Abstract
Recently, methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant Staphylococcus pseudintermedius (MRSP) have been increasingly isolated from veterinarians and companion animals. With a view to preventing the spread of MRSA and MRSP, we evaluated the occurrence and molecular characteristics of each in a veterinary college. MRSA and MRSP were isolated from nasal samples from veterinarians, staff members, and veterinary students affiliated with a veterinary hospital. Using stepwise logistic regression, we identified two factors associated with MRSA carriage: (i) contact with an identified animal MRSA case (odds ratio [OR], 6.9; 95% confidence interval [95% CI], 2.2 to 21.6) and (ii) being an employee (OR, 6.2; 95% CI, 2.0 to 19.4). The majority of MRSA isolates obtained from individuals affiliated with the veterinary hospital and dog patients harbored spa type t002 and a type II staphylococcal cassette chromosome mec (SCCmec), similar to the hospital-acquired MRSA isolates in Japan. MRSA isolates harboring spa type t008 and a type IV SCCmec were obtained from one veterinarian on three different sampling occasions and also from dog patients. MRSA carriers can also be a source of MRSA infection in animals. The majority of MRSP isolates (85.2%) carried hybrid SCCmec type II-III, and almost all the remaining MRSP isolates (11.1%) carried SCCmec type V. MRSA and MRSP were also isolated from environmental samples collected from the veterinary hospital (5.1% and 6.4%, respectively). The application of certain disinfection procedures is important for the prevention of nosocomial infection, and MRSA and MRSP infection control strategies should be adopted in veterinary medical practice.
Methicillin-resistant Staphylococcus aureus (MRSA) is an important cause of nosocomial infections in human hospitals. The prevalence of hospital-acquired MRSA (HA-MRSA) infection among inpatients in intensive care units (ICUs) continues to increase steadily in Japan. Recently, cases of community-acquired MRSA (CA-MRSA) have been documented in persons without an established risk factor for HA-MRSA infection (14, 32, 36, 49).
There has also been an increase in the number of reports of the isolation of MRSA from veterinarians and companion animals (5, 21, 23-26, 28, 31, 34, 38, 44, 50, 51, 53). Values reported for the prevalence of MRSA among veterinary staff include 17.9% in the United Kingdom (21), 10% in Japan (38), 3.9% in Scotland (13), and 3.0% in Denmark (28). Loeffler et al. reported that the prevalence of MRSA among dog patients and healthy dogs owned by veterinary staff members was 8.9% (21). In Japan, an MRSA isolate was detected in only one inpatient dog (3.8%) and could not be detected in any of 31 outpatient dogs (38). In the United States, MRSA isolates were detected in both dog (0.1%) and cat (0.1%) patients (31). The prevalence of MRSA among healthy dogs has been reported to be 0.7% (5). Hanselman et al. suggested that MRSA colonization may be an occupational risk for large-animal veterinarians (12). Recently, Burstiner et al. reported that the frequency of MRSA colonization among companion-animal veterinary personnel was equal to the frequency among large-animal veterinary personnel (6).
In addition, other methicillin-resistant coagulase-positive staphylococci (MRCPS), such as methicillin-resistant Staphylococcus pseudintermedius (MRSP) and methicillin-resistant Staphylococcus schleiferi (MRSS), isolated from dogs, cats, and a veterinarian have been reported (11, 31, 38, 40, 52). MRSP isolates have also been detected among inpatient dogs (46.2%) and outpatient dogs (19.4%) in a Japanese veterinary teaching hospital (38). In Canada, however, MRSP and MRSS isolates were detected in only 2.1% and 0.5% of dog patients, respectively (11).
Methicillin-resistant staphylococci produce penicillin-binding protein 2′, which reduces their affinity for β-lactam antibiotics. This protein is encoded by the mecA gene (48), which is carried on the staphylococcal cassette chromosome mec (SCCmec). SCCmec is a mobile genetic element characterized by the combination of the mec and ccr complexes (16), and it is classified into subtypes according to differences in the junkyard regions (43). SCCmec typing can be used as a molecular tool (22, 27, 30, 33, 36, 55) for examining the molecular epidemiology of methicillin-resistant staphylococci.
In this study, we investigated the occurrence and characteristics of MRCPS isolates in a veterinary hospital in order to establish the transmission route of MRCPS in a veterinary hospital and with a view to preventing the spread of MRCPS infection. In addition, we evaluated the factors associated with MRCPS. Further, as Heller et al. have reported the distribution of MRSA within veterinary hospital environments and suggested the necessity to review cleaning protocols of hospital environments (13), we also attempted to isolate MRCPS from environmental samples collected in a veterinary hospital for an evaluation of MRSA transmission cycle though environmental surfaces in the veterinary hospital.
MATERIALS AND METHODS
Sampling.
