Abstract
Salmonella enterica serovars Enteritidis and Typhimurium are the leading causative agents of salmonellosis in the United States. S. Enteritidis is predominantly associated with contamination of shell eggs and egg products, whereas S. Typhimurium is frequently linked to tainted poultry meats, fresh produce, and recently, peanut-based products. Chlorine is an oxidative disinfectant commonly used in the food industry to sanitize the surfaces of foods and food processing facilities (e.g., shell eggs and poultry meats). However, chlorine disinfection is not always effective, as some S. enterica strains may resist and survive the disinfection process. To date, little is known about the underlying mechanisms of how S. enterica responds to chlorine-based oxidative stress. In this study, we designed a custom bigenome microarray that consists of 385,000 60-mer oligonucleotide probes and targets 4,793 unique gene features in the genomes of S. Enteritidis strain PT4 and S. Typhimurium strain LT2. We explored the transcriptomic responses of both strains to two different chlorine treatments (130 ppm of chlorine for 30 min and 390 ppm of chlorine for 10 min) in brain heart infusion broth. We identified 209 S. enterica core genes associated with Fe-S cluster assembly, cysteine biosynthesis, stress response, ribosome formation, biofilm formation, and energy metabolism that were differentially expressed (>1.5-fold; P < 0.05). In addition, we found that serovars Enteriditis and Typhimurium differed in the responses of 33 stress-related genes and 19 virulence-associated genes to the chlorine stress. Findings from this study suggest that the oxidative-stress response may render S. enterica resistant or susceptible to certain types of environmental stresses, which in turn promotes the development of more effective hurdle interventions to reduce the risk of S. enterica contamination in the food supply.
Salmonella enterica is the causative agent of salmonellosis, the second most common bacterial food-borne illness in the United States (11, 29, 45, 57). The reported incidence of salmonellosis is about 14 cases per 100,000 population, resulting in approximately 30,000 confirmed cases annually in the United States (11, 29, 57). Considering only 3% of the infections are reported nationwide, the U.S. Centers for Disease Control and Prevention (CDC) has estimated that over 1.4 million cases of infection and 600 deaths related to salmonellosis may occur every year, accounting for about 31% of all food-related deaths in the nation (11, 29, 57).
Among the >2,463 S. enterica serotypes that have been identified, we are most interested in two serotypes, S. enterica serovar Typhimurium and S. enterica serovar Enteritidis, because they are the leading causes of food-borne enteric illnesses in the United States (11, 29, 57). According to the CDC Food-borne Disease Active Surveillance Network, S. Typhimurium and S. Enteritidis caused 22.1% and 17.7% of all salmonellosis cases in 2001, respectively; in 2008, S. Enteritidis and S. Typhimurium accounted for 20.1% and 16.0% of all Salmonella infections in the nation, respectively (12). Becker et al. reported that the number of infections caused by S. Enteritidis between 1985 and 1999 more than tripled (4). A comparison of the preliminary surveillance data for 2005 with the baseline data for 1996 to 1998 on the five most common Salmonella serotypes revealed that the estimated incidence of S. Enteritidis alone increased 25%, which was in contrast to the overall decrease of the Salmonella incidence in the nation (12).
It is notable that S. Enteritidis and S. Typhimurium seem to be associated with different types of foods. For instance, shell eggs and egg products account for 77% of S. Enteritidis infections (57). S. Enteritidis can contaminate shell eggs through the transovarian route as a result of intestinal carriage by hens. Entering the egg contents and staying alive in the albumen is a unique ability of S. Enteritidis (22). In contrast, most other S. enterica serotypes are found on the shell surface as a result of fecal contamination; some may also contaminate the inside of the egg as a consequence of shell damage or removal of egg contents (29). S. Typhimurium, on the other hand, is a frequent contaminant of poultry meats and drinking water. S. Typhimurium has also been implicated in several multistate food-borne outbreaks associated with peanut butters (11) and fresh produce (13). However, S. Typhimurium has not been linked to egg contamination, for unclear but intriguing reasons.
To reduce the total microbial load, including pathogens like S. enterica that may be attached to the surfaces of foods and food processing equipment, the food industry often uses sodium hypochlorite solutions (or chlorine) for disinfection purposes. Chlorine may generate hydroxyl radicals via a Fenton-type reaction, and these reactive oxidative species can disrupt cellular structures and metabolic processes. Our recent study of Escherichia coli O157:H7 showed that sublethal concentrations of chlorine can trigger stress resistance mechanisms in bacteria (66). However, it is not yet clear how S. Enteritidis or S. Typhimurium responds to chlorine oxidation or how such a response may compare to responses to other environmental or host stresses, such as pH shifts, lack of oxygen, osmotic shock, and starvation, during the establishment of infection (54). For this reason, our aim in this study was to explore the transcriptomic responses of fully sequenced strains S. Typhimurium LT2 and S. Enteritidis PT4 under sublethal chlorine exposure.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. enterica strains LT2 (ATCC 19585) and PT4 (ATCC 13076) were obtained from the American Type Culture Collection (Manassas, VA). For growth studies, a single colony of each strain was inoculated into 10 ml brain heart infusion (BHI; Difco, Detroit, MI) broth, and the cultures were incubated at 37°C with aeration for 18 h. The bacteria were then harvested by centrifugation at 4,000 × g for 10 min at room temperature and washed with 10 ml Butterfield phosphate buffer (BPB). The cell density was adjusted in BPB by using a Spectronic 21 spectrophotometer to an optical density at 600 nm (OD600) of 0.8. Dilutions of 1:100 were made in BHI broth without and with sodium hypochlorite solution (13%; Acros Organics, NJ) added at selected concentrations of 130, 260, and 390 ppm.
The Bioscreen C automatic microbiology growth curve analysis system was used to monitor the growth of the diluted cultures. An aliquot of 2 μl of each culture was loaded in five replicate wells of a Bioscreen C honeycomb plate, each of which was preloaded with 200 μl BHI. The Bioscreen C instrument was programmed to measure cell turbidity (OD600) every 10 min for a total of 18 h at 37°C with 5 s of shaking prior to each measurement. For each experiment, BHI broth without bacterial inoculum was used as a negative control. Growth curves were generated using Microsoft Excel after importing the Bioscreen C cell turbidity data. The lag phase (in hours) was defined as the time between the initial inoculation and the time at which cell turbidity reached a value of 0.2. The lag phase extension (in hours) was calculated as the lag phase of the strain grown in BHI containing chlorine minus that without chlorine. All growth experiments were performed in at least three independent trials for statistical analysis.
Bigenome DNA array design.
The sequences and annotations for the LT2 and PT4 genomes were obtained from GenBank under accession numbers NC_003197 (45) and NC_011294 (12), respectively. Orthologous and strain-specific genes were identified by whole-genome alignment using Nucmer (34). Strain-specific genes were identified as those without an alignment covering at least 90% of the gene length at greater than 90% nucleotide identity. A high-density DNA array consisting of 385,000 oligonucleotide probes was designed to target all predicted genes in the LT2 genome plus all PT4-specific genes, thus covering all genes of both serotypes. Probes (60-mer each) were synthesized in situ on the array by using a maskless array synthesizer (Roche NimbleGen, Madison, WI). A total of 4,793 genes were targeted by this array, including 4,527 predicted genes in the LT2 genome and an additional 266 genes specific to the PT4 genome. On average, each gene was targeted by 19 different probes. Each probe was printed in four replicates at random locations on the array to avoid positional bias of hybridization and image analysis. Unused features were filled with randomly generated negative control probes with a comparable GC content of 52%.
RNA isolation and DNA array hybridization.
