Abstract
A novel biotechnological process has been developed for the isolation of desiccation-tolerant microorganisms and their xeroprotectants, i.e., compatible solutes involved in long-term stability of biomolecules in the dry state. Following exposure of soil samples to chloroform, we isolated a collection of desiccation-tolerant microorganisms. This collection was screened for the production of xeroprotectants by a variation of the bacterial milking (osmotic downshock) procedure and by a novel air-drying/rehydration (“dry milking”) incubation method. The resultant solutes were shown to protect both proteins and living cells against desiccation damage, thereby validating them as xeroprotectants. Nuclear magnetic resonance (NMR) analytical studies were performed to identify the xeroprotectants; synthetic mixtures of these compounds were shown to perform similarly to natural isolates in drying experiments with proteins and cells. This new approach has biotechnological and environmental implications for the identification of new xeroprotectants of commercial and therapeutic value.
Some microorganisms accumulate small organic compounds in response to changing extracellular osmolarity, as with desiccation or addition of salts (2, 8). These compatible solutes, e.g., the disaccharide trehalose or the hydroxypyrimidine hydroxyectoine, act as osmoprotectants but under laboratory conditions can also stabilize enzymes, DNA, membranes, and even whole cells against different kinds of stress, such as freezing, drying, and heating (8, 18, 21, 37). A further benefit of nonreducing sugars such as trehalose is that they do not undergo damaging Maillard reactions with amino acids or proteins in the dry state; they therefore offer excellent protection to biomolecules and even living cells during desiccation (9, 12, 15). In consequence, there is great biotechnological interest in these solutes, since many different biomolecules and cells require stabilization to allow long-term storage for commercial use (4, 24, 35). Those compatible solutes that ensure long-term storage stability in the dry state (10, 11, 13) we term “xeroprotectants.” Different biomolecules or cells are protected to different degrees depending on the xeroprotectants that are used (21). Therefore, new xeroprotectants together with new formulations are likely to be needed to preserve a wide range of desiccation-sensitive biomolecules and cell types, such as some vaccines, mammalian cells, or even whole tissues (30).
In order to obtain large amounts of some compatible solutes (ectoine and hydroxyectoine), Sauer and Galinski (25) developed a technology termed bacterial milking. This technology is based on incubation of appropriate cells (i.e., Halomonas elongata) under hyperosmotic conditions followed by a transfer of the cells to low-salinity medium (osmotic downshock), which results in the release of the compatible solutes into the medium. Here we have used a variation of the bacterial milking method and have also developed a new technique for extracting xeroprotectants by slowly air drying newly isolated desiccation-tolerant microorganisms, followed by rapid rehydration; we term this “dry milking.”
We also have previously shown how drying living cells in the presence of glass-forming protective molecules, such as trehalose or hydroxyectoine, results in extremely high solvent tolerance (23, 31). Thus, bacterial cells of Escherichia coli or Pseudomonas putida dried in the presence of xeroprotectants can be exposed to organic solvents such as pure chloroform or acetone without a detrimental effect on their viability. We hypothesize that naturally desiccation-tolerant microorganisms show similar solvent tolerances, and we therefore used a combination of organic solvent treatment of soil samples followed by milking of the bacterial isolates to identify new desiccation-tolerant microorganisms and their xeroprotectants. We obtained a collection of xerotolerant strains and demonstrated potential applications for the xeroprotectants extracted from those strains as stabilizers of lipase enzyme and living cells of E. coli MC4100 in the dry state. The chemical composition of the extracts was examined by nuclear magnetic resonance (NMR), and synthetic mixtures based on such analysis were tested for their xeroprotective potential.
MATERIALS AND METHODS
Soil samples.
Soil samples were taken from Nerium oleander rhizosphere subjected to seasonal drought at Granada (Spain) (37.182 N, 3.624 W) after a period of 3 months with no registered rainfall or any exposure to water. The soil samples were collected in plastic bags, air dried at room temperature, homogenized, and sieved (2-mm mesh).
Microorganisms, media, and culture conditions.
The type strain of H. elongata (DSM 2581T) was obtained from Deutsche Sammlung Von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany. The strains used in this study are shown in Table 1. The organisms described in this study will be made available upon request. Bacteria were grown in tryptic soy broth (TSB) or M9 minimal medium (Sigma M6030) with glucose or fructose (50 mM) as the sole carbon source at 30°C for the different isolates and at 37°C for E. coli as previously described (15). To generate hypersaline minimal medium (HMM), NaCl was added to M9 medium at the specified concentration as previously described (21).
TABLE 1.
Strains used in this work and sequence analysis of the isolated strains based on partial 16S rRNA gene sequencing
| Representative clone (GenBank accession no.) | GenBank accession no. of nearest species (similarity [%]) | Subgroup | Reference |
|---|---|---|---|
| KT2440 | P. putida | 14 | |
| MC4100 | E. coli | 28 | |
| AC68 | A. calcoaceticus | Unpublished | |
| DSM 2581T | H. elongata | 32 | |
| 3J1 (GU815136) | EU714371.1 (99) | Microbacterium | This study |
| 4J2A2 (GU815137) | U27579.1 (100) | Rhodococcus | This study |
| 4J7B1 (GU815138) | DQ406732.1 (99) | Leucobacter | This study |
| 4J27 (GU815139) | GU391465.1 (99) | Arthrobacter | This study |
| 5J12 (GU815140) | DQ486130.1 (99) | Arthrobacter | This study |
Isolation of bacteria by a standard isolation method.