This study was approved by the Ethics Committee of the Graduate School of Dairy Science, Rakuno Gakuen University (RGU). Volunteers were recruited to provide the nasal swabs for the isolation of MRCPS from staff affiliated with the Department of Veterinary Companion Animals (DVCA) of the veterinary hospital and with three nonclinical laboratories in the School of Veterinary Medicine, RGU. Nasal swab samples were collected using BBL culture swab EZ (Becton Dickinson Japan, Co., Ltd., Tokyo, Japan) in March and April 2007 from 20 of the 46 veterinarians (43.5%) affiliated with the DVCA, all 21 staff members (including veterinary technicians) affiliated with the DVCA, and 51 of 60 veterinary students (85%) affiliated with the DVCA. In addition, 5 staff members and 31 veterinary students of the 42 personnel (85.7%) affiliated with nonclinical laboratories also provided nasal samples. Ten nasal samples from the 31 veterinary students affiliated with nonclinical laboratories were collected in October 2007.
On a single day, we also collected 75 environmental surface samples (e.g., from floors, top parts of medical examination stands, and cages) from 30 areas in the veterinary hospital, including the following areas: waiting area; consulting, procedure, and operation rooms; dispensary; animal wards; administrative office; and a veterinarian's office. Sampling areas were selected from areas frequented by individuals affiliated with the veterinary hospital and by animal patients. These samples were collected using single-use cotton swabs moistened with sterile physiological saline. In cases where presumptive MRCPS isolates were obtained from individuals who had companion animals at home, samples of buccal mucosa from the companion animals were also tested.
In the second sampling period from February to April 2008, the nasal samples were collected from 31 of 50 clinical veterinarians (62%), all 18 staff members, and 65 of 89 veterinary students (73%) affiliated with the DCVA of the veterinary hospital. In addition, three veterinarians, one staff member, and nine veterinary students affiliated with the Department of Large Animal Clinical Sciences provided nasal samples. Additionally, a total of 81 environmental surface samples were collected from areas similar to those described above.
Eight presumed MRSA strains from dog patients were also collected for use in this study for comparison with MRSA isolates from veterinarians, and veterinary students. These strains were isolated from the affected parts of dog patients during the period from 2007 to 2008. These strains were identified genetically and characterized by SCCmec and molecular typing in this study.
Fact-finding inquiry.
In order to establish the factors associated with MRCPS carriage in humans, information on the following was gathered from subjects: hospitalization and surgery received within the previous year, antibiotics taken within the previous month, known contact with MRSA-identified animal patients, career, affiliations, and the presence of companion animals at home. During the 2008 investigation, we questioned volunteers about their participation in the previous investigation in 2007 and their MRSA-positive or -negative status as determined by the previous test. Human samples and questionnaire answers were coded to protect anonymity.
Isolation of MRCPS.
Swabs were directly inoculated onto CHROMagar MRSA medium (Kanto Kagaku Co., Ltd., Tokyo, Japan) and incubated at 35°C for 24 h. For enrichment, the same swabs were inoculated into 5-ml portions of heart infusion broth (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) containing 7.5% sodium chloride and incubated at 35°C for 48 h. Enrichment cultures were streaked out on CHROMagar MRSA medium and incubated as described above. Representative colonies colored light purple (presumptive MRSA), blue, or white were selected from each sample and subcultured on nutrient agar (Nissui Pharmaceutical Co., Ltd.).
For testing, we selected one isolate from each sample that was a catalase-positive, Gram-positive coccus and coagulase positive in tube tests with rabbit plasma (Eiken Chemistry Co., Ltd., Tokyo, Japan). The isolates were stored in a Microbank (Pro-Lab Diagnostics Inc., Richmond Hill, Canada) at −80°C.
Following the detection of MRSA carriage, 10 MRSA carriers requested consultation with a school doctor for treatment. They were treated with mupirocin ointment for 3 days. The nasal samples were again tested for the presence of MRSA after 1 month of treatment.
Detection of mecA and pvl, SCCmec typing, and identification of S. aureus and S. pseudintermedius.
Staphylococcal genomic DNA was extracted from cultures using a QIAamp DNA minikit (Qiagen Co., Ltd., Tokyo, Japan) or an InstaGene matrix (Bio-Rad Laboratories Inc., Tokyo, Japan).
The Panton-Valentine leukocidin (pvl), mecA, and femA genes were investigated using multiplex PCR 1 (M-PCR 1) (Table 1); the primers for femA gene are specific to S. aureus. M-PCR 1 was performed in a 50-μl reaction mixture containing 1× HF PCR buffer (containing 1.5 mM MgCl2), 200 μM (each) deoxynucleoside triphosphate (dNTP), 200 nM (each) primer (Table 1), 1 U Phusion DNA polymerase (Finnzymes, Espoo, Finland), and 1 μl of sample DNA. The samples were subjected to an initial denaturation at 98°C for 30 s, followed by 30 amplification cycles, with each cycle consisting of 98°C for 5 s, 57°C for 10 s, and 72°C at 10 s. A final primer extension was performed at 72°C for 5 min, and the samples were then maintained at 4°C.
TABLE 1.