An aliquot of 1 ml overnight culture of LT2 or PT4 was inoculated in 9 ml BHI broth and grown at 37°C for 4 h to reach early exponential phase. Cells were washed with 0.1% buffered peptone water at room temperature and incubated in 10 ml BHI broth supplemented with 30 μl sodium hypochlorite stock solution (13%) at 37°C for 10 min (equivalent to 390 ppm) or 10 μl sodium hypochlorite stock solution (13%) at 37°C for 30 min (equivalent to 130 ppm). A 5-ml sample was then mixed with 10 ml RNAprotect (Qiagen, Valencia, CA), vortexed, incubated at room temperature for 5 min, and centrifuged at 3,220 × g for 15 min to precipitate the cells. Cell pellets were resuspended in 200 μl lysis buffer (30 mM Tris-HCl, pH 8.0, 1 mM EDTA, 15 mg/ml lysozyme, 10 μl proteinase K). Total bacterial RNA was extracted using an RNeasy Midi kit (Qiagen) according to manufacturer's instructions. The RNA quantity and integrity were analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Hybridization, staining, and processing of arrays were performed at Roche NimbleGen (Madison, WI). Briefly, the cDNA was synthesized from the total RNA pool by using the SuperScript double-stranded cDNA synthesis kit (Invitrogen). Hybridizations were carried out at 45°C for 16 h by using the Roche NimbleGen hybridization system. After washing and drying, arrays were scanned at a 5-μm resolution with a NimbleGen MS 200 microarray scanner. All experiments were repeated three times on different days for statistical analysis.
DNA array data analysis.
The acquisition and initial quantification of array images were performed using the NimbleScan 2.5 software (Roche NimbleGen, Madison, WI). Raw array data were normalized per chip and per gene and filtered based on raw signal intensity and detection call. A Pearson correlation coefficient (r value) was calculated to assess the reproducibility of duplicate array data sets (via Free Statistics Software version 1.1.23-r3 [P. Wessa, Office for Research Development and Education]). Subsequent data comparison was performed using ArrayStar 3 (DNASTAR, Madison, WI). Briefly, normalized signal intensities of S. enterica genes on replicate arrays were averaged, converted to log2 values, and compared between experiments using the F test (analysis of variance [ANOVA]). The P values were adjusted to correct for false-positive errors by using the Benjamini-Hochberg false discovery rate approach (5). Genes with an expression fold change of ≥1.5 (P < 0.05) between a treatment and a control were considered to be significant in this study. The hierarchical clustering analysis was performed using the MultiExperiment Viewer module of the TM4 microarray software suite, version 4.5.1 (55).
Quantitative RT-PCR analysis.
Twelve genes that showed significant up- or downregulation (P < 0.05) in the microarray experiments were selected for quantitative reverse transcription-PCR (qRT-PCR) validation. Forward and reverse PCR primers (Table 1) were designed using Primer 3 software to produce an amplicon size of approximately 150 to 200 bp for each gene. RNA samples were extracted as described above and treated with DNase I (Invitrogen, Carlsbad, CA) to eliminate genomic DNA contamination. qRT-PCR was performed in a LightCycler 480 (Roche Applied Science, Indianapolis, IN) with cDNA reverse transcribed from 1 μg of purified total RNA by using the Transcriptor First Strand kit (Roche Applied Science) as previously described (66). The 16S rRNA gene was used as an internal reference for data normalization. All experiments included a minimum of at least three independent replicates for statistical analysis.
TABLE 1.
Primer pairs used for qRT-PCR analysis
| GeneID | Gene name | Protein annotation | Primer typea | Primer sequence |
|---|---|---|---|---|
| STM1572 | nmpC | Putative outer membrane porin precursor | F | 5′-CGA CCA GGA TCT GGT TGA GT-3′ |
| R | 5′-CTT TAG CCG CTT TGG TGA AG-3′ | |||
| STM2543 | nifS | Cysteine desulfurase | F | 5′-ATC GCG AAA GAA GAG ATG GA-3′ |
| R | 5′-TCG CCG TTC AGG TAA ACT TC-3′ | |||
| STM2542 | nifU | NifU-like protein | F | 5′-AAC GAC GAT AAC GTG GGA AG-3′ |
| R | 5′-GCA GCC GTA AGT CTT GAA GC-3′ | |||
| STM2544 | yfhP | Putative iron-sulfur cluster regulatory protein | F | 5′-TTA CCT TAG GCG AGC TGG TG-3′ |
| R | 5′-GCG CGT AAT TTA ACG TCG AT-3′ | |||
| STM0823 | ybiJ | Putative periplasmic protein | F | 5′-TGC ATA AAA TCG GCG TAG TG-3′ |
| R | 5′-TGC CGC TCA TTT TGT CAT TA-3′ | |||
| STM1214 | ycfR | Putative outer membrane protein | F | 5′-ACG CCA GAA GGT CAA CAG AA-3′ |
| R | 5′-GGG CCG GTA ACA GAG GTA A-3′ | |||
| STM2430 | cysK | O-Acetylserine sulfhydrolase A | F | 5′-CGC TAT TCA GAA AGC CGA AG-3′ |
| R | 5′-CAT CGG TGT CTT CCC AGA TT-3′ | |||
| STM2338 | pta | Phosphate acetyltransferase | F | 5′-AGC CAC GTT GAA TCT CTG CT-3′ |
| R | 5′-AGA CCT TCA ACC AGC ACC AC-3′ | |||
| STM3867 | atpA | ATP synthase subunit A | F | 5′-GGC GAC GTA TTC TAC CTC CA-3′ |
| R | 5′-CGG TTT TCC CTT TCA CTT CA-3′ | |||
| STM1378 | pykF | Pyruvate kinase | F | 5′-ATG AAC GTG ATG CGT CTG AA-3′ |
| R | 5′-TAA TGG TGC GGA TTT CTG GT-3′ | |||
| SEN2085D | rfbS | Paratose synthase | F | F:5′-TGGCTTAGCAAGGAAGAGGA-3′ |
| R | 5′-TGGCAGTGATGTTCCACAAT-3′ | |||
| SEN2085C | rfbE | CDP-tyvelose-2-epimerase | F | F:5′-CAAAAGGTGCTGCAGATCAA-3′ |
| R | 5′-CAGCATGCAAAACATCCCTA-3′ |
F, forward; R, reverse.
Microarray data accession number.
The microarray data from this study were deposited in the NCBI Gene Expression Omnibus under accession number GSE20701.
RESULTS AND DISCUSSION
Adaptive gene expression allows S. enterica to respond to a wide variety of environmental and host stresses. Despite the extensive studies on the stress response and pathogenesis of S. enterica in the host, relatively little is known about the stress response of S. enterica to chlorine, an oxidative reagent used by the food industry to disinfect food surfaces and sanitize food processing equipment. In this study, we focused our study on examining the transcriptomic responses of two representative S. enterica strains, LT2 and PT4, to chlorine-based stress. We employed a newly designed bigenome DNA array that covered all predicted genes in both LT2 and PT4 genomes and identified differentially expressed genes that potentially mediate the stress response of S. enterica to chlorine oxidation.
Comparison of PT4 and LT2 genomes.
For the bigenome array design, we first compared the main chromosomes of S. Typhimurium LT2 and S. Enteritidis PT4 in order to identify core sequences (shared by both genomes) and strain-specific sequences (present in one genome but absent in the other). We found that strain-specific sequences comprised about 8% of the 4.9-Mbp LT2 chromosome and about 5% of the 4.7-Mbp PT4 chromosome. Of the remaining shared sequences, the two chromosomes exhibited an average of 99% nucleotide identity. An alignment of the two chromosomes indicated a single large (∼900-kbp) inversion between the 1- and 2-Mbp marks, along with small insertions and deletions scattered throughout the chromosomal regions. With a cutoff of 90% nucleotide identity over 90% of the gene length, totals of 399 and 266 strain-specific genes were identified in the LT2 and PT4 chromosomes, respectively. The majority of the PT4-specific genes were prophage-like elements, whereas 69% of the LT2-specific genes encoded putative and hypothetical proteins (61).
Chlorine treatments.