One gram of air-dried soil was added to 10 ml TSB and thoroughly mixed. After the soil particles settled, serial dilutions in saline buffer were made and 100-μl aliquots from each dilution were plated on tryptic soy agar (TSA) plates. After 48 h of incubation at 30°C, individual colonies were randomly picked and streaked out to obtain pure cultures. Soil samples were mixed with 3 ml chloroform and incubated for different times (30, 60, and 120 min) with occasional vortexing. After incubation, the mixtures were transferred to sterile glass plates and incubated under sterile conditions for 30 min to allow complete evaporation of the chloroform. The chloroform-treated dry soil was mixed with TSB and diluted as described above. Tests were performed in triplicate.
Sequencing of 16S rRNA genes and phylogenetic analysis.
All strains isolated in this study were identified by analysis of the partial sequence of the gene encoding 16S rRNA. The primers fD1, fD2, rD1, and rD2 (36) were synthesized by Sigma Genosys (United Kingdom) and used to amplify and sequence almost the full length of the 16S rRNA gene.
Air drying: determination of survival rates.
A colony of each pure culture containing 107 to 109 cells was resuspended in 1 ml of M9 minimal medium. Aliquot fractions (100 μl) were placed on sterile petri dishes and dried under a current of sterile air for 24 h. Cells were resuspended in 1 ml sterile saline buffer, and serial dilutions of the cells before and after drying were plated on TSA plates. All manipulations were performed at room temperature. The survival rate was calculated as cells/ml after drying with reference to cells/ml before drying, expressed as a percentage. The assays were performed in triplicate.
Sporulation test.
To determine if the isolated strains were sporulant, a 4-day-old colony was resuspended in 1 ml of sterile saline M9 buffer and incubated at 72°C for 30 min as described previously (31). Aliquot volumes (100 μl) were plated before and after the incubation. Strains able to grow under both conditions were discarded as sporulant or thermotolerant. Therefore, only temperature-sensitive strains were selected.
Modified bacterial milking.
Halotolerance tests were first made in M9 medium with glucose or fructose (50 mM) as a carbon source, supplemented with NaCl at concentrations of 8.5 mM and 0.2, 0.4, 0.8, and 1.6 M (Table 2). Depending on the above results, fresh cultures were started at the optimal salt concentration (hypersaline minimal medium) where the maximal optical density at 600 nm (OD600) was observed and incubated for 48 h. An additional increase in NaCl in the cultures was made up to that registered as the maximal tolerated NaCl concentration where some growth had been detected (supersaline minimal medium). At such salinities, an additional incubation of 48 h was made to allow cells to accumulate their respective xeroprotectants. Then, cells were centrifuged for 15 min at 15,000 × g and cell pellets were resuspended in 30 ml sterile deionized water. After 20 min of shaking at 30°C, the suspensions were centrifuged again and supernatants were filtered through a 0.22-μm filter, lyophilized, and resuspended in deionized water to obtain a 10% (wt/vol) concentration. Alternatively, exudates were obtained using an air-drying protocol (“dry milking”). Cells from a saturated culture in minimal medium were collected and placed on a 47-mm-diameter filter membrane with a pore size of 0.22 μm, purchased from Millipore (EZGSWG474), placed over a minimal medium plate without a lid. The plate was incubated under a flow of sterile air for 8 h, and the filter and cells were removed from the plate and subjected to an additional incubation under the sterile air flow for a further 16 h. Finally, the cell-containing filter was incubated with shaking for 20 min in 30 ml sterile deionized water. Cells were separated from the supernatant by centrifugation, and supernatants were filtered through a 0.22-μm filter, lyophilized, and resuspended in deionized water to obtain a 10% (wt/vol) concentration.
TABLE 2.
Optimum and maximal NaCl concentrations tolerated by the different isolates
| Strain | NaCl concn (M) |
|
|---|---|---|
| Optimum | Maximal | |
| Microbacterium sp. 3J1 | 0.2 | 1.6 |
| Rhodococcus sp. 4J2A2 | 0.2 | 0.4 |
| Leucobacter sp. 4J7B1 | 0.2 | 0.2 |
| Arthrobacter sp. 4J27 | 0.8 | 1.6 |
| Arthrobacter sp. 5J12A | 0.2 | 0.8 |
Xeroprotection assay.
Lipase from Burkholderia cepacia was purchased from Sigma-Aldrich (62309) and was used without further purification. High-grade trehalose was obtained from Alchemy International Ltd., Hambrook, United Kingdom. Lipase (5.5 U) was suspended in 15 μl of the relevant exudates. As a positive control, 15 μl of 10% (wt/vol) trehalose was used, and as negative controls, 15 μl of deionized water was used. The 15-μl lipase solution was dried at 50°C for 120 min to remove most of the water. Additional incubation for 5 min at 100°C was included to remove the remaining water. Dry samples were maintained in the dry state for 24 h at 37°C in a desiccator. Then, 1 ml of Tris-HCl (50 mM, pH 8) was added to resuspend the enzyme and was used for the lipase assay as described by Gupta et al. (17). After 22 min, resuspended lipase was mixed with 1 ml substrate solution (0.3 mg p-nitrophenyl palmitate, 0.1 ml isopropanol, 50 μl Tris-HCl [50 mM, pH 8], 4 μl Triton-X, and 1 mg gum arabic, in water). The mixture was incubated at 30°C for 30 min with shaking. Reactions were stopped by a 4-min incubation at 100°C, and the colorimetric change to yellow was measured at 410 nm. One unit of lipase-specific activity was defined as the amount of that enzyme that hydrolyzed 1 μmol of p-nitrophenyl palmitate per minute at 30°C. Each assay was performed in triplicate.
NMR spectroscopy.
