, 3). This causal relationship between reductant concentration and hydrogen production has been proposed in heterotrophic bacteria (9), and Enterobacter aerogenes displays redox-linked hydrogen production with increased NAD(P)H/NAD(P)+ ratios driving increased hydrogen production (19). Further, the temporal link between NAD(P)H pool size and hydrogen production has been demonstrated previously in A. maxima (1). However, the possibility of engineering the metabolic pathways, which produce or consume intracellular reductant (NADH, NADPH, reduced ferredoxin, etc.), toward the goal of increasing the total reductant available for H2 production, had not been explored extensively with cyanobacteria until this report.
Wild-type Synechococcus 7002 excreted lactate as the predominant fermentative end product. NADH (E0 NAD+/NADH = −315 mV), the main product of carbohydrate oxidation, is expected to reduce pyruvate preferentially over protons, as the redox potential of pyruvate/lactate (E0 = −185 mV) is significantly more positive than that of H+/H2 (E0 = −421 mV at pH 7) under standard conditions. Consequently, redirection of the reductant flux into H+ reduction for increased H2 production is feasible only if the competition for NADH oxidation or NAD+ reduction can be altered. The intracellular ratio of NADH/NAD+ was indeed observed to increase 1.3- and 2.3-fold for the ldhA mutant cells after 1 and 2 days of fermentation, respectively. The concentrations of NADH and NAD+ were identical in both WT and ldhA mutant cells prior to the onset of fermentation, as would be expected for a mutation that acts only on anaerobic metabolism and which was also consistent with the equal photoautotrophic growth rates that were observed. The concentration of NADPH remained below detection throughout fermentation, indicating that NADH is the major pyridine nucleotide carrier during fermentation in Synechococcus 7002.
As predicted, the ldhA mutant cells produced hydrogen at a significantly higher rate (∼5-fold) than wild-type cells (Fig. 2). This increased rate occurred continuously over 4 days, with no noticeable decrease in production rate over time. The total amount of H2 that was produced from the WT and ldhA mutant over 4 days was found to be 11 and 54 mol per 1017 cells, respectively. As normalized to the total reduced carbohydrate catabolized, the yield of hydrogen in the ldhA mutant increased 5-fold from 0.23 to 1.15 mol H2 per mol hexose equivalent consumed. Because neither protein nor lipids were degraded in Synechococcus 7002 during 3 days of fermentation, the anthrone-detectable sugars are assumed to be the sole source of carbon and reductant. To estimate the energy conversion efficiency (ECE) of stored carbohydrate into hydrogen during fermentation, the enthalpies of combustion of hydrogen (286 kJ/mol) and glucose (2,805 kJ/mol) were compared. WT cells produced 0.23 mol hydrogen per mol of glucose equivalent catabolized, which gives a fermentative ECE of 2.3%. The ldhA mutant cells produced 1.15 mol hydrogen per mol of glucose equivalent catabolized, which gives a 5-fold improved fermentative ECE of 12%. The maximum obtainable fermentative ECE via acetogenic fermentation (glycolysis, including acetyl-CoA formation) would be 41%, corresponding to 4 mol hydrogen per mol glucose equivalent. Higher reductant yields are potentially achievable by diverting carbohydrate catabolism to the oxidative pentose phosphate pathway, for example, which would provide about 12 mol hydrogen per mol glucose, although this has been demonstrated only in vitro (32). Nonphototrophs display corresponding ECEs of about 20% and 30% for Enterobacter and E. coli, respectively (6, 16). However, the glucose substrate in these studies is exogenously supplied, not endogenously formed from atmospheric CO2 as it is in our study.
Table 1 accounts for carbon equivalents and reductant mobilized during fermentation. The endogenous sugar that is catabolized during fermentation in WT cells is closely (>90%) accounted for by the carbon and reductant excreted as hydrogen, acetate, lactate, alanine, succinate, and CO2. In contrast, almost 60% of the carbon and reductant derived from catabolized sugar in the ldhA mutant cells is not accounted for by the excreted products of fermentation. This difference can be explained only by the accumulation of other reduced intracellular metabolites or macromolecules that were not monitored. The amount of catabolized carbohydrate that is not excreted is equivalent to 165 carbon equivalents (28 mol glucose per 1017 cells [Table 1]). Pyruvate, the oxidized metabolic precursor to lactate, was monitored but did not increase in ldhA mutant cells (see Fig. S4 in the supplemental material) and therefore does not account for the nonexcreted product. Additionally, polyhydroxybutyrate, which is a reduced polymer formed by some strains of cyanobacteria (30), cannot be formed as a fermentative product in this strain due to the lack of the poly(3-hydroxybutyrate) polymerase gene phaC in the genome of Synechococcus 7002. We conclude that other unmonitored products are responsible for the catabolized carbon and reductant under dark/anaerobic conditions, possibly multiple products derived from the reversible reactions of gluconeogenesis/glycolysis, the pentose phosphate pathway, CO2 fixation, or biosynthetic pathway intermediates. We note that some CO2 fixation occurs under dark anaerobic conditions in A. maxima (1).