Fifty- to 200-ppm chlorine solutions are commonly used by the industry for food surface disinfection (6). We conducted a growth study for both LT2 and PT4 strains challenged with low (130 ppm), medium (260 ppm), and high (390 ppm) concentrations of chlorine in BHI broth. The levels of resistance of LT2 and PT4 to different concentrations of chlorine were compared based on the extension of the lag phase under the chlorine stress (see Fig. S1 in the supplemental material). Strains grown in BHI broth supplemented with 130 ppm of chlorine showed a lag phase of approximately 4 h. At 260 ppm, the lag phases of both LT2 and PT4 extended to 5 to 6 h. At 390 ppm, both strains showed no growth in the first 18 h of incubation. These results suggested that 130 ppm of chlorine in BHI broth imposed weak oxidative stress to S. enterica whereas 390 ppm of chlorine in BHI broth inhibited the growth of the bacterial cells. We realized that the organic compounds in BHI broth can rapidly neutralize the free chlorine (ClO−), as we have shown that the oxidation reduction potential (ORP) of chlorine decreased 49% in BHI broth compared to that in a water solution after 5 min (66). We treated cells in BHI broth supplemented with 130 ppm of chlorine for 30 min and with 390 ppm of chlorine for 10 min prior to RNA extraction; under these conditions, we characterized the transcriptomic responses of LT2 and PT4 to weak and strong oxidative stresses, respectively.
Global gene expression profiles under the chlorine treatment.
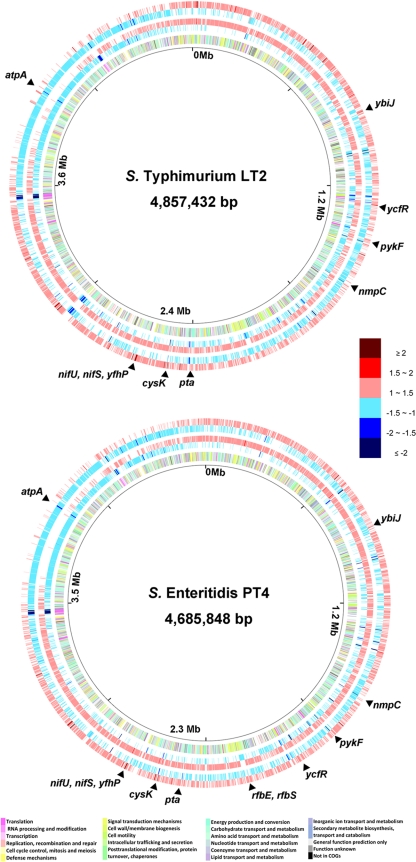
Before comparison of gene expression profiles, we first evaluated the data reproducibility for the replicate experiments on different days by using the pairwise linear correlation analysis. The correlation coefficients (r) between two replicate experiments ranged from 0.709 to 0.993, suggesting satisfactory reproducibility of the arrays under all experimental conditions (Table 2). Based on the fold changes relative to control experiments without chlorine, we constructed circular maps (Fig. 1) to show the global gene expression profiles of 4,423 genes in the LT2 genome (43) and 4,206 genes in the PT4 genome (58) under the two different chlorine treatments. Overall, under 130 ppm for 30-min chlorine treatment, 2,013 (45.4%) of the LT2 genes and 2,092 (49.7%) of the PT4 genes were upregulated (>1-fold) and 2,400 (54.3%) of the LT2 genes and 2,058 (48.9%) of the PT4 genes were downregulated (>1-fold). Under 390 ppm for 10-min chlorine treatment, 2,707 (61.2%) of the LT2 genes and 2,589 (61.6%) of the PT4 genes were upregulated (>1-fold) and 1,712 (38.7%) LT2 genes and 1,571 (37.4%) PT4 genes were downregulated (>1-fold). With a cutoff of ≥1.5-fold (P < 0.05), 44 and 156 genes were found to be significantly up- and downregulated, respectively, in at least one of the S. enterica strains, representing about 2.2% of all LT2 genes and about 3% of all PT4 genes (Table 3).
TABLE 2.
Linear correlation coefficients between replicate array experiments
| Strain | Treatment | Correlation coefficienta |
||
|---|---|---|---|---|
| Array 1 vs 2 | Array 1 vs 3 | Array 2 vs 3 | ||
| S. Enteritidis PT4 | Control | 0.733 | 0.741 | 0.977 |
| 130 ppm, 30 min | 0.993 | 0.874 | 0.869 | |
| 390 ppm, 10 min | 0.940 | 0.936 | 0.956 | |
| S. Typhimurium LT2 | Control | 0.920 | 0.929 | 0.977 |
| 130 ppm, 30 min | 0.981 | 0.773 | 0.709 | |
| 390 ppm, 10 min | 0.975 | 0.929 | 0.912 | |
Arrays 1, 2, and 3 were three independent biological replicates prepared and hybridized on different days. Pairwise comparisons were made between each pair of biological replicates to assess the reproducibility of array data.
FIG. 1.
Circular maps that compare the global gene expression profiles of S. Typhimurium strain LT2 and S. Enteritidis strain PT4 under the chlorine treatments. The innermost circle gives the genomic coordinates of the LT2 or PT4 chromosome. From the inside out, the second circle shows all protein-encoding genes color coded based on clusters of orthologous groups (COGs) of proteins (see the color codes at the bottom). The third and forth circles show the downregulated (blue) and upregulated (red) genes, respectively, under strong chlorine oxidation (390 ppm for 10 min). The fifth and sixth circles show the downregulated (blue) and upregulated (red) genes, respectively, under weak chlorine oxidation (130 ppm for 30 min). All differentially expressed genes are color coded based on the fold change relative to the control experiment (see the color code on the right side). Genes selected for quantitative RT-PCR are marked at the corresponding locations on the chromosome. The circular maps were constructed using the GenomeViz 1.2 software.
TABLE 3.
Numbers of differentially expressed genes in LT2 and PT4 under the chlorine treatmentsa
| Expression type | No. of differentially expressed genes |
|||
|---|---|---|---|---|
| 130 ppm, 30 min |
390 ppm, 10 min |
|||
| S. Typhimurium LT2 | S. Enteritidis PT4 | S. Typhimurium LT2 | S. Enteritidis PT4 | |
| Upregulated | 35 | 21 | 2 | 8 |
| Downregulated | 70 | 104 | 99 | 135 |
Differential expression characterized by ≥1.5-fold expression (P < 0.05).
Table 4 lists some differentially expressed genes with functional annotations. The upregulated genes were mostly related to stress response, iron-sulfur (Fe-S) cluster formation, cysteine biosynthesis, and biofilm formation. The downregulated genes were predominantly associated with general cellular metabolism, ribosomal proteins, flagellum biosynthesis, and virulence.
TABLE 4.