Before NMR spectroscopy, bacterial milking products from standard and dry milking samples, approximately 10 mg, were diluted with 600 μl of deuterium oxide (D2O). Spectra were acquired at 400 MHz on a Bruker Avance 400 spectrometer (Bruker, Madrid, Spain). One-dimensional spectra consisted of the accumulation of 64 scans with a pulse-and-acquire sequence preceded by WATERGATE 3-9-19 (33) to remove the residual water signal. To further help metabolite identification, two-dimensional 1H-1H correlation spectroscopy and 1H-13C heteronuclear single-quantum coherence spectra were also acquired from the same samples.
Nucleotide sequence accession numbers.
Sequences for 16S rRNA genes of strains used in this work have been submitted to GenBank under the accession numbers listed in Table 1.
RESULTS
Isolation of a collection of desiccation-tolerant strains.
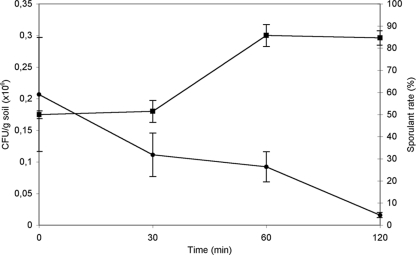
Bacterial cells dried in xeroprotectants that are insoluble in organic solvents are not damaged by exposure to chloroform or acetone (23, 31). Therefore, we developed a procedure to isolate novel, highly desiccation-tolerant strains from soil using chloroform as a selective agent, assuming that the microorganisms are protected by endogenous xeroprotectants. Accordingly, dry soil samples were treated with chloroform for 0 (nontreated), 30, 60, or 120 min as described in Materials and Methods. After complete evaporation of the chloroform and subsequent rehydration and resuspension of the soil sample, serial dilutions were plated out. Multiple strains from each treatment time were selected, and pure cultures were obtained, resulting in a total of 223 isolates. A general reduction in total CFU/g of soil with time of treatment was found from more than 2 × 105 CFU/g soil in negative controls (time zero) to less than 2 × 103 CFU/g soil after 120 min of exposure to chloroform (Fig. 1). To differentiate those microorganisms that tolerate desiccation and organic solvents from those that withstand these stressors due to spore formation, we performed a test to identify sporulant strains based on the tolerance of spores for high temperatures (27, 31). We observed an increase in the proportion of strains able to withstand 72°C with the time of solvent treatment from 50% for nontreated samples to more than 80% after 60 min (Fig. 1). Due to the generally low numbers of thermotolerant strains present in soil, we considered the survivors to be sporulant strains. This is consistent with spore resistance to both temperature and organic solvents. Therefore, we observed a decrease in the overall survival rate of the population of microorganisms as a whole, and an increase in the proportion of sporulant microorganisms within the population, with treatment time. Thus, 77 isolates (from the original 223) were selected that tolerated treatment with organic solvent but were temperature sensitive, and we considered these to be chloroform-resistant, nonsporulating strains.
FIG. 1.
Changes in CFU and sporulant rate per gram of soil over chloroform treatment time. Squares show values of CFU in millions, and circles show percentages of sporulant bacteria from corresponding CFU. Error bars show the standard deviations for at least three replicates.
Desiccation-tolerant strains.
In order to study the degree of desiccation tolerance of the isolates, the 77 selected isolates were air dried. A strain of Acinetobacter calcoaceticus (AC68) from a previous study (M. Manzanera and A. Tunnacliffe, unpublished results) was used as a positive control. Cells of P. putida KT2440 were used as desiccation-sensitive controls (22). Five strains (3J1, 4J2A2, 4J7B1, 4J27, and 5J12A) were shown to have significantly higher (P > 0.05) levels of desiccation tolerance than A. calcoaceticus and were chosen for further study. Table 3 shows the survival rates of those strains. They were identified by nucleotide sequencing of the near-complete 16S rRNA gene (1,417 bp), which showed that all of them belonged to the Actinobacteria class (Table 1).
TABLE 3.
Viability of different bacterial isolates after 24 h of air drying
| Strain (GenBank accession no.) | Survival rate (%)a |
|---|---|
| AC68 | 5.91 ± 1.90 |
| KT2440 | 0 |
| 3J1 (GU815136) | 48.73 ± 1.73 |
| 4J2A2 (GU815137) | 21.99 ± 5.52 |
| 4J7B1 (GU815139) | 24.81 ± 3.53 |
| 4J27 (GU815139) | 23.26 ± 2.60 |
| 5J12 (GU815140) | 34.39 ± 4.68 |
The data are the means and standard deviations of three independent determinations.
Halotolerance and xeroprotection tests.