Primers used in this study
| PCR method (purpose), locus, and primera | Primer sequence (5′→3′) | Size of product (bp) | Reference |
|---|---|---|---|
| M-PCR 1 (for identifying S. aureus and detecting mecA and pvl) | |||
| mecA1 | TGT CCG TAA CCT GAA TCA GC | 519 | 47 |
| mecA2 | TGC TAT CCA CCC TCA AAC AG | ||
| femA-2F | AAC TGT TGG CCA CTA TGA GT | 308 | 35 |
| femA-2R | CCA GCA TTA CCT GTA ATC TCG | ||
| luk-PV-1 (modified) | TCA TTA GGT AAA ATG TCT GGA CAT GAT CCA | 433 | 20 |
| luk-PV-2 | GCA TCA AST GTA TTG GAT AGC AAA AGC | ||
| M-PCR 2 (for amplication of mec gene complex class) | |||
| mI6 | CAT AAC TTC CCA TTC TGC AGA TG | 1,963 | 18 |
| IS7 | ATG CTT AAT GAT AGC ATC CGA ATG | 2,827 | |
| IS2 | TGA GGT TAT TCA GAT ATT TCG ATG T | 804 | |
| mA7 | ATA TAC CAA ACC CGA CAA CTA CA | ||
| M-PCR 3 (for amplification of ccr gene complex type) | |||
| ccrAB-β2 | ATT GCC TTG ATA ATA GCC ITC T | ||
| ccrAB-α2 | AAC CTA TAT CAT CAA TCA GTA CGT | 695 | 16 |
| ccrAB-α3 | TAA AGG CAT CAA TGC ACA AAC ACT | 937 | |
| ccrAB-α4 | AGC TCA AAA GCA AGC AAT AGA AT | 1,791 | |
| M-PCR 4 (for amplication of elements on SCCmec) | |||
| Locus A | |||
| CIF2 F2 | TTC GAG TTG CTG ATG AAG AAG G | 495 | 33 |
| CIF2 R2 | ATT TAC CAC AAG GAC TAC CAG C | ||
| Locus B | |||
| KDP F1 | AAT CAT CTG CCA TTG GTG ATG C | 284 | 33 |
| KDP R1 | CGA ATG AAG TGA AAG AAA GTG G | ||
| Locus C | |||
| MECI P2 | ATC AAG ACT TGC ATT CAG GC | 209 | 33 |
| MECI P3 | GCG GTT TCA ATT CAC TTG TC | ||
| Locus D | |||
| DCS F2 | CAT CCT ATG ATA GCT TGG TC | 342 | 33 |
| DCS R1 | CTA AAT CAT AGC CAT GAC CG | ||
| Locus E | |||
| RIF4 F3 | GTG ATT GTT CGA GAT ATG TGG | 243 | 33 |
| RIF4 R9 | CGC TTT ATC TGT ATC TAT CGC | ||
| Locus F | |||
| RIF5 F10 | TTC TTA AGT ACA CGC TGA ATC G | 414 | 33 |
| RIF5 R13 | GTC ACA GTA ATT CCA TCA ATG C | ||
| Locus G | |||
| IS431 P4 | CAG GTC TCT TCA GAT CTA CG | 381 | 33 |
| pUB110 R1 | GAG CCA TAA ACA CCA ATA GCC | ||
| Locus H | |||
| IS431 P4 | CAG GTC TCT TCA GAT CTA CG | 303 | 33 |
| pT181 R1 | GAA GAA TGG GGA AAG CTT CAC | ||
| mecA | |||
| MECA P4 | TCC AGA TTA CAA CTT CAC CAG G | 162 | 33 |
| MECA P7 | CCA CTT CAT ATC TTG TAA CG | ||
| PCR 1 (for identification of S. pseudintermedius by restriction fragment length polymorphism using MboI) | |||
| pta_f1 | AAA GAC AAA CTT TCA GGT AA | 320 | 4 |
| pta_r1 | GCA TAA ACA AGC ATT GTA CCG | ||
| PCR 2 (for amplification of the ccrC gene) | |||
| ccrC F2 | GTA CTC GTT ACA ATG TTT GG | 449 | 27 |
| ccrC R2 | ATA ATG GCT TCA TGC TTA CC | ||
| PCR 3 (for amplification of the J1 region) | |||
| SCCmec III J1 F | CAT TTG TGA AAC ACA GTA CG | 243 | 27 |
| SCCmec III J1 R | GTT ATT GAG ACT CCT AAA GC |
Forward and reverse primers are indicated by F and R, respectively.
To identify the species of isolates other than S. aureus, direct sequencing of the sodA and hsp60 genes was performed as previously described (19, 37, 39). Sequencing reactions were performed using a Big Dye Terminator version 1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI Prism 310 genetic analyzer (Applied Biosystems). Additionally, restriction patterns produced by MboI (Takara Bio, Ltd., Shiga, Japan) digestion of PCR 1-amplified pta were also analyzed for the identification of S. pseudintermedius by the method of Bannoehr et al. (4).
SCCmec typing was performed by amplification of the mec regions (classes A, B, and C) and the ccr regions (types 1, 2, 3, and 5). The class of mec complex was determined by M-PCR 2 (18). The type of ccr complex was determined by M-PCR 3 (which detects types 1, 2, and 3) (16) or PCR 2 (which detects type 5). Moreover, M-PCR 4 was performed to determine the structure of SCCmec using the strategy of Oliveira and de Lencastre (33) with Phusion DNA polymerase (Finnzymes). Finally, the structure of the J1 region was confirmed for SCCmec type III isolates by PCR 3. The primer sequences are listed in Table 1.
Antimicrobial susceptibility testing.