Differentially expressed genes in LT2 and PT4 under the chlorine treatmentsa
| Function and gene | Annotation | Fold change |
|||
|---|---|---|---|---|---|
| 130 ppm, 30 min |
390 ppm, 10 min |
||||
| LT2 | PT4 | LT2 | PT4 | ||
| Fe-S cluster formation and cysteine synthesis/transport protein | |||||
| hscA | Chaperone protein HscA | 1.59 | 1.16 | 1.12 | 1.09 |
| hscB | Cochaperone HscB | 1.56 | 1.25 | 1.23 | 1.26 |
| iscA | Iron-sulfur cluster assembly protein | 2.07 | 1.84 | 1.17 | 1.20 |
| iscR/yfhP | DNA-binding transcriptional regulator IscR | 3.45 | 3.45 | 1.61 | 1.39 |
| fdx | Electron carrier protein | 1.81 | 1.39 | 1.19 | 1.10 |
| sufE/ynhA | Cysteine desulfuration protein SufE | 1.16 | 1.06 | 1.04 | −1.03 |
| sufS | Bifunctional cysteine desulfurase/selenocysteine lyase | 1.06 | −1.06 | 1.24 | 1.06 |
| sufD | Cysteine desulfurase activator complex subunit SufD | 1.07 | −1.02 | 1.25 | 1.11 |
| sufC | Cysteine desulfurase ATPase component | 1.09 | 1.02 | 1.16 | 1.16 |
| sufB | Cysteine desulfurase activator complex subunit SufB | 1.15 | 1.04 | 1.14 | 1.02 |
| sufA | Iron-sulfur cluster assembly scaffold protein | 1.14 | −1.05 | 1.24 | 1.13 |
| yfhJ | Hypothetical protein | 1.61 | 1.34 | 1.16 | 1.16 |
| nifS | Cysteine desulfurase | 3.31 | 3.04 | 1.26 | 1.57 |
| nifU | NifU-like protein | 3.49 | 2.92 | 1.29 | 1.23 |
| yfhP | Putative iron-sulfur cluster regulatory protein | 3.45 | 3.45 | 1.39 | 1.61 |
| sbp | Sulfate transport protein | 1.47 | 1.81 | −1.10 | −1.11 |
| cysC | Adenylylsulfate kinase | 1.93 | 1.56 | 1.35 | 1.20 |
| cysN | Sulfate adenylyltransferase subunit 1 | 2.28 | 2.90 | 1.25 | 1.45 |
| cysD | Sulfate adenylyltransferase subunit 2 | 2.09 | 2.99 | 1.18 | 1.44 |
| cysH | Phosphoadenosine phosphosulfate reductase | 1.72 | 1.66 | 1.28 | 1.12 |
| cysI | Sulfite reductase subunit beta | 2.05 | 2.06 | 1.14 | 1.32 |
| cysJ | Sulfite reductase subunit alpha | 1.71 | 1.92 | 1.37 | 1.38 |
| cysA | Sulfate/thiosulfate transporter subunit | 1.52 | 1.44 | 1.23 | 1.11 |
| cysP | Thiosulfate transport protein | 2.69 | 2.31 | 1.58 | 1.37 |
| cysK | Cysteine synthase A | 2.72 | 3.71 | 1.23 | 1.42 |
| Phosphotransferase system | |||||
| manX | Mannose-specific enzyme IIAB | −1.64 | −2.71 | −2.32 | −3.36 |
| manY | Mannose-specific enzyme IIC | −1.72 | −2.47 | −2.01 | −2.50 |
| manZ | Mannose-specific enzyme IID | −1.31 | −1.76 | −1.49 | −1.83 |
| ptsH | Phosphohistidinoprotein-hexosephosphotransferase | −1.56 | −2.26 | −2.35 | −2.62 |
| ptsI | PEP-protein phosphotransferaseb | −1.47 | −1.92 | −2.11 | −2.12 |
| ptsG | Glucose-specific IIBC component | −1.52 | −1.74 | −1.52 | −1.72 |
| Cell envelope | |||||
| ompF | Outer membrane protein F precursor | −1.25 | −1.63 | −1.32 | −2.03 |
| ompA | Putative hydrogenase membrane component precurosr | 1.65 | 1.33 | −1.23 | −1.45 |
| nmpC | Putative outer membrane porin precursor | −3.19 | −2.52 | −3.56 | −3.21 |
| ompC | Outer membrane protein C precursor | −1.25 | −1.69 | −1.10 | −1.41 |
| fljB | Flagellin | 1.07 | 1.02 | −1.82 | −1.65 |
| flgB | Flagellar basal body rod protein | −1.57 | −1.50 | −1.74 | −1.81 |
| flgC | Flagellar basal body rod protein | −1.89 | −1.81 | −1.86 | −1.75 |
| flgD | Flagellar basal body rod modification protein | −1.87 | −1.71 | −1.78 | −1.62 |
| flgE | Flagellar hook protein | −1.66 | −1.77 | −1.86 | −1.88 |
| flgF | Cell-proximal portion of basal body rod | −1.54 | −1.65 | −1.52 | −1.71 |
| flgG | Flagellar basal body rod protein | −1.47 | −1.68 | −1.54 | −1.58 |
| fliA | Flagellar biosynthesis sigma factor FliA | −1.19 | −1.53 | −1.44 | −1.48 |
| fliR | Flagellar biosynthesis protein | −1.08 | 1.13 | 1.02 | 1.54 |
| Nitrosative-stress-sensitive proteins | |||||
| atpA | ATP synthase subunit A | −1.53 | −2.09 | −1.86 | −2.20 |
| fabB | 3-Oxoacyl-(acyl carrier protein) synthase | 2.08 | 1.19 | 1.01 | −1.20 |
| fabF | 3-Oxoacyl-(acyl carrier protein) synthase | −1.24 | −1.93 | −1.52 | −1.91 |
| gapA | Glyceraldehyde-3-phosphate dehydrogenase | −1.13 | −1.41 | −2.04 | −2.01 |
| tuf | Elongation factor Tu | −1.17 | −1.39 | −1.72 | −1.58 |
| gltA | Citrate synthase | 1.55 | 1.49 | 1.27 | 1.26 |
| Metabolic genes | |||||
| atpD | ATP synthase subunit B | −1.36 | −1.97 | −1.39 | −1.92 |
| atpF | ATP synthase subunit B | −1.29 | −1.76 | −1.57 | −1.95 |
| atpG | ATP synthase subunit C | −1.46 | −2.04 | −1.63 | −2.13 |
| atpH | ATP synthase subunit D | −1.51 | −2.16 | −1.80 | −2.38 |
| lamB | Maltoporin precursor | −1.00 | −2.31 | −1.17 | −2.13 |
| malE | Periplasmic maltose-binding protein | 1.02 | −1.59 | −1.12 | −1.51 |
| malM | Periplasmic protein precursor | −1.88 | −1.16 | −1.81 | |
| malK | Maltose transport protein/repressor | −1.03 | −2.04 | −1.13 | −1.86 |
| malP | Maltodextrin phosphorylase | −1.04 | −1.74 | −1.05 | −1.65 |
| malQ | 4-Alpha-glucanotransferase | −1.02 | −1.53 | −1.05 | −1.46 |
| pta | Phosphate acetyltransferase | −1.72 | −2.28 | −1.67 | −1.82 |
| Regulatory genes | |||||
| dksA | DnaK suppressor protein | 1.58 | 1.21 | 1.10 | 1.04 |
| yfiA | Ribosome stabilization factor | 2.04 | 1.58 | 1.14 | 1.02 |
| Biofilm formation | |||||
| bssS | Biofilm formation regulatory protein BssS | 1.57 | 1.37 | 1.07 | 1.04 |
| ycfR | Putative outer membrane protein | 2.76 | 2.30 | 1.44 | 1.54 |
| Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | |||||
| rfbH | CDP-6-deoxy-d-xylo-4-hexulose-3-dehydrase | −1.30 | −1.45 | −1.56 | −1.68 |
| rfbF | Glucose-1-phosphate cytidylyltransferase | −1.40 | −1.46 | −1.73 | −1.69 |
| rfbC | dTDP-4,deoxyrhamnose-3,5-epimerase | −1.18 | −1.20 | −1.52 | −1.55 |
| rfbA | dTDP-glucose pyrophosphorylase | −1.09 | −1.13 | −1.37 | −1.52 |
| rfaJ | Lipopolysaccharide glucosyltransferase | −1.10 | −1.02 | −1.26 | −1.50 |
Differential expression characterized by ≥1.5-fold expression (P < 0.05).
PEP, phosphoenolpyruvate.
Transcriptomic response of the S. enterica core genes to chlorine treatment.
Strains PT4 and LT2 share 3,991 “core genes” in their respective genomes (61). E. coli also shares about 83% homologous genes with S. enterica subspecies I, including genes associated with metabolism, transcription, translation, cell motility, and signal transduction (2). To identify the common stress response mechanisms of Gram-negative bacterial pathogens to chlorine-based oxidation, we first compared the transcriptomic profiles of the core genes between the two S. enterica strains and then with those in the invariant gene pool between S. enterica and E. coli.