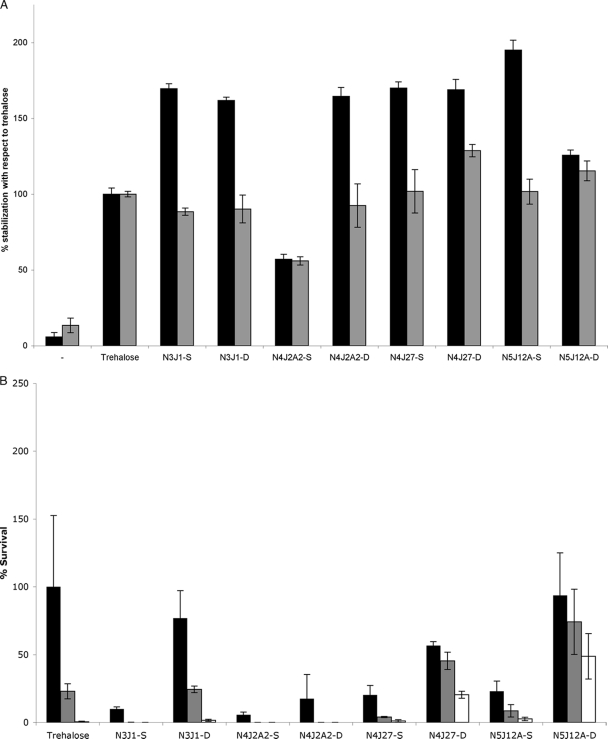
To stimulate the intracellular production of compatible solutes in the newly isolated xerotolerant strains, we first simulated desiccation signals by increasing the osmotic pressure of growth media by addition of NaCl (Table 2). Tests of halotolerance were made to assess the optimal and maximal salt concentrations tolerated by the different strains. To extract any compatible solutes produced, a modification of the bacterial milking method (25) was developed. Cells were grown on minimal medium in the presence of their optimal NaCl concentration, and then the NaCl concentration was increased to the maximum tolerated. Then, cells were subjected to osmotic downshock to provoke the release of their compatible solutes into the incubation media, from which these solutes were purified as described in Materials and Methods. Using this method, we extracted the exudates N3J1-S, N4J2A2-S, N4J27-S, and N5J12A-S from the Microbacterium sp. 3J1, Rhodococcus sp. 4J2A2, Arthrobacter sp. 4J27, and Arthrobacter sp. 5J12A strains, respectively. The Leucobacter sp. 4J7B1 strain was not included in this test because it did not grow in M9 medium under the conditions used. To determine whether these exudates conferred xeroprotection, 5.5 U of lipase was dried in the presence or absence of 10% (wt/vol) of the corresponding exudates as described in Materials and Methods. Lipase dried in 10% trehalose (wt/vol) was included as a positive control (3, 10), while lipase dried from solution without added excipients was included as an internal negative control. We observed xeroprotection of lipase by the exudates from all xerotolerant strains, with especially high values for the exudate N5J12A-S, produced by Arthrobacter sp. 5J12A (Fig. 2 A). Similarly, we decided to compare the xeroprotective effects of these compounds at 10% (wt/vol) with that of 10% (wt/vol) trehalose on microbial cells (21, 22). We found that E. coli MC4100 cells were protected against desiccation when extracts were used but generally with a markedly lower survival rate than that with trehalose (Fig. 2B).
FIG. 2.
(A) Lipase activity after dehydration of the enzyme in the presence of 10% (wt/vol) of the different exudates at day 1 (black bars) or day 150 (gray bars). Names of different exudates comprise the name of the isolate that produced them, preceded by the letter N for “naturally synthesized by microorganisms” and followed by -S if extracted by standard bacterial milking procedure and by -D if extracted by dry milking procedure. Bars indicate percentages with reference to the lipase activity given by trehalose-stabilized lipase. (B) Viability of E. coli MC4100 after vacuum drying in 10% (wt/vol) of the different exudates at day 1 (black bars), day 15 (gray bars), or day 30 (white bars) after drying.
An alternative, “dry milking” method was developed for the recovery of compatible solutes from dried cells. The different isolates were grown on minimal medium and then collected and transferred to a filter placed on a minimal medium plate to allow slow drying of the cells, during which time they are expected to accumulate xeroprotectants. Cells were then incubated in hypoosmotic media, and exudates were obtained from cells as described in Materials and Methods. This method allowed us to collect the exudates N3J1-D, N4J2A2-D, N4J27-D, and N5J12A-D from Microbacterium sp. 3J1, Rhodococcus sp. 4J2A2, Arthrobacter sp. 4J27, and Arthrobacter sp. 5J12A strains, respectively. These dry milking exudates were also used for lipase and bacterial stabilization, and most showed higher levels of stabilization of dried enzyme than the corresponding exudates obtained by osmotic downshock. The cell stabilization levels were also higher for dry milking exudates from strains Arthrobacter sp. 4J27 (N4J27-D) and Arthrobacter sp. 5J12A (N5J12A-D), especially in the long term (Fig. 2A and B). These experiments demonstrate that all extracts have xeroprotectant activity, although these vary in degree, particularly regarding cell stabilization.
Compositional analysis of xeroprotectants.
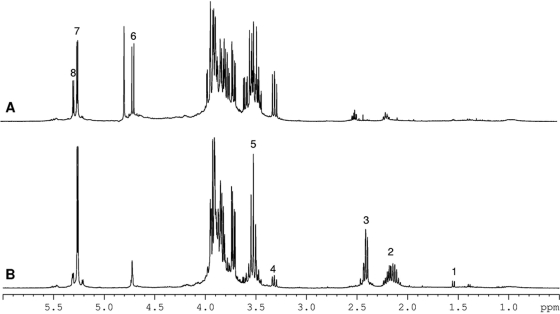
To identify the composition of the xeroprotectants assessed in Fig. 2, NMR spectroscopy was used. For these experiments, lyophilized samples of xeroprotectants were dissolved in phosphate-buffered saline prepared with D2O (600 μl). Interestingly, the spectra obtained showed that there was no single common molecule present in exudates of all four species; indeed, extracts produced from the same bacterial strain usually differed according to whether osmotic downshock or dry milking was used (Fig. 3). The chemical composition of the different exudates is shown in Table 4. For example, in the case of the osmotic downshock extract from Arthrobacter sp. 4J27, glucose, glutamine, glutamic acid, oxoglucuronic acid, and β-hydroxybutyrate were identified in a molar ratio of 6.8:4:2:1.3:1, respectively, while the dry milking extract from the same species comprised trehalose, glucose, and glutamine in a ratio of 5.8:2.8:1, respectively. In the case of Rhodococcus sp. 4J2A2, the identified composition of the exudates included fructose, glutamic acid, and acetate in a ratio of 41:2.4:1 when obtained by the osmotic downshock method. However, dry milking produced fructose, glutamic acid, lactate, acetate, and β-hydroxybutyrate in a ratio of 16:4:1.4:1:0.8, respectively. Arthrobacter sp. 5J12A was the only example of the four isolates where identical components of the xeroprotectant mixtures were obtained by both the osmotic downshock method and the dry milking method, albeit in different ratios. The compounds identified were glucose, acetate, lactate, and valine in a ratio of 1:0.1:0.25:0.37 with the former method and in a ratio of 1:0.3:0.12:0.04 with the latter.