In E. coli, increases in hydrogen of 13% and 63% (after 6 and 10 h) were obtained by mutationally inactivating the pathways for the production of lactate or both lactate and acetate, respectively (26, 33). Mutants of Enterobacter sp. strains displayed increases in hydrogen production of 63% and 93% (after 10 and 48 h), with nitroglycerin inhibition of alcohol- and acid-forming pathways (17, 22). Here, we show an ∼500% sustained increase in hydrogen production yield and rate, with a knockout of lactate dehydrogenase in Synechococcus 7002. Despite this significant increase, the overall rate of fermentative hydrogen produced in this marine cyanobacterium is relatively low at 1.2 μmol per liter culture per h (ldhA mutant cells) compared to that of the most prolific nondiazotrophic H2-producing cyanobacteria, such as Arthrospira maxima, which we have observed to evolve hydrogen in the dark at maximum rates approaching 400 μmol per liter culture per h (1). The genetic elimination of metabolic pathways consuming NAD(P)H, such as that demonstrated here, may be applied to other strains for which a more reducing intracellular redox poise is the critical kinetic limitation to H2 production. The genome of A. maxima was recently sequenced (A. M. Garcia Costas, Z. Li, and D. A. Bryant, unpublished data), and tools for genetic modification of this strain are currently being developed.
In addition to lactate production, pathways that consume intracellular reductant via the production and excretion of ethanol and formate from acetyl-CoA are potential targets that could be eliminated to improve the fermentative yield of NADH available for H2 production in other AMOPs. Besides increasing the available reductant, other targets for metabolic and genetic engineering could increase proton availability to the hydrogenase as well as increase the activity of the enzyme itself.
Supplementary Material
Acknowledgments
This work was supported by the Air Force Office of Scientific Research (MURI grant FA9550-05-1-0365), which is gratefully acknowledged.
L. Tiago Guerra is acknowledged for discussions. Miguel Frada and Kevin Wyman are gratefully acknowledged for their assistance with lipid measurements.
Footnotes
Published ahead of print on 11 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ananyev, G., D. Carrieri, and G. C. Dismukes. 2008. Optimization of metabolic capacity and flux through environmental cues to maximize hydrogen production by the cyanobacterium “Arthrospira (Spirulina) maxima.” Appl. Environ. Microbiol. 74:6102-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, A., M. Schmidt, W. Jäger, and A. Pühler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 3.Carrieri, D., G. Ananyev, A. M. Garcia Costas, D. A. Bryant, and G. C. Dismukes. 2008. Renewable hydrogen production by cyanobacteria: nickel requirements for optimal hydrogenase activity. Int. J. Hydrogen Energy 33:2014-2022. [Google Scholar]
- 4.Carrieri, D., K. McNeely, A. C. DeRoo, N. Bennette, I. Pelczer, and G. C. Dismukes. 2009. Identification and quantification of water-soluble metabolites by cryoprobe-assisted nuclear magnetic resonance spectroscopy applied to microbial fermentation. Magn. Reson. Chem. 47:S138-S146. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee, R., C. S. Millard, K. Champion, D. P. Clark, and M. I. Donnelly. 2001. Mutation of the ptsG gene results in increased production of succinate in fermentation of glucose by Escherichia coli. Appl. Environ. Microbiol. 67:148-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chittibabu, G., K. Nath, and D. Das. 2006. Feasibility studies on the fermentative hydrogen production by recombinant Escherichia coli BL-21. Process Biochem. 41:682-688. [Google Scholar]
- 7.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Lett. 63:223-234. [DOI] [PubMed] [Google Scholar]
- 8.Das, D., and T. N. Vezireoglu. 2001. Hydrogen production by biological processes: a survey of literature. Int. J. Hydrogen Energy 26:13-28. [Google Scholar]
- 9.Datta, R., and J. G. Zeikus. 1985. Modulation of acetone-butanol-ethanol fermentation by carbon monoxide and organic acids. Appl. Environ. Microbiol. 49:522-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dismukes, G. C., D. Carrieri, N. Bennette, G. M. Ananyev, and M. C. Posewitz. 2008. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 19:235-240. [DOI] [PubMed] [Google Scholar]
- 11.Gutthann, F., M. Egert, A. Marques, and J. Appel. 2007. Inhibition of respiration and nitrate assimilation enhances photohydrogen evolution under low oxygen concentrations in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1767:161-169. [DOI] [PubMed] [Google Scholar]