A group of genes commonly induced under the chlorine treatments in both S. enterica and E. coli include those involved in the Fe-S cluster biosynthesis (8). The assembly of Fe-S clusters is a key step in the posttranslational maturation of Fe-S proteins in Gram-negative bacteria. This process involves two independent systems, the iron-sulfur cluster (ISC) and the sulfur utilization factor (SUF). The ISC system plays a primary role in Fe-S cluster biosynthesis, whereas the SUF system serves as an accessory pathway (58). The iscRSUA-hscBA-fdx operon encoding the ISC system showed >2-fold upregulation in both PT4 and LT2. The sufABCDSE operon encoding the SUF system was upregulated in LT2 but not in PT4. Similarly, the E. coli O157:H7 ISC and SUF operons were both upregulated under the chlorine stress, but genes encoding the ISC system exhibit a fold change greater than those of genes encoding the SUF system (66).
The majority of cysteine biosynthesis genes, including cysCND, cysJIH, and cysZK, were upregulated >2-fold in both PT4 and LT2 under the 130-ppm chlorine treatment, similar to what we have observed in E. coli O157:H7 (66). This is likely because cysteine residues play a role in bacterial oxidative stress response by serving as indispensable components in the Fe-S clusters and other oxidation-related proteins. For example, one of the six cysteine residues (Cys-199) is critical for the redox sensitivity of the transcriptional regulator OxyR (33). It is notable that the cysteine biosynthesis operons were less upregulated (1.2- to 1.5-fold) under the 390-ppm chlorine treatment.
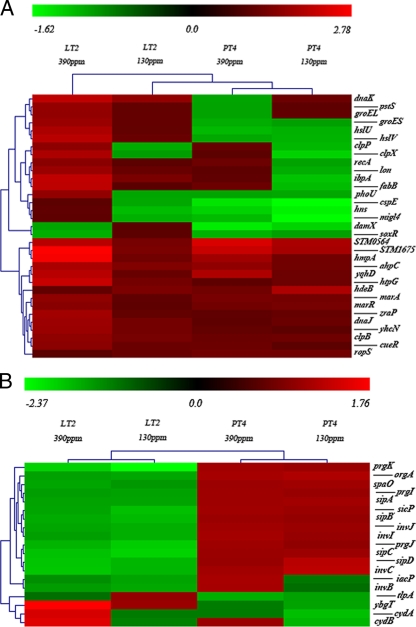
Reactive nitrogen species (RNS) function as powerful antimicrobials in mammalian hosts, but bacteria have developed certain mechanisms to survive under the nitrosative stress. This process involves RNS-mediated protein modifications, including the reversible binding of NO to multiple Fe-S clusters or thiol groups (8). Six of the 10 previously identified nitrosative-stress-sensitive protein-encoding genes (atpA, fabB, fabF, gapA, gltD, and tuf) were differentially regulated under the chlorine treatment (Fig. 2A), challenging some previous speculations that genes which respond to reactive oxygen are distinct from RNS response genes in Gram-negative bacteria (8).
FIG. 2.
Hierarchical cluster plots that show the gene expression patterns of 33 stress-related genes (A) and 19 virulence-associated genes (B) in LT2 and PT4 under the weak (130 ppm) and strong (390 ppm) chlorine treatments. The pseudo-color bar indicates the relative fold change.
We found that a number of genes involved in bacterial biofilm formation were upregulated under chlorine oxidation. For instance, ycfR, which encodes a multiple-stress resistance protein and regulates biofilm formation by changing cell surface hydrophobicity (67), was induced >2-fold in both PT4 and LT2. Gene bssS (or yceP), which regulates E. coli biofilm formation by controlling indole transportation (16), was also upregulated. Deletion of bssS or ycfR can lead to increased biofilm production in E. coli K-12 (16, 67), but the functions of their homologs in S. enterica have not been reported. Cell motility is intrinsically correlated to biofilm formation (28). Deletion of bssS leads to the induction of flagellum biosynthesis genes in E. coli K-12 (16). It is yet to be determined how oxidative stress affects biofilm formation and cell motility in S. enterica. DksA inhibits the flagellar expression during stationary phase or under starvation in E. coli (38). We observed the upregulation of dksA in both PT4 and LT2 under chlorine oxidation, along with downregulation of flagellum biosynthesis and assembly genes. The flagellar basal body operon flgBCDEFG and flagellar biosynthesis factors such as fliA and fijB were downregulated. Inhibition of the synthesis and assembly of flagella, which are large macromolecular complexes of the bacterial cell, may serve as an energy conservation strategy for S. enterica under stress. A ribosome stabilization factor, yfiA, was induced under chlorine treatments. The yfiA gene is typically induced when ribosomal structure changes drastically, leading to the formation of inactive forms of the ribosome (63). Upregulation of yfiA suggests that S. enterica may initiate stabilization mechanisms to protect the ribosome from degradation and to maintain cell function under oxidative stress.
The manXYZ operon, encoding a phosphotransferase system (PTS) transporter, was downregulated >2-fold under chlorine treatment in both S. enterica strains. The expression of this operon in E. coli was found to enhance bacterial tolerance to organic solvents (47). Meanwhile, genes encoding the PTS in S. enterica, such as ptsG and ptsHI, were also significantly downregulated. The PTS is necessary in the process of glycolysis, an integral part of Salmonella infection in the host (7). Downregulation of the PTSs may lead to a reduced pathogenic potential. Besides the PTS, gapA, which encodes the glyceraldehyde-3-phosphate dehydrogenase in the glycolysis pathway, was also downregulated as a result of chlorine oxidation.
The chlorinated BHI broth has a neutral pH, approximately 7.0. We noticed that a small number of metabolic genes responsive to basic or acidic stresses showed downregulation. This was expected, because the bacterial response to oxidative stress may overlap with responses to acid or alkaline stress (68). For example, a group of genes in the atp operon encoding the FoF1 proton-translocating ATPase complex were downregulated. This complex is involved in the electron transport-linked phosphorylation by importing protons during oxidative respiration (43). The maltose regulons (lamB, malEKM, and malPQ) that were found to be inhibited by acidic stress (14) were repressed under chlorine treatment. This is possibly because chlorine-based oxidation can decrease the amount of proton import as well as the metabolic rate. Several osmolarity- and pH-regulated outer membrane porins, such as those encoded by ompA, ompC, ompF, and nmpC were also downregulated under the chlorine treatments.
Differential regulation of strain-specific genes.
Among the 266 PT4- and 399 LT2-specific genes, only four PT4-specific genes and three LT2-specific genes were found to be differentially regulated under the chlorine treatments (Table 5). Two of the PT4-specific genes, SEN1135 and SEN1992, are prophage-like elements, and both were upregulated under chlorine oxidation. SEN1135 encodes a hok protein-like membrane protein (61). This small polypeptide shares 87% amino acid sequence homology with a cell toxin protein, Hok, in E. coli K-12. Several hok homologs encode the peptides that mediate plasmid maintenance by killing plasmid-free cells (50). Hok peptides kill bacterial cells by depolarizing the cytoplasmic membrane when it is electroporated into or synthesized in the cytosol (49). It remains to be elucidated if the hok expression triggered by oxidative stress is related to plasmid maintenance in S. Enteritidis.
TABLE 5.
Differentially expressed genes specific to LT2 and PT4
| Strain and gene | Protein annotation | Gene expression fold change under chlorine treatment |
|
|---|---|---|---|
| 130 ppm, 30 min | 390 ppm, 10 min | ||
| PT4 | |||
| SEN1135 | Phage-encoded Hok-like membrane protein | 1.51 | 1.23 |
| SEN1992 | Hypothetical protein | 1.80 | 1.86 |
| rfbE | CDP-tyvelose-2-epimerase | −1.96 | −1.31 |
| rfbS | Paratose synthase | −1.95 | −1.56 |
| LT2 | |||
| yigG | Putative inner membrane protein | −1.51 | −1.07 |
| sirA | Invasol SirA | 1.618 | 1.16 |
| STM2666 | Hypothetical protein | −1.52 | −1.06 |
Two PT4-specific lipopolysaccharide (LPS) O chain genes, rfbE and rfbS, were downregulated under the chlorine treatment, similar to rfbH, rfbF, and the rfbABC operon shared by LT2 and PT4. LPS is the major component of the outer membrane of Gram-negative bacteria and a primary target by the host innate immune system. The composition of LPS is important for the survival of S. Enteritidis in avian macrophages (27). Gene rfbH is required for S. Enteritidis to survive in egg albumen (23), which also plays a role in the adaptive resistance of S. Enteritidis biofilms to benzalkonium chloride (41). Mizumoto et al. (46) suggested that LPS structure in S. Enteritidis may contribute to the attachment to chicken vaginal explants. Downregulation of LPS-associated genes may negatively impact S. Enteritidis survival in eggs.