FIG. 3.
1H NMR spectra of xeroprotectants obtained from 4J27D (A) or 4J27 (B), showing that the extraction/milking protocol influences the xeroprotectant mixture composition. Peak assignments are as follows: 1, alanine; 2, glutamate; 3, glutamine; 4, 6, and 8, glucose; 5 and 7, trehalose. The area between 3.7 and 4 ppm corresponds mainly to protons from glucose and trehalose.
TABLE 4.
Relative chemical compositions of artificial synthetic xeroprotectants
| Component | Relative amt (% mol)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| S4J27-S | S4J27-D | S4J2A2-S | S4J2A2-D | S5J12A-S | S5J12A-D | S3J1-S | S3J1-D | |
| Fructose | 41 | 16 | ||||||
| Glutamic acid | 2 | 2.4 | 4 | 0.14 | 0.1 | |||
| Acetate | 1 | 1 | 0.1 | 0.03 | ||||
| β-Hydroxybutyrate | 1 | 0.8 | ||||||
| Lactate | 1.4 | 0.25 | 0.12 | 1.18 | 0.62 | |||
| Glucose | 6.8 | 2.8 | 1 | 1 | ||||
| Valine | 0.37 | 0.04 | ||||||
| Trehalose | 5.8 | 1 | 1 | |||||
| Oxoglucuronic | 1.3 | 0.31 | 0.06 | |||||
| Glutamine | 4 | 1 | 0.28 | 0.1 | ||||
| Fucose | 2.26 | |||||||
| Pyruvate | 0.23 | 0.21 | ||||||
Relative to the reference component, which was assigned a value of 1.
Stabilization of lipase and bacterial cells using synthetic xeroprotectant mixtures.
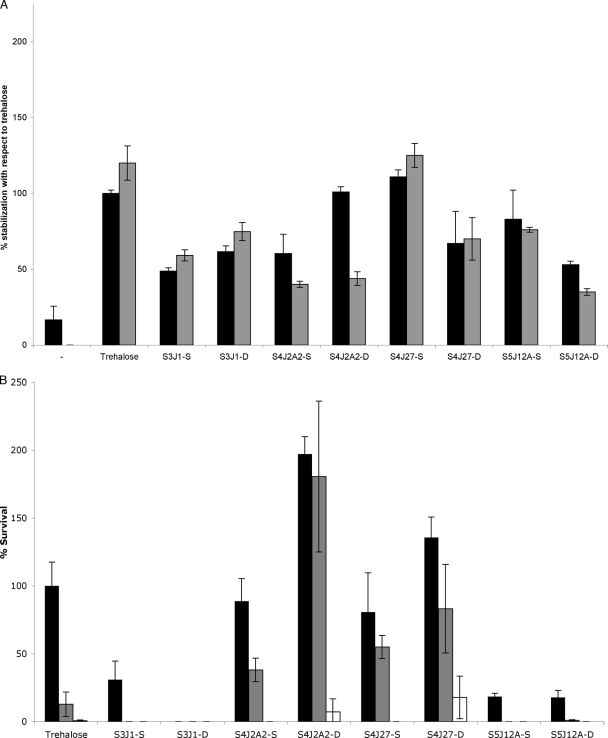
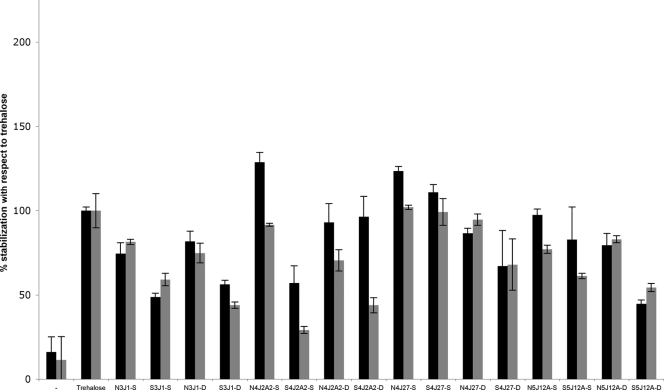
To show whether the compounds identified in the various exudates could act as xeroprotectants, we decided to test the ability of synthetic chemical mixtures corresponding to the naturally derived formulations to stabilize lipase or living cells (E. coli MC4100). These synthetic mixtures were made up in the same molar ratios as their natural counterparts and were termed S3J1-S, S4J2A2-S, S4J27-S, S5J12A-S, S3J1-D, S4J2A2-D, S4J27-D, and S5J12A-D. The results show that the synthetic mixtures used at 10% (wt/vol) can protect lipase against desiccation damage, in some cases as well as 10% (wt/vol) trehalose (Fig. 4 A). With regard to bacterial cells (Fig. 4B), although the synthetic mixtures S3J1-S, S3J1-D, S5J12A-S, and S5J12A-D showed little protection in the short term and close to no effect after 30 days of desiccation, the S4J2A2-S, S4J2A2-D, S4J27-S, and S4J27-D synthetic mixtures gave excellent results. The xeroprotective effect was especially high for S4J2A2-D and S4J27-D, with values significantly higher than those for the trehalose positive control at all times tested. In general, however, natural exudates performed slightly better than their synthetic counterparts when tested alongside each other in the same experiment (Fig. 5). This suggests that there are other components of the natural mixtures that have not yet been identified and that make some contribution to the stabilizing effect of the exudate.