We performed MIC determinations for 14 antimicrobial agents (7). For the isolates obtained in 2007, MIC tests were conducted using the agar dilution method with Mueller-Hinton agar (Oxoid Ltd., Hampshire, United Kingdom). The following antimicrobial agents were tested: oxacillin, ampicillin, cefazolin, kanamycin, gentamicin, erythromycin, chloramphenicol, minocycline, oxytetracycline, vancomycin, and enrofloxacin (all from Sigma-Aldrich Japan K. K., Tokyo, Japan); imipenem (Banyu Pharmaceutical Co., Ltd., Tokyo, Japan); arbekacin (Meiji Seika Ltd., Tokyo, Japan); and teicoplanin (Astellas Pharma Inc., Tokyo, Japan). For the isolates obtained in 2008, we used the broth microdilution method with an Eiken frozen plate (Eiken Chemistry Co. Ltd.). Again, we tested 14 antimicrobial agents, 12 of which were the same as those tested for the 2007 isolates. This time, however, we used tetracycline and ciprofloxacin instead of oxytetracycline and enrofloxacin.
Molecular typing.
Pulsed-field gel electrophoresis (PFGE) analysis for MRSA isolates was performed as previously described (2, 15). Briefly, bacterial cultures were grown overnight in Mueller-Hinton broth (Oxoid Ltd.) and resuspended in TN buffer (10 mM Tris, 1 M NaCl, pH 8.0). The cultures were mixed with the same volume of 2% UltraPure low-melting-point (LMP) agarose (Invitrogen Japan Ltd., Tokyo, Japan), dissolved in TN buffer, and pipetted into a plug mold. After bacterial lysis in a lysis buffer containing 5 mg/ml lysozyme (Wako Pure Chemical Industries, Ltd., Osaka, Japan) and 20 μg/ml lysostaphin (Sigma-Aldrich Japan K. K.), genomic DNA was digested with SmaI (Takara Bio, Ltd.). PFGE was performed using a CHEF-DRIII system (Bio-Rad Laboratories Inc.), under the following conditions: switch time of 5.3 to 34.9 s and running time of 20 h for MRSA (15); included angle, 120°; voltage, 6 V/cm; and set temperature, 14°C.
All MRSA isolates were typed by DNA sequence analysis of the X region of the protein A gene (spa) as previously described (42). The spa types of MRSA isolates were determined by using Ridom SpaServer (http://www.spaserver.ridom.de/).
Statistical analysis.
Categorical comparisons were performed using Fisher's exact test. A P value of less than 0.05 was considered to be significant. The associations between MRSA and MRSP carriage and each inquiry shown in Table 3 were evaluated with a stepwise forward logistic regression using SPSS 15.0J software (SPSS Japan Inc., Tokyo, Japan). Variables with a significance level of P of <0.05 in the univariate analysis were considered for inclusion in the multivariate model. Variables with a P value of <0.05 in the final model were also considered significant, and odds ratios (ORs) with 95% confidence intervals (95% CIs) were calculated.
TABLE 3.
Factors associated with methicillin-resistant S. aureus and methicillin-resistant S. pseudintermedius carriage by univariable analysisa
| Inquiry | Answer to inquiry | MRSA |
MRSP |
||||
|---|---|---|---|---|---|---|---|
| Positive rateb (%) | ORc | P value | Positive rate (%) | OR | P value | ||
| Contact with MRSA-positive animal patients | Yes | 12/61 (19.7) | 6.1 | <0.01 | 5/61 (8.2) | 11.5 | <0.05 |
| No | 5/130 (3.8) | 1/130 (0.8) | |||||
| Employee | Yes | 12/65 (18.5) | 5.5 | <0.01 | 2/65 (3.1) | 0.97 | 1.0 |
| No | 5/126 (4.0) | 4/126 (3.2) | |||||
| Affiliated with the veterinary hospital | Yes | 17/155 (11.0) | <0.05 | 6/155 (3.9) | 0.6 | ||
| No | 0/36 | 0/36 | |||||
| Hospitalization or surgery within the previous year | Yes | 0/10 | 0 | 0.60 | 0/10 | 0 | 1.0 |
| No | 17/180 (9.4) | 6/180 (3.3) | |||||
| Intake of antibiotics within the previous month | Yes | 3/17 (17.6) | 2.4 | 0.18 | 0/17 | 0 | 1.0 |
| No | 14/173 (8.1) | 6/173 (3.5) | |||||
| Presence of companion animals at home | Yes | 10/96 (10.4) | 1.4 | 0.61 | 1/96 (1.0) | 0.2 | 0.11 |
| No | 7/93 (7.5) | 5/93 (5.4) | |||||
MRSA, methicillin-resistant S. aureus; MRSP, methicillin-resistant S. pseudintermedius.
The positive rate is the number of individuals positive for methicillin-resistant staphylococci/number of those who answered “yes” or “no” for each inquiry.
OR, odds ratio.
RESULTS
Identification and frequencies of MRSA, MRSP, and MRSS.