Three LT2-specific genes, STM2666, sirA, and yigG, were downregulated under the 130-ppm chlorine treatment. STM2666 encodes a protein identical to the E. coli K-12 PheA leader peptide. Gene pheA is conserved in both S. Enteritidis and S. Typhimurium and was shown to be responsive to oxidative stress in Mycobacterium species (17). However, only LT2 has this leader peptide gene located upstream of pheA. It remains to be determined if this leader peptide is essential for a functional PheA in S. Typhimurium. LT2-specific gene sirA belongs to the BarA/SirA two-component regulatory system critical for bacterial biofilm formation and virulence (60). SirA can activate the noncoding small RNAs, such as csrB and csrC, and the fim operon to promote biofilm formation and inhibit the translation of flagellar proteins (59). Another LT2-specific gene, yigG, is located downstream of an oxidative-stress regulator corA, but its function is not known.
Other stress-related genes.
Environmental stimuli can alter the expression of bacterial stress-related genes. In turn, the expression of stress-related genes may allow bacterial cells to adapt to and survive better under chorine-based oxidation. In this study, we identified a number of stress-related regulatory genes that were induced by the chlorine stress. The expression of central stress regulator rpoS was slightly upregulated 1.2-fold in both strains. Gene pta, which encodes a phosphotransferase, was downregulated about 2-fold. Deletion of this gene in E. coli was shown to activate the rpoS regulon (30). The oxidative-stress regulator oxyR was induced 1.4-fold under the 130-ppm/30-min chlorine treatment but was not induced by the 390-ppm/10-min treatment. Genes in the oxyR regulon, such as ahpC, were upregulated 1.8-fold. Gene soxS, a component of the oxidative-stress regulator operon soxRS, was slightly repressed (1.2-fold) by the 130-ppm/30-min chlorine treatment. The differential regulation of the oxyR and soxRS regulons suggested that they respond differently to chlorine-based oxidation in S. enterica. Most oxidoreductase-encoding genes, such as hmpA, yqhD, STM0564, and STM1675, were induced in both PT4 and LT2. However, upregulation of several other oxidative-stress-related genes, such as pstS, phoU, and recA, was observed only in LT2.
The adaptive responses of E. coli and Salmonella to phosphate limitation are coordinated by the pho regulon (64, 65). In this regulon, the PstSCAB2 transporter can sense phosphate levels and communicate with the two-component signaling protein PhoRB through PhoU (53). The pho regulon, including phoU and pstS, was induced in both E. coli O157:H7 (66) and LT2 under chlorine oxidation. However, the (pstSCAB)2-phoU operon did not respond to chlorine-based oxidative stress in PT4. Similarly, another putative membrane protein gene, yhcN, was upregulated only in LT2 and not in PT4. This gene was recently found to be associated with oxidative- and acid stress responses as well as biofilm formation in E. coli (36). The multidrug resistance operon marRAB was induced in both strains under the chlorine treatment. The induction of marRAB was found to activate other stress resistance genes through a single and uniquely configured marbox (42).
Environmental stresses such as heat, oxidation, and extreme pH can lead to the destabilization and unfolding of proteins in microorganisms (26). Bacteria have developed two major strategies to deal with protein unfolding and aggregation: molecular chaperones and general proteolysis (18). In this study, genes encoding chaperones (dnaKJ, groESL, and htpG) were significantly upregulated by the chlorine treatments in LT2 but not in PT4, similar to protease-encoding genes hlsVU and lon. An ATP-dependent protease gene, clpXP, was induced in LT2 but slightly repressed in PT4. The clpXP operon plays an important role in bacterial survival under osmotic or oxidative stress and cold shock. The loss of clpX could also reduce biofilm formation (21). It has been shown that clpXP controls the RpoS concentration in E. coli (4). The cold shock protein-encoding gene cspE was downregulated only in PT4, and this gene has also been found to be involved in the regulation of RpoS (51). In contrast, clpB, which encodes a protein disaggregation chaperone (44), and hdeB, which encodes an acid stress chaperone (40), were upregulated in both strains. Several other stress-related genes, such as cueR (responsive to copper toxicity) (52), zraP (regulating zinc tolerance) (37), and fabB (mediating the nitrosative-stress response) (9), were induced in LT2. Genes damX (involved in bile resistance) (39), mig14 (related to antimicrobial peptide resistance) (56), and hns (central metabolism and stress tolerance regulator) (19) were repressed in PT4. In summary, many stress-related genes were upregulated in LT2 but not in PT4, although the two strains exhibited similar growth patterns under chlorine treatments.
Virulence-related genes.
Virulence genes are often clustered in various pathogenicity islands in Salmonella chromosomes. Salmonella pathogenicity island 1 (SPI1) enables the entry of Salmonella into nonphagocytic cells by triggering invasion and the penetration of gastrointestinal epithelium (25). We found that about half of the genes in SPI1, including orgA, prgJIK, iacP, sipABCD, spaO, and invJICB, were downregulated in LT2 under the chlorine treatments (Fig. 2B). Similarly, we found that the prgJIK operon of the type III secretion system was downregulated in LT2. Gene prgK is involved in the formation of the membrane-localized basal substructure of the complex (31). Gene prgI forms the needle portion of the complex (32). Chicken egg albumen contains various antimicrobial compounds, such as ovotransferrin and lysozyme, and is inhibitory for bacterial growth (3). Deletion of prgJ can lead to increased susceptibility of S. Enteritidis to egg albumen (15).
Other downregulated virulence factors included sipB, sipC, and sipD, which encode a protein translocase (35), sipA, which encodes an SPI1 effector protein (10), invB, which encodes the chaperone for sipA (35), invC, which encodes a general ATPase (1), invI, which is essential for the type III export pathway (20), and invJ, which encodes a nonflagellar type III secretion ATPase (48). All of these genes are essential for Salmonella invasion in the host. Collectively, we speculate that the oxidative stress may affect the biosynthesis and assembly of the type III secretion system, which lead to reduced virulence of LT2. In contrast, no significant change in the expression of the above SPI virulence factors was observed in PT4 under the same conditions. It is worth mentioning that the cydAB operon, which encodes the cytochrome D oxidase that generates the proton gradient across the cytoplasmic membrane (24), was downregulated 1.7-fold in LT2 but upregulated 1.5-fold in PT4. Deletion of cydA in S. Typhimurium and S. Gallinarum led to significant attenuation in virulence for chickens (62). The potential correlation of cydAB to the different host specificities of S. Typhimurium and S. Enteritidis remains to be determined.
qRT-PCR validation of DNA microarray gene expression results.
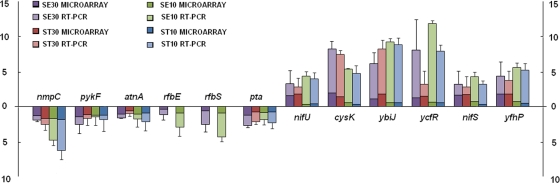
Analysis of six upregulated genes and six downregulated genes, including two PT4-specific genes, was used to evaluate the DNA microarray expression data in this study. As shown in Fig. 3, qRT-PCR data correlated well with the microarray expression data in identifying gene up- or downregulation. For expression levels for the majority of the genes, however, the fold changes detected by qRT-PCR were higher than those detected by DNA microarray. This is not unusual, since qRT-PCR is more sensitive than DNA microarray in detecting gene expression fold changes. No expression was detected for the two PT4-specific genes in LT2.