FIG. 4.
Xeroprotective effect of 10% (wt/vol) synthetic xeroprotectants on lipase protein at day 1 (black bars) or day 30 (gray bars) (A) or of 34% (wt/vol) mixtures on E. coli MC4100 at day 1 (black bars), day 15 (gray bars), or day 30 (white bars) after drying (B). Names of different mixtures comprise the name of the isolate that was used for their identification, preceded by the letter S since they were synthetically produced and followed by -S or -D depending on the used source of the sample (standard bacterial milking or dry milking procedure, respectively) for their chemical composition.
FIG. 5.
Xeroprotective effect of 10% (wt/vol) natural and synthetic xeroprotectants on lipase at day 1 (black bars) or day 30 (gray bars). Names of different mixtures comprise the names of the bacterial isolates that produced them, preceded by the letter N when naturally synthesized or the letter S when synthetically produced and followed by -S if extracted by the standard bacterial milking procedure or by -D if extracted by the dry milking procedure.
To determine whether all components of a specific mixture were required for stabilization, we tested the individual components of the synthetic mixtures and combinations of two components, including the reducing sugar (glucose or fructose) and one of the acids from samples S4J2A2-S, S4J2A2-D, S4J27-S, and S4J27-D. We found that although a degree of stabilization was observed, particularly for dried lipase, the effect was dramatically reduced for these mixtures (Table 5). This suggests that the individual components of the exudates themselves have limited xeroprotectant activity and therefore that the composition and proportion of the chemicals in these mixtures are of great importance.
TABLE 5.
Xeroprotective effect of 10% (wt/vol) individual components of the synthetic mixtures and combinations of two components, including the reducing sugar and one additional component from samples S4J2A2-S, S4J2A2-D, S4J27-S, and S4J27-D
| Component | Xeroprotective effect relative to that of trehalose (%) |
|---|---|
| MilliQ water | 23.11 ± 13.07 |
| Trehalose | 100 ± 13.97 |
| S4J2A2-S | 85.40 ± 18.04 |
| S4J2A2-D | 106.13 ± 18.34 |
| S4J27-S | 126.22 ± 16.46 |
| Glucose | 14.08 ± 6.13 |
| Fructose | 23.32 ± 4.04 |
| Glutamic acid | 46.87 ± 44.78 |
| Acetate | 36.19 ± 20.58 |
| Glucuronic acid | 25.69 ± 17.20 |
| Glutamine | 41.06 ± 11.95 |
| Lactate | 39.56 ± 2.24 |
| β-Hydroxybutyrate | 34.39 ± 4.77 |
| Glucose + glucuronic acid | 0 |
| Glucose + glutamine | 0 |
| Glucose + β-hydroxybutyrate | 3.08 ± 2.19 |
| Glucose + glutamic acid | 6.73 ± 3.53 |
| Fructose + glutamic acid | 0 |
| Fructose + β-hydroxybutyrate | 0 |
DISCUSSION
In the present work, we describe a novel and simple methodology for the isolation and identification of desiccation-tolerant microorganisms and xeroprotectants and demonstrate potential biotechnological applications of the xeroprotectants characterized. Taxonomical studies of the desiccation-tolerant isolates showed that all of them belonged to the order Actinomycetales. Two of them (4J27 and 5J12) were assigned to the genus Arthrobacter, and despite similar tolerances for desiccation, these isolates seem to differ not only in their osmotolerance but also in the variable region of their 16S rRNA genes, suggesting that they represent different species. The Arthrobacter genus has been found previously in dry environments, and some species produce, accumulate, and release large amounts of carbohydrates, including trehalose (6, 7, 38). Our results showed that one of the desiccation-tolerant strains (Arthrobacter sp. 4J27) does accumulate trehalose, in addition to glutamine and glucose, when a desiccation protocol (dry milking) is used to obtain the exudates but not when an osmotic downshock protocol is used. This might explain why the dry milking exudate from 4J27 gives better protection of cells than that obtained by osmotic downshock. However, the other Arthrobacter isolate (5J12A) produced a rather different mixture, consisting of glucose, acetate, lactate, and valine, suggesting that trehalose is not essential in this genus for protection against desiccation. Therefore, it is likely that several alternative strategies based on accumulation of xeroprotectants have been developed by Arthrobacter spp. to withstand long periods of heat, frost, salinity, and desiccation (39).
The other isolates characterized were members of the Rhodococcus (4J2A2), Microbacterium (3J1), and Leucobacter (4J7B1) genera. Previous studies have found species of the genus Rhodococcus among desiccation-tolerant microorganisms (26). Since we have found fructose, glutamic acid, β-hydroxybutyrate, acetate, and lactate in dry milking exudates from Rhodococcus sp. 4J2A2, these solutes might represent (part of) its natural protection system. The absence of trehalose was surprising, since other desiccation-tolerant Rhodococcus species are reported to accumulate trehalose, ectoine, and hydroxyectoine in response to desiccation (1, 19). It is possible that the milking procedures we used are selective and that these compounds were simply not released from Rhodococcus sp. 4J2A2; this could be tested by examining whole extracts of dried cells. In addition, other species of Rhodococcus could be tested for production of fructose, glutamic acid, acetate, β-hydroxybutyrate, and lactate under conditions of low water activity to determine whether common mechanisms against desiccation are employed within the genus. Since strains Leucobacter sp. 4J7B1 and Microbacterium sp. 3J1 showed remarkably high desiccation tolerance, it is very surprising that, to our knowledge, there are no published studies of desiccation tolerances of these genera. Trehalose accumulation by Microbacterium ammoniaphilum has been described, but no reference was made to desiccation tolerance (33).