The occurrence of methicillin-resistant Staphylococcus aureus (MRSA), methicillin-resistant Staphylococcus pseudintermedius (MRSP), and methicillin-resistant Staphylococcus schleiferi (MRSS) among each sample type are shown in Table 2. MRSA isolates were obtained from 11 of 92 individuals (12.0%; 95% CI, 6.1% to 20.4%) and 10 of 127 individuals (7.9%; 95% CI, 3.8% to 14.0%) affiliated with the veterinary hospital in 2007 and 2008, respectively. In addition, MRSA isolates were also obtained from 2 of 10 MRSA carriers even after they underwent treatment. Analysis of the environmental surface samples showed that MRSA had contaminated the floors of rooms used for X-ray computed tomography (CT) scans and administration of anesthetics and the cat ward, a cat cage in the cat ward room, and the top part of an anesthetics stand in 2007. The following areas were identified as infected: the floors of rooms in the cat ward, a preliminary meeting room for veterinary technicians, and an intern veterinarian's office in 2008.
TABLE 2.
Carriage of methicillin-resistant S. aureus, methicillin-resistant S. pseudintermedius, and methicillin-resistant S. schleiferi
| Source of sample | Yr | No. of samples | No. of samples in which methicillin-resistant staphylococcia were isolated (%) [95% confidence interval] |
||
|---|---|---|---|---|---|
| MRSA | MRSP | MRSS | |||
| Individuals in the veterinary hospital | |||||
| Veterinarians | 2007 | 20 | 5 (25.0)c [8.7-49.1] | 1 (5.0) [0.1-24.9] | 0 [0-13.9] |
| 2008 | 34 | 8 (23.5)d,e [10.7-41.2] | 3 (8.8) [1.9-23.7] | 1 (2.9) [0.1-15.3] | |
| Staff members | 2007 | 21 | 3 (14.3) [3.0-36.3] | 1 (4.8) [0.1-23.8] | 0 [0-13.3] |
| 2008 | 19 | 0e [0-14.6] | 0 [0-14.6] | 0 [0-14.6] | |
| Students | 2007 | 51 | 3 (5.9)c [1.2-16.2] | 1 (2.0) [0.1-10.4] | 0 [0-5.7] |
| 2008 | 74 | 2 (2.7)d [0.3-9.4] | 7 (9.5) [3.9-18.5] | 0 [0-4.0] | |
| Staff and students in nonclinical laboratories | 2007 | 36 | 0 [0-8.0] | 0 [0-8.0] | 0 [0-8.0] |
| Companion animalsb | 2007 | 5 | 0 [0-45.1] | 3 (60) [14.7-94.7] | 0 [0-45.1] |
| Environment in the veterinary hospital | 2007 | 75 | 5 (6.7) [2.2-14.9] | 10 (13.3) [6.6-23.2] | 0 [0-3.9] |
| 2008 | 81 | 3 (3.7) [0.8-10.4] | 0 [0-3.6] | 4 (4.9) [1.4-12.2] | |
| MRSA carriers after treatment for MRSA colonization | 2007 | 10 | 2 (20) [1.2-31.7] | ||
MRSA, methicillin-resistant S. aureus; MRSP, methicillin-resistant S. pseudintermedius; MRSS, methicillin-resistant S. schleiferi.
These were kept by MRSA- or MRSP-positive persons in 2007.
The percentages of MRSA carriers among veterinarians and students in 2007 were not significantly different (P = 0.08).
The percentage of MRSA carriers among veterinarians in 2008 was significantly higher than that among students in 2008 (P < 0.01).
The percentage of MRSA carriers among veterinarians in 2008 was significantly higher than that among staff members in 2008 (P < 0.01).
The values for the incidence of MRSP carriage among individuals affiliated with the veterinary hospital were 3.3% (3/92) (95% CI, 0.7% to 9.2%) and 7.9% (10/127) (95% CI, 3.8% to 14.0%) in 2007 and 2008, respectively. MRSP isolates were obtained from three buccal mucosa samples from five companion animals owned by methicillin-resistant coagulase-positive staphylococci (MRCPS)-positive individuals (60%). In 2007, MRSP was found to have contaminated the following areas: the cages for large dogs, the top surfaces of an X-ray CT scan stand, a stand in the cat ward, the ICU, and the floor of a magnetic resonance imaging (MRI) room. These isolates were identified as MRSP by the high degree of similarity between their sodA and hsp60 sequences and those of the S. pseudintermedius strain LMG 22219T and by the MboI restriction pattern of the PCR-amplified pta gene. Although the aggregates of MRSP colonies cultured on the CHROMagar MRSA medium were light purple (similar to the MRSA colonies), most of the single colonies of MRSP were white and very small. The diameter of the MRSP single colonies cultured on the CHROMagar MRSA medium after incubation for 24 h was approximately 0.5 mm. On the other hand, the diameter of MRSA single colonies after the same incubation period was approximately 1 mm.
In 2008, MRSS isolates were obtained from a nasal swab sample from a veterinarian (1/127 [0.4%]) and from four environmental surface samples (4.9%). MRSS was found to have contaminated the floor of a consulting room, the reception area, an X-ray CT scan preparation room, and the top surface of an examination stand. These MRSS isolates were identified by the high degree of similarity between their sodA and hsp60 sequences with those of the S. schleiferi subsp. coagulans strain CIP 104370T. The colonies of these isolates cultured on CHROMagar MRSA medium were also white and appeared to be very small.