FIG. 3.
A bar graph that compares the gene expression fold changes identified by DNA microarrays and quantitative RT-PCR analysis. Six upregulated genes and six downregulated genes were compared. The fold changes were converted into log2 values for comparison. Standard deviation error bars for each mean are presented. SE, S. Enteritidis PT4; ST, S. Typhimurium LT2; 30, the weak chlorine treatment (130 ppm for 30 min); 10, the strong chlorine treatment (390 ppm for 10 min); microarray, the fold change identified by microarray; RT-PCR, the fold change identified by qRT-PCR.
Concluding remarks.
This is the first transcriptomic study that compares the two major food-borne S. enterica serovars, Typhimurium and Enteritidis, under chlorine-based oxidative stress. Our data showed that S. Enteritidis and S. Typhimurium respond to oxidative stress through coordinated regulation of a variety of genes associated with stress response, gene regulation, metabolism, and virulence. Similar to the case for E. coli O157:H7 (66), genes involved in Fe-S cluster formation and cysteine biosynthesis were most significantly upregulated in S. enterica under chlorine stress. In contrast, genes involved in LPS biosynthesis were downregulated in S. enterica. We found that genes in SPI1, type III secretion systems, and the pho regulon and genes involved in the heat shock response in LT2 and PT4 responded differently to the chlorine treatments. It remains to be further elucidated if the difference in transcriptional responses of these genes correlates to their respective prevalence in contaminating different types of food vehicles, through which these pathogens transmit to humans and consequently cause infectious disease. Studies have also been undertaken in our laboratories to compare the transcriptomic responses of bacterial cells present in various food matrices or attached to food and food contact surfaces under industrial settings, as such studies may provide further insights into the bacterial stress response pertinent to food contamination events.
Supplementary Material
Acknowledgments
This study was supported in part by USDA Agriculture and Food Research Initiative grant no. 2010-65201-20593 to W.Z. and an FDA cooperative agreement research fund to the National Center for Food Safety and Technology of the Illinois Institute of Technology.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akeda, Y., and J. E. Galan. 2005. Chaperone release and unfolding of substrates in type III secretion. Nature 437:911-915. [DOI] [PubMed] [Google Scholar]
- 2.Anjum, M. F., C. Marooney, M. Fookes, S. Baker, G. Dougan, A. Ivens, and M. J. Woodward. 2005. Identification of core and variable components of the Salmonella enterica subspecies I genome by microarray. Infect. Immun. 73:7894-7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, F., R. Storb, B. E. Storer, M. B. Maris, D. Niederwieser, J. A. Shizuru, T. R. Chauncey, B. Bruno, S. J. Forman, P. A. McSweeney, R. T. Maziarz, M. A. Pulsipher, E. D. Agura, J. Wade, M. Sorror, D. G. Maloney, and B. M. Sandmaier. 2006. Factors associated with outcomes in allogeneic hematopoietic cell transplantation with nonmyeloablative conditioning after failed myeloablative hematopoietic cell transplantation. J. Clin. Oncol. 24:4150-4157. [DOI] [PubMed] [Google Scholar]
- 4.Becker, G., E. Klauck, and R. Hengge-Aronis. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. U. S. A. 96:6439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57:289-300. [Google Scholar]
- 6.Beuchat, L. R. 1998. Surface decontamination of fruits and vegetables eaten raw: a review. WHO/FSF/FOS publication 98.2. World Health Organization, Geneva, Switzerland.
- 7.Bowden, S. D., G. Rowley, J. C. Hinton, and A. Thompson. 2009. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar Typhimurium. Infect. Immun. 77:3117-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd, J. M., J. A. Lewis, J. C. Escalante-Semerena, and D. M. Downs. 2008. Salmonella enterica requires ApbC function for growth on tricarballylate: evidence of functional redundancy between ApbC and IscU. J. Bacteriol. 190:4596-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandes, N., A. Rinck, L. I. Leichert, and U. Jakob. 2007. Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Mol. Microbiol. 66:901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bronstein, P. A., E. A. Miao, and S. I. Miller. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CDC. 2009. Multistate outbreak of Salmonella infections associated with peanut butter and peanut butter-containing products—United States, 2008-2009. MMWR Morb. Mortal. Wkly. Rep. 58:1-6. [PubMed] [Google Scholar]
- 12.CDC. 2009. Preliminary foodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2008. MMWR Morb. Mortal. Wkly. Rep. 58:333-337. [PubMed] [Google Scholar]
- 13.CDC. 2006. Salmonellosis—outbreak investigation, October 2006. CDC, Atlanta, GA.
- 14.Chagneau, C., M. Heyde, S. Alonso, R. Portalier, and P. Laloi. 2001. External-pH-dependent expression of the maltose regulon and ompF gene in Escherichia coli is affected by the level of glycerol kinase, encoded by glpK. J. Bacteriol. 183:5675-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clavijo, R. I., C. Loui, G. L. Andersen, L. W. Riley, and S. Lu. 2006. Identification of genes associated with survival of Salmonella enterica serovar Enteritidis in chicken egg albumen. Appl. Environ. Microbiol. 72:1055-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domka, J., J. Lee, and T. K. Wood. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dosanjh, N. S., M. Rawat, J. H. Chung, and Y. Av-Gay. 2005. Thiol specific oxidative stress response in mycobacteria. FEMS Microbiol. Lett. 249:87-94. [DOI] [PubMed] [Google Scholar]
- 18.Dougan, D. A., A. Mogk, and B. Bukau. 2002. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell. Mol. Life Sci. 59:1607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erol, I., K. C. Jeong, D. J. Baumler, B. Vykhodets, S. H. Choi, and C. W. Kaspar. 2006. H-NS controls metabolism and stress tolerance in Escherichia coli O157:H7 that influence mouse passage. BMC Microbiol. 6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, L. D., and C. Hughes. 2009. Selective binding of virulence type III export chaperones by FliJ escort orthologues InvI and YscO. FEMS Microbiol. Lett. 293:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 22.Gantois, I., R. Ducatelle, F. Pasmans, F. Haesebrouck, R. Gast, T. J. Humphrey, and F. Van Immerseel. 2009. Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol. Rev. 33:718-738. [DOI] [PubMed] [Google Scholar]
- 23.Gantois, I., R. Ducatelle, F. Pasmans, F. Haesebrouck, and F. Van Immerseel. 2009. The Salmonella Enteritidis lipopolysaccharide biosynthesis gene rfbH is required for survival in egg albumen. Zoonoses Public Health 56:145-149. [DOI] [PubMed] [Google Scholar]
- 24.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Maganasik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 25.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 26.Hartl, F. U., and M. Hayer-Hartl. 2009. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16:574-581. [DOI] [PubMed] [Google Scholar]
- 27.He, H., K. J. Genovese, D. J. Nisbet, and M. H. Kogut. 2006. Involvement of phosphatidylinositol-phospholipase C in immune response to Salmonella lipopolysacharide in chicken macrophage cells (HD11). Int. Immunopharmacol. 6:1780-1787. [DOI] [PubMed] [Google Scholar]
- 28.Houry, A., R. Briandet, S. Aymerich, and M. Gohar. 2010. Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 156:1009-1018. [DOI] [PubMed] [Google Scholar]
- 29.Humphrey, T. 2004. Salmonella, stress responses and food safety. Nat. Rev. Microbiol. 2:504-509. [DOI] [PubMed] [Google Scholar]