The stabilization of a wide range of biomolecules by some compatible solutes has practical implications for a number of areas, ranging from basic science through health care (e.g., stabilization of labile vaccines), agriculture (e.g., phytohormones), and bioelectronics (e.g., nanocircuits) to environmental sciences (e.g., bioremediation of arid and semiarid regions) (5, 11, 29, 34). The identification of new xeroprotectants, and in particular the effectiveness of complex mixtures of solutes, would therefore be of potential biotechnological significance. The different chemical compositions of the exudates could reflect differences both in the physiology of the microorganisms involved and in their individual responses to different and independent environmental conditions, i.e., desiccation or salt stress in the experiments described here. The latter effect is particularly clear for Rhodococcus sp. 4J2A2, where xeroprotectant mixtures of different effectivenesses were produced depending on the methodology (osmotic downshock or dry milking) used to obtain the exudates; between two and three times more effective stabilization of lipase was observed when dry milking was used instead of the osmotic downshock method. These differences in lipase stabilization might reflect the effect of β-hydroxybutyrate and lactate in the dry milking exudates. A surprising feature of the exudates is the presence of reducing sugars such as glucose or fructose, which normally give rise to high levels of detrimental Maillard (browning) activity with amino groups of proteins in the dry state (20). One possibility is that some of the other substances in the mixture would reduce Maillard activity and therefore allow successful stabilization, but this remains to be tested.
The composition and proportion of the chemicals in these mixtures seem to be of great importance for their xeroprotective effect. Incomplete mixtures do not show the xeroprotective levels of the complete synthetic mixtures. Thus, after acetate was removed from the S4J2A2-S synthetic mixture, stabilization of lipase was not found. In most cases, the synthetic mixtures did not attain the xeroprotective levels of the bacterially produced mixtures, except for N4J27-S in comparison to S4J27-S, where no significant differences were found. The reason for these differences may lie in the fact that not all chemical components have been fully identified and/or that proteins or nucleic acids present in the natural exudates play an important role. A xeroprotective function for macromolecules cannot be ruled out, since such an effect has been described for the hydrophilic LEA (late embryogenesis abundant) proteins either alone or in combination with trehalose (16), for example.
In summary, the approach we describe here is an efficient means of isolating desiccation-tolerant microorganisms and the xeroprotectants these organisms produce. This new methodology has environmental and biotechnological implications for the identification of new xeroprotectants of potential commercial and therapeutic interest.
Acknowledgments
We thank the Junta de Andalucia (Spain) for funding this study through project reference P07-RNM-02588 and the Spanish Ministry of Health (Red Enfermedades Cardiovasculares RECAVA). M. Manzanera and I. Barba received grants from the Programa Ramón y Cajal, and J. J. Narvaez-Reinaldo received funding from the FPU program (Ministerio de Educación y Ciencia MEC, Spain, and ERDF, European Union).
J.J.N.-R. and M.M. thank the members of the Tunnacliffe laboratory for generous technical advice.
Footnotes
Published ahead of print on 18 June 2010.
REFERENCES
- 1.Alvarez, H. M., R. A. Silva, A. C. Cesari, A. L. Zamit, S. R. Peressutti, R. Reichelt, U. Keller, U. Malkus, C. Rasch, T. Maskow, F. Mayer, and A. Steinbüchel. 2004. Physiological and morphological responses of the soil bacterium Rhodococcus opacus strain PD630 to water stress. FEMS Microbiol. Ecol. 50:75-86. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, T., and S. N. Timasheff. 1982. Stabilization of protein structure by sugars. Biochemistry 21:6536-6544. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa, T., and S. N. Timasheff. 1985. The stabilization of proteins by osmolytes. Biophys. J. 47:411-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arakawa, T., K. Tsumoto, Y. Kita, B. Chang, and D. Ejima. 2007. Biotechnology applications of amino acids in protein purification and formulations. Amino Acids 33:587-605. [DOI] [PubMed] [Google Scholar]
- 5.Bashan, Y. 1986. Alginate beads as synthetic inoculant carriers for slow release of bacteria that affect plant growth. Appl. Environ. Microbiol. 51:1089-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boylen, C. W. 1973. Survival of Arthrobacter crystallopoietes during prolonged periods of extreme desiccation. J. Bacteriol. 113:33-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylen, C. W., and J. C. Ensign. 1970. Intracellular substrates for endogenous metabolism during long-term starvation of rod and spherical cells of Arthrobacter crystallopoietes. J. Bacteriol. 103:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, A. D. 1976. Microbial water stress. Bacteriol. Rev. 40:803-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter, J. F., L. M. Crowe, and J. H. Crowe. 1987. Stabilization of phosphofructokinase with sugars during freeze-drying: characterization of enhanced protection in the presence of divalent cations. Biochim. Biophys. Acta 923:109-115. [DOI] [PubMed] [Google Scholar]