Seven of eight presumed MRSA strains isolated from dog patients were confirmed as MRSA. Another isolate from a dog patient was identified as MRSP. MRSA, MRSP, and MRSS were not isolated from any of the nasal swab samples obtained from individuals affiliated with nonclinical laboratories.
Characteristics of MRSA- and MRSP-positive humans.
A total of 64 individuals (20 veterinarians, 14 staff members, and 30 students) provided samples in both 2007 and 2008. To avoid duplication, we undertook an analysis of the factors associated with MRSA and MRSP colonization by assessing the results from individuals who had first provided samples and answered the questionnaire. Research students and graduate students were categorized as employees because they diagnosed and treated animal patients in the capacity of veterinarians. One person failed to answer questions regarding hospitalization, surgery, and antibiotic intake, whereas an additional two people failed to answer questions regarding the presence of companion animals in the home; these individuals were therefore excluded from the analysis. The univariate analysis results for MRSA and MRSP carriage are shown in Table 3.
According to the stepwise forward logistic regression, both contact with MRSA-identified animal patients and employment were associated with MRSA carriage. The predictive accuracy was 91.0%, as determined by the Hosmer-Lemeshow test. The incidence of MRSA carriers among those who had had contact with MRSA-identified animal patients (19.7%) was higher than those who had had no contact (3.8%; OR, 6.88; 95% CI, 2.20 to 21.57) (P < 0.01). The incidence of MRSA carriers among employees of the veterinary hospital or of nonclinical laboratories (18.5%) was higher than that among students (4.0%; OR, 6.18; 95% CI, 1.97 to 19.41). Contact with MRSA-identified animal patients was also associated with MRSP carriage. The predictive accuracy was 96.8%, as determined by the Hosmer-Lemeshow test. The incidence of MRSP carriers among those who had had contact with MRSA-identified animal patients (8.2%) was higher than the incidence in those who had had no contact with MRSA-identified animal patients (0.8%; OR, 11.55; 95% CI, 1.32 to 101.14) (P < 0.05).
Molecular characterization of MRCPS.
Two MRSA isolates from MRSA carriers after treatment, seven MRSA isolates from dog patients, 21 MRSA isolates from individuals affiliated with the veterinary hospital, and eight MRSA isolates from environmental surface samples were characterized by SCCmec typing, PFGE, and spa typing. Twenty-seven MRSA isolates were classified as SCCmec type II. The absence of pUB110 and kdp was detected in seven isolates and one isolate of SCCmec type II, respectively. Eight MRSA isolates were classified as SCCmec type IV. All eight SCCmec type IV MRSA isolates harbored pUB110, and three of the eight isolates also harbored pls. Three of the MRSA isolates were classified as SCCmec type I.
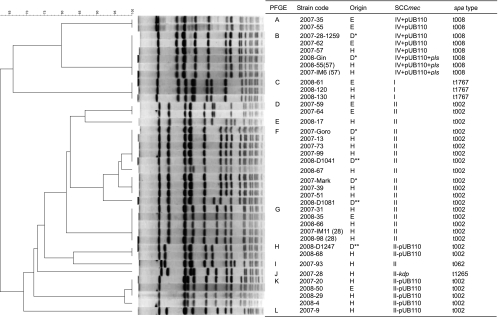
The genetic relationship among the MRSA isolates, based on PFGE, is shown in Fig. 1. Analysis using the Dice coefficient, followed by the unweighted-pair group method with arithmetic means (UPGMA), revealed 12 clusters (clusters A to L) of MRSA. Band position tolerance and optimization were both set at 1.0%. Similarities among the SCCmec type II isolates that harbored both pUB110 and kdp were higher than those among isolates that had lost either pUB110 or kdp. MRSA isolates with SCCmec type II in clusters I and J had spa type t062 and t1265, respectively. The remaining 25 MRSA isolates with SCCmec type II had spa type t002. All eight MRSA isolates with SCCmec type IV had spa type t008. All three MRSA isolates with SCCmec type I had spa type t1767.
FIG. 1.
PFGE dendrogram showing percentage similarities calculated by Dice coefficient (tolerance of 1% and optimization of 1%) and SCCmec and spa types of MRSA isolates. The MRSA isolates were taken from the environment (E), dogs (dog patients at the RGU hospital [D*] and dog patients at the primary veterinary hospital [D**]), and humans (H).
Two spa type t002 MRSA isolates in cluster G (2007-IM11 [28] and 2008-98 [28]) were isolated from a nasal sample from a veterinarian after treatment for MRSA carriage in November 2007 and in 2008; however, an MRSA isolate from the same veterinarian before treatment was grouped in cluster J and did not harbor kdp. The three spa type t008 isolates in cluster B were obtained from the same veterinarian, before and after treatment (March and November 2007 and February 2008).
Twenty-three MRSP isolates carried a class A mec complex and a type 3 ccr complex. The absence of insertion Tn554 (loci E and F were negative, according to M-PCR 4) and pT181 (locus H) was found in 23 MRSP isolates. These MRSP isolates harbored the dcs region and a gene in the J1 region present in SCCmec type III; the latter was confirmed by PCR 3. The SCCmec structure in these 23 MRSP isolates matched the DNA sequence of SCCmec type II-III (GenBank accession no. AM904732); therefore, the SCCmec type of these MRSP isolates was presumed to be type II-III. Three MRSP isolates were classified as SCCmec type V and harbored Tn554 in the chromosomal right junction (locus F was positive). In one MRSP isolate, ccr was not amplified; therefore, the SCCmec type was not determined.