- 30.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 32.Kubori, T., A. Sukhan, S. I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U. S. A. 97:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kullik, I., M. B. Toledano, L. A. Tartaglia, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177:1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lara-Tejero, M., and J. E. Galan. 2009. Salmonella enterica serovar Typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 77:2635-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, J., S. R. Hiibel, K. F. Reardon, and T. K. Wood. 2010. Identification of stress-related proteins in Escherichia coli using the pollutant cis-dichloroethylene. J. Appl. Microbiol. 108:2088-2102. [DOI] [PubMed] [Google Scholar]
- 37.Lee, L. J., J. A. Barrett, and R. K. Poole. 2005. Genome-wide transcriptional response of chemostat-cultured Escherichia coli to zinc. J. Bacteriol. 187:1124-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemke, J. J., T. Durfee, and R. L. Gourse. 2009. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol. Microbiol. 74:1368-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.López-Garrido, J., N. Cheng, F. Garcia-Quintanilla, F. Garcia-del Portillo, and J. Casadesus. 2010. Identification of the Salmonella enterica damX gene product, an inner membrane protein involved in bile resistance. J. Bacteriol. 192:893-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malki, A., H. T. Le, S. Milles, R. Kern, T. Caldas, J. Abdallah, and G. Richarme. 2008. Solubilization of protein aggregates by the acid stress chaperones HdeA and HdeB. J. Biol. Chem. 283:13679-13687. [DOI] [PubMed] [Google Scholar]
- 41.Mangalappalli-Illathu, A. K., and D. R. Korber. 2006. Adaptive resistance and differential protein expression of Salmonella enterica serovar Enteritidis biofilms exposed to benzalkonium chloride. Antimicrob. Agents Chemother. 50:3588-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, R. G., W. K. Gillette, S. Rhee, and J. L. Rosner. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431-441. [DOI] [PubMed] [Google Scholar]
- 43.Maurer, L. M., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczewski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClelland, M., K. E. Sanderson, S. W. Clifton, P. Latreille, S. Porwollik, A. Sabo, R. Meyer, T. Bieri, P. Ozersky, M. McLellan, C. R. Harkins, C. Wang, C. Nguyen, A. Berghoff, G. Elliott, S. Kohlberg, C. Strong, F. Du, J. Carter, C. Kremizki, D. Layman, S. Leonard, H. Sun, L. Fulton, W. Nash, T. Miner, P. Minx, K. Delehaunty, C. Fronick, V. Magrini, M. Nhan, W. Warren, L. Florea, J. Spieth, and R. K. Wilson. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268-1274. [DOI] [PubMed] [Google Scholar]
- 45.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 46.Mizumoto, N., K. Sasai, H. Tani, and E. Baba. 2005. Specific adhesion and invasion of Salmonella Enteritidis in the vagina of laying hens. Vet. Microbiol. 111:99-105. [DOI] [PubMed] [Google Scholar]
- 47.Okochi, M., M. Kurimoto, K. Shimizu, and H. Honda. 2007. Increase of organic solvent tolerance by overexpression of manXYZ in Escherichia coli. Appl. Microbiol. Biotechnol. 73:1394-1399. [DOI] [PubMed] [Google Scholar]
- 48.Paul, K., M. Erhardt, T. Hirano, D. F. Blair, and K. T. Hughes. 2008. Energy source of flagellar type III secretion. Nature 451:489-492. [DOI] [PubMed] [Google Scholar]
- 49.Pecota, D. C., G. Osapay, M. E. Selsted, and T. K. Wood. 2003. Antimicrobial properties of the Escherichia coli R1 plasmid host killing peptide. J. Biotechnol. 100:1-12. [DOI] [PubMed] [Google Scholar]
- 50.Pedersen, K., and K. Gerdes. 1999. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 32:1090-1102. [DOI] [PubMed] [Google Scholar]
- 51.Phadtare, S., and M. Inouye. 2001. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pontel, L. B., and F. C. Soncini. 2009. Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol. Microbiol. 73:212-225. [DOI] [PubMed] [Google Scholar]
- 53.Rice, C. D., J. E. Pollard, Z. T. Lewis, and W. R. McCleary. 2009. Employment of a promoter-swapping technique shows that PhoU modulates the activity of the PstSCAB2 ABC transporter in Escherichia coli. Appl. Environ. Microbiol. 75:573-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rychlik, I., and P. A. Barrow. 2005. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 29:1021-1040. [DOI] [PubMed] [Google Scholar]
- 55.Saeed, A. I., N. K. Bhagabati, J. C. Braisted, W. Liang, V. Sharov, E. A. Howe, J. Li, M. Thiagarajan, J. A. White, and J. Quackenbush. 2006. TM4 microarray software suite. Methods Enzymol. 411:134-193. [DOI] [PubMed] [Google Scholar]
- 56.Spencer, H., M. H. Karavolos, D. M. Bulmer, P. Aldridge, S. R. Chhabra, K. Winzer, P. Williams, and C. M. Khan. 2010. Genome-wide transposon mutagenesis identifies a role for host neuroendocrine stress hormones in regulating the expression of virulence genes in Salmonella. J. Bacteriol. 192:714-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.St. Louis, M. E., D. L. Morse, and M. E. Potter. 1988. The emergence of grade A eggs as a major source of Salmonella enteritidis infections: new implications for the control of salmonellosis. JAMA 259:2103-2107. [PubMed] [Google Scholar]
- 58.Takahashi, Y., and U. Tokumoto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277:28380-28383. [DOI] [PubMed] [Google Scholar]
- 59.Teplitski, M., A. Al-Agely, and B. M. Ahmer. 2006. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology 152:3411-3424. [DOI] [PubMed] [Google Scholar]
- 60.Teplitski, M. A., and B. M. M. Ahmer. 2004. The control of secondary metabolism, motility, and virulence by the two-component regulatory system BarA/SirA of Salmonella and other γ-proteobacteria, p. 107-133. In B. M. Pruss and P. Matsumura (ed.), Global regulatory networks in enteric bacteria. Research Signpost, Kerala, India.
- 61.Thomson, N. R., D. J. Clayton, D. Windhorst, G. Vernikos, S. Davidson, C. Churcher, M. A. Quail, M. Stevens, M. A. Jones, M. Watson, A. Barron, A. Layton, D. Pickard, R. A. Kingsley, A. Bignell, L. Clark, B. Harris, D. Ormond, Z. Abdellah, K. Brooks, I. Cherevach, T. Chillingworth, J. Woodward, H. Norberczak, A. Lord, C. Arrowsmith, K. Jagels, S. Moule, K. Mungall, M. Sanders, S. Whitehead, J. A. Chabalgoity, D. Maskell, T. Humphrey, M. Roberts, P. A. Barrow, G. Dougan, and J. Parkhill. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turner, A. K., L. Z. Barber, P. Wigley, S. Muhammad, M. A. Jones, M. A. Lovell, S. Hulme, and P. A. Barrow. 2003. Contribution of proton-translocating proteins to the virulence of Salmonella enterica serovars Typhimurium, Gallinarum, and Dublin in chickens and mice. Infect. Immun. 71:3392-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueta, M., H. Yoshida, C. Wada, T. Baba, H. Mori, and A. Wada. 2005. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells 10:1103-1112. [DOI] [PubMed] [Google Scholar]
- 64.Vershinina, M. Y., A. N. Narovlyansky, P. G. Deryabin, A. M. Amchenkova, A. M. Ivanova, V. E. Scherbenko, E. V. Nagurskaya, V. A. Bechalo, T. Y. Timofeeva, A. V. Sanin, and F. I. Ershov. 2002. Regulation of cytokine mRNAs by interferon and interferon inducers. Russ. J. Immunol. 7:161-166. [PubMed] [Google Scholar]
- 65.Vershinina, O. A., and L. V. Znamenskaia. 2002. The Pho regulons of bacteria. Mikrobiologiia 71:581-595. (In Russian.) [PubMed] [Google Scholar]
- 66.Wang, S., K. Deng, S. Zaremba, X. Deng, C. Lin, Q. Wang, M. L. Tortorello, and W. Zhang. 2009. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 75:6110-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, X. S., R. Garcia-Contreras, and T. K. Wood. 2007. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J. Bacteriol. 189:3051-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.