- 10.Carpenter, J. F., B. Martin, L. M. Crowe, and J. H. Crowe. 1987. Stabilization of phosphofructokinase during air-drying with sugars and sugar/transition metal mixtures. Cryobiology 24:455-464. [DOI] [PubMed] [Google Scholar]
- 11.Colaco, C., S. Sen, M. Thangavelu, S. Pinder, and B. Roser. 1992. Extraordinary stability of enzymes dried in trehalose: simplified molecular biology. Biotechnology (NY) 10:1007-1011. [DOI] [PubMed] [Google Scholar]
- 12.Crowe, J. H., J. F. Carpenter, and L. M. Crowe. 1998. The role of vitrification in anhydrobiosis. Annu. Rev. Physiol. 60:73-103. [DOI] [PubMed] [Google Scholar]
- 13.Crowe, J. H., L. M. Crowe, and R. Mouradian. 1983. Stabilization of biological membranes at low water activities. Cryobiology 20:346-356. [DOI] [PubMed] [Google Scholar]
- 14.Franklin, F. C. H., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta-cleavage pathway. Proc. Natl. Acad. Sci. U. S. A. 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia de Castro, A., H. Bredholt, A. R. Strom, and A. Tunnacliffe. 2000. Anhydrobiotic engineering of gram-negative bacteria. Appl. Environ. Microbiol. 66:4142-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goyal, K., L. J. Walton, and A. Tunnacliffe. 2005. LEA proteins prevent protein aggregation due to water stress. Biochem. J. 388:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, N., P. Rathi, and R. Gupta. 2002. Simplified para-nitrophenyl palmitate assay for lipases and esterases. Anal. Biochem. 311:98-99. [DOI] [PubMed] [Google Scholar]
- 18.Knapp, S., R. Ladenstein, and E. A. Galinski. 1999. Extrinsic protein stabilization by the naturally occurring osmolytes beta-hydroxyectoine and betaine. Extremophiles 3:191-198. [DOI] [PubMed] [Google Scholar]
- 19.LeBlanc, J. C., E. R. Goncalves, and W. W. Mohn. 2008. Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 74:2627-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maillard, L. C. 1912. The action of amino acids on sugar; the formation of melanoidin by a methodic route. C. R. Hebd. Seances Acad. Sci. 154:66-68. [Google Scholar]
- 21.Manzanera, M., A. Garcia de Castro, A. Tondervik, M. Rayner-Brandes, A. R. Strom, and A. Tunnacliffe. 2002. Hydroxyectoine is superior to trehalose for anhydrobiotic engineering of Pseudomonas putida KT2440. Appl. Environ. Microbiol. 68:4328-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manzanera, M., S. Vilchez, and A. Tunnacliffe. 2004. High survival and stability rates of Escherichia coli dried in hydroxyectoine. FEMS Microbiol. Lett. 233:347-352. [DOI] [PubMed] [Google Scholar]
- 23.Manzanera, M., S. Vilchez, and A. Tunnacliffe. 2004. Plastic encapsulation of stabilized Escherichia coli and Pseudomonas putida. Appl. Environ. Microbiol. 70:3143-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patro, S. Y., E. Freund, and B. S. Chang. 2002. Protein formulation and fill-finish operations. Biotechnol. Annu. Rev. 8:55-84. [DOI] [PubMed] [Google Scholar]
- 25.Sauer, T., and E. A. Galinski. 1998. Bacterial milking: a novel bioprocess for production of compatible solutes. Biotechnol. Bioeng. 57:306-313. [PubMed] [Google Scholar]
- 26.Shukla, M., R. Chaturvedi, D. Tamhane, P. Vyas, G. Archana, S. Apte, J. Bandekar, and A. Desai. 2007. Multiple-stress tolerance of ionizing radiation-resistant bacterial isolates obtained from various habitats: correlation between stresses. Curr. Microbiol. 54:142-148. [DOI] [PubMed] [Google Scholar]
- 27.Shull, J. J., G. T. Cargo, and R. R. Ernst. 1963. Kinetics of heat activation and of thermal death of bacterial spores. Appl. Microbiol. 11:485-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Trevors, J. T., J. D. Vanelsas, H. Lee, and A. C. Wolters. 1993. Survival of alginate-encapsulated Pseudomonas fluorescens cells in soil. Appl. Microbiol. Biotechnol. 39:637-643. [DOI] [PubMed] [Google Scholar]
- 30.Tunnacliffe, A., A. Garcia de Castro, and M. Manzanera. 2001. Anhydrobiotic engineering of bacterial and mammalian cells: is intracellular trehalose sufficient? Cryobiology 43:124-132. [DOI] [PubMed] [Google Scholar]
- 31.Vilchez, S., A. Tunnacliffe, and M. Manzanera. 2008. Tolerance of plastic-encapsulated Pseudomonas putida KT2440 to chemical stress. Extremophiles 12:297-299. [DOI] [PubMed] [Google Scholar]
- 32.Vreeland, R. H., C. D. Litchfield, E. L. Martin, and E. Elliot. 1980. Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int. J. Syst. Bacteriol. 30:485-495. [Google Scholar]
- 33.Walker, T. E., C. H. Han, V. H. Kollman, R. E. London, and N. A. Matwiyoff. 1982. 13C nuclear magnetic resonance studies of the biosynthesis by Microbacterium ammoniaphilum of L-glutamate selectively enriched with carbon-13. J. Biol. Chem. 257:1189-1195. [PubMed] [Google Scholar]
- 34.Wang, W. 2000. Lyophilization and development of solid protein pharmaceuticals. Int. J. Pharm. 203:1-60. [DOI] [PubMed] [Google Scholar]
- 35.Wang, W. 2005. Protein aggregation and its inhibition in biopharmaceutics. Int. J. Pharm. 289:1-30. [DOI] [PubMed] [Google Scholar]
- 36.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yancey, P. H., M. E. Clark, S. C. Hand, R. D. Bowlus, and G. N. Somero. 1982. Living with water stress: evolution of osmolyte systems. Science 217:1214-1222. [DOI] [PubMed] [Google Scholar]
- 38.Zevenhuizen, L. P. 1966. Formation and function of the glycogen-like polysaccharide of Arthrobacter. Antonie Van Leeuwenhoek 32:356-372. [DOI] [PubMed] [Google Scholar]
- 39.Zevenhuizen, L. P. 1992. Levels of trehalose and glycogen in Arthrobacter globiformis under conditions of nutrient starvation and osmotic stress. Antonie Van Leeuwenhoek 61:61-68. [DOI] [PubMed] [Google Scholar]