Five MRSS isolates were classified as SCCmec type IV. These isolates harbored pls and the dcs region (loci A and D were positive). No pvl genes were found in any of the MRCPS isolates.
Antimicrobial susceptibility testing.
The frequencies of resistance of MRSA, MRSP, and MRSS and the range of the MICs for each SCCmec type are shown in Table 4. All isolates were resistant to both oxacillin and ampicillin. All MRSA and MRSS isolates were resistant to cefazolin; however, more than half of the MRSP isolates were susceptible to it. Moreover, almost all MRSA SCCmec type I and II isolates were resistant to imipenem, whereas almost all MRSA SCCmec type IV isolates, all MRSP isolates, and MRSS isolates were susceptible to this antibiotic. The frequencies of MRSP resistance to kanamycin and gentamicin were higher than those for MRSA and MRSS.
TABLE 4.
Distribution of MIC for methicillin-resistant coagulase-positive staphylococci in each SCCmec typea
| Antimicrobial agent | Breakpoint (mg/liter) | MRSA (n = 33) |
MRSP (n = 27) |
MRSS (n = 5) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of resistant isolates (%) | Range of MICs for the following SCCmec typeb: |
No. of resistant isolates (%) | Range of MICs for the following SCCmec type: |
No. of resistant isolates (%) | Range of MICs for SCCmec type IV (n = 5) | |||||||||
| I (n = 3) | II (n = 16) | II − pUB110 (n = 6) | II − kdp (n = 1) | IV + pUB110 (n = 5) | IV + pUB110 + pls (n = 2) | II-III (n = 23) | V (n = 3) | UT (n = 1) | ||||||
| Oxacillin | ≧4 | 33 (100) | >128 | ≧128 | ≧128 | 128 | ≧128 | ≧128 | 27 (100) | ≧128 | 16-≧128 | >128 | 5 (100) | >128 |
| Ampicillin | ≧0.25 | 33 (100) | 32-64 | 32-64 | 32-64 | 32 | 16-64 | 32-64 | 27 (100) | 0.5-64 | 16 | 32 | 5 (100) | 8-16 |
| Cefazolin | ≧32 | 33 (100) | 256 | ≧256 | 256 | 64 | 128-256 | 64-128 | 13 (48.1) | 0.25-256 | 0.5-2 | 32 | 5 (100) | 128-256 |
| Imipenem | ≧16 | 27 (81.8) | 16 | ≧16 | ≧16 | 1 | 0.5->16 | 2-16 | 0 | ≦0.5 | ≦0.5 | ≦0.5 | 0 | ≦0.5 |
| Kanamycin | ≧32 | 27 (81.8) | >512 | 64->512 | 2-16 | 64 | 256 | >512 | 27 (100) | 256->512 | 256 | >512 | 0 | 1 |
| Gentamicin | ≧8 | 22 (66.7) | 64-128 | 0.5-128 | ≦0.25-2 | ≦0.25 | 32-256 | 32->512 | 26 (96.3) | 8-64 | 4-16 | 16 | 0 | ≦0.25 |
| Arbekacin | ≥16 | 0 | ≦0.5-1 | ≦0.5-1 | ≦0.5 | ≦0.5 | ≦0.5-1 | ≦0.5-4 | 0 | ≦0.5-2 | ≦0.5 | ≦0.5 | 0 | ≦0.5 |
| Erythromycin | ≧8 | 32 (97.0) | >128 | 8->128 | >128 | >128 | >128 | >128 | 27 (100) | >128 | >128 | >128 | 0 | ≦0.125 |
| Chloramphenicol | ≧32 | 8 (24.2) | 8 | 8-64 | 8 | 8 | 1 | 8-128 | 21 (77.8) | 1-256 | 1-64 | 64 | 3 (60) | 4-64 |
| Minocycline | ≧16 | 8 (24.2) | 8 | 4-16 | 4->16 | ≦0.5 | ≦0.5 | ≦0.5 | 0 | ≦0.5-2 | 2 | ≦0.5 | 0 | ≦0.5 |
| Tetracyclinec or oxytetracyclined | ≧32 | 25 (75.8) | 64 | 32-64 | 32-128 | 4 | 1 | 0.25 | 17 (63.0) | 1->64 | 1-64 | ≦0.125 | 0 | 1 |
| Vancomycin | >4 | 0 | 0.5 | 0.5-2 | 1-2 | 1 | 1-2 | 0.5-1 | 0 | 0.25-2 | 1 | 0.25 | 0 | 1 |
| Teicoplanin | >8 | 0 | 0.5-1 | 0.5-4 | 1-2 | 2 | 0.5-2 | 0.5-1 | 0 | 0.5-2 | 0.5-4 | 0.5 | 0 | 0.5-1 |
| Ciprofloxacinc | ≧4 | 14 (100) | ≧128 | 16->128 | 16-64 | 16 | 11 (40.7) | 8-32 | ≦0.125 | 32 | 5 (100) | 16-32 | ||
| Enrofloxacin | ≧4 | 18 (94.7) | ≧128 | 32 | 1 | 8 | 14 (87.5) | 32-64 | <0.125-0.5 | |||||
MRSA, methicillin-resistant S. aureus; MRSP, methicillin-resistant S. pseudintermedius; MRSS, methicillin-resistant S. schleiferi; UT, untypeable.
Range of MICs for SCCmec types I, II, and IV with (+) and without (−) pUB110 and the kdp or pls gene. The number of isolates is shown in the parentheses after the SCCmec type.
Tetracycline and ciprofloxacin were used to test isolates obtained in 2008.
Oxytetracycline and enrofloxacin were used to test isolates obtained in 2007.
DISCUSSION
Our study revealed that two factors were independently associated with MRSA carriage: (i) contact with an identified animal MRSA case, and (ii) being an employee of a veterinary hospital or a nonclinical laboratory (versus being a student). It is suggested that animal patients spread MRSA infection among human individuals in veterinary hospitals. Contact with MRSA-infected animals has previously been identified as a risk factor among equine veterinarians (1); however, it has never been identified among companion-animal veterinarians and personnel attending a veterinary surgery conference (6, 12, 28). The higher risk of MRSA carriage among employees might be attributed to their frequency of contact with each animal patient, their closeness of contact during treatment, and the duration of their experience in terms of contact with animal patients.
Since contact with MRSA-infected animal patients is a factor considered to be associated with MRSA carriage, it can be inferred that MRSA carriers may originally have been infected by animal patients. However, a study conducted on 31 outpatient dogs in Japan reported that MRSA isolates were not detected in any of the dogs (38); similar previous studies have also reported that the prevalence of MRSA among dogs and cats was low (5, 31). Manian reported a case in which MRSA colonization was detected in a dog, which was believed to be the source of recurrent MRSA infection in its owner, a diabetes patient, and his wife; the dog was believed to have contracted the infection through contact with the owner (24). Another previous study reported MRSA colonization in a dog whose owner was a nurse caring for MRSA patients; it was concluded that the dog contracted the infection from the nurse, who was found to be a carrier of MRSA infection (50). Thus, human MRSA carriers can also be a source of MRSA infection in animals encountered in veterinary medical practice. In the present study, the MRSA isolate with SCCmec type IV and spa type t008 was obtained from a dog patient in March 2008 after one veterinarian was identified to be a carrier of MRSA spa type t008 on three different sampling occasions. The dog patient had undergone medical examinations in the clinical department to which the veterinarian belonged. This clone might, therefore, have spread from the veterinarian to the dog patient by direct contact or through the medium of contaminated environments.
In this study, the frequencies of MRSA infection among veterinarians and staff members in the veterinary hospital was found to be 19.5% (8/41) and 15.1% (8/53) in 2007 and 2008, respectively. These frequencies are comparable to the values reported in a previous study in the United Kingdom (17.9%) (21) and that reported in a study of veterinary personnel attending a veterinary surgery conference (17.3%) (6); however, these values were higher than those reported in a Japanese veterinary teaching hospital (10%) (38), in Scotland (3.9%) (13), and in Denmark (3.0%) (28). Moreover, the frequency of MRSA carriers has been reported to be only 0.54% among dog owners (5).
SCCmec type II was prevalent among the MRSA isolates obtained from individuals affiliated with the veterinary hospital and from dog patients. Most of the MRSA isolates with SCCmec type II also harbored spa type t002. A majority of the hospital-acquired MRSA (HA-MRSA) isolates identified in Japan also harbor SCCmec type II and are derived from the New York/Japan clone (multilocus sequence type 5 [ST5]) (17, 22, 36, 54). The MRSA of the Japan clone is generally known to have spa type t002 (http://www.spaserver.ridom.de/). MRSA isolates with SCCmec type II have been isolated from patients without established risk factors for HA-MRSA (36). Moreover, MRSA isolates with SCCmec type II have also been isolated from healthy children (14); thus, infection of this MRSA clone has already spread from hospital environments to a wider community, including veterinary staff and animal patients, in Japan.
Zaraket et al. reported that a MRSA clone with SCCmec type II has continued to colonize hospitals in Japan and has undergone genomic divergence (54). In the present study, all but two MRSA isolates with SCCmec type II had spa type t002; the remaining two isolates had spa type t062 and t1265. Unlike spa type t002, the spa type t062 has a deletion in the middle three repeats (34-17-20) of t002, whereas the spa type t1265 has a double repeat of r12 instead of r17. Moreover, the MRSA isolates of spa type t002 were grouped into seven clusters by PFGE, and the SCCmec types therein were classified into different subtypes. It is not known whether the SCCmec type II clone varies in veterinary medical practice or if some clones with a small genetic diversity have been introduced into the veterinary hospital.
The PFGE method can help divide MRSA isolates into more groups than spa typing, as shown in a previous study (44). Although spa types depend on the DNA sequence of only region X of the spa gene, PFGE profiles depend on the numbers and positions of the restriction sites in the DNA sequence of the entire genome. Therefore, PFGE enables better discrimination between strains. On the other hand, spa typing enabled comparison of the MRSA isolates obtained in this study with those reported in other