- 12.Hassid, W. Z., and S. Abraham. 1957. [7] Chemical procedures for analysis of polysaccharides. Methods Enzymol. 3:34-50. [Google Scholar]
- 13.Hwang, T. L., and A. J. Shaka. 1995. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. A 112:275-279. [Google Scholar]
- 14.Ingram, L. O., P. F. Gomez, X. Lai, M. Moniruzzaman, B. E. Wood, L. P. Yomano, and S. W. York. 1998. Metabolic engineering of bacteria for ethanol production. Biotechnol. Bioeng. 58:204-214. [DOI] [PubMed] [Google Scholar]
- 15.Kengen, S. W. M., and A. J. M. Stams. 1994. Formation of l-alanine as a reduced end product in carbohydrate fermentation by the hyperthermophilic archaeon Pyrococcus furiosus. Arch. Microbiol. 161:168-175. [Google Scholar]
- 16.Kumar, N., and D. Das. 2000. Enhancement of hydrogen production by Enterobacter cloacae IIT-BT 08. Process Biochem. 35:589-593. [Google Scholar]
- 17.Kumar, N., A. Ghosh, and D. Das. 2001. Redirection of biochemical pathways for the enhancement of H2 production by Enterobacter cloacae. Biotechnol. Lett. 23:537-541. [Google Scholar]
- 18.Maeda, T., V. Sanchez-Torres, and T. Wood. 2007. Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77:879-890. [DOI] [PubMed] [Google Scholar]
- 19.Nakashimada, Y., M. A. Rachman, T. Kakizono, and N. Nishio. 2002. Hydrogen production of Enterobacter aerogenes altered by extracellular and intracellular redox states. Int. J. Hydrogen Energy 27:1399-1405. [Google Scholar]
- 20.Nomura, C., S. Persson, G. Shen, K. Inoue-Sakamoto, and D. Bryant. 2006. Characterization of two cytochrome oxidase operons in the marine cyanobacterium Synechococcus sp. PCC 7002: inactivation of ctaDI affects the PS I:PS II ratio. Photosynth. Res. 87:215-228. [DOI] [PubMed] [Google Scholar]
- 21.Örlygsson, J., R. Anderson, and B. H. Svensson. 1995. Alanine as an end product during fermentation of monosaccharides by Clostridium strain P2. Antonie Van Leeuwenhoek 68:273-280. [DOI] [PubMed] [Google Scholar]
- 22.Rachman, M. A., Y. Furutani, Y. Nakashimada, T. Kakizono, and N. Nishio. 1997. Enhanced hydrogen production in altered mixed acid fermentation of glucose by Enterobacter aerogenes. J. Ferment. Bioeng. 83:358-363. [Google Scholar]
- 23.Rodríguez-Ruiz, J., E.-H. Belarbi, J. L. G. Sánchez, and D. L. Alonso. 1998. Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol. Tech. 12:689-691. [Google Scholar]
- 24.Sakamoto, T., and D. A. Bryant. 1997. Growth at low temperature causes nitrogen limitation in the cyanobacterium Synechococcus sp. PCC 7002. Arch. Microbiol. 169:10-19. [DOI] [PubMed] [Google Scholar]
- 25.Sheremetieva, M. E., O. Y. Troshina, L. T. Serebryakova, and P. Lindblad. 2002. Identification of hox genes and analysis of their transcription in the unicellular cyanobacterium Gloeocapsa alpicola CALU 743 growing under nitrate-limiting conditions. FEMS Microbiol. Lett. 214:229-233. [DOI] [PubMed] [Google Scholar]
- 26.Sode, K., M. Watanabe, H. Makimoto, and M. Tomiyama. 1999. Construction and characterization of fermentative lactate dehydrogenase Escherichia coli mutant and its potential for bacterial hydrogen production. Appl. Biochem. Biotechnol. 77:317-323. [Google Scholar]
- 27.Stal, L. J., and R. Moezelaar. 1997. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21:179-211. [Google Scholar]
- 28.Stevens, S. E., C. O. P. Patterson, and J. Myers. 1973. The production of hydrogen peroxide by blue-green algae: a survey. J. Phycol. 9:427-430. [Google Scholar]
- 29.Sticklen, M. 2006. Plant genetic engineering to improve biomass characteristics for biofuels. Curr. Opin. Biotechnol. 17:315-319. [DOI] [PubMed] [Google Scholar]
- 30.Vincenzini, M., C. Sili, R. de Philippis, A. Ena, and R. Materassi. 1990. Occurrence of poly-beta-hydroxybutyrate in Spirulina species. J. Bacteriol. 172:2791-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, X., D. L. Falcone, and F. R. Tabita. 1993. Reductive pentose phosphate-independent CO2 fixation in Rhodobacter sphaeroides and evidence that ribulose bisphosphate carboxylase/oxygenase activity serves to maintain the redox balance of the cell. J. Bacteriol. 175:3372-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodward, J., M. Orr, K. Cordray, and E. Greenbaum. 2000. Biotechnology: enzymatic production of biohydrogen. Nature 405:1014-1015. [DOI] [PubMed] [Google Scholar]
- 33.Yang, Y., K. San, and G. N. Bennett. 1999. Redistribution of metabolic fluxes in Escherichia coli with fermentative lactate dehydrogenase overexpression and deletion. Metab. Eng. 1:141-152. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida, A., T. Nishimura, H. Kawaguchi, M. Inui, and H. Yukawa. 2006. Enhanced hydrogen production from glucose using ldh- and frd-inactivated Escherichia coli strains. Appl. Microbiol. Biotechnol. 73:67-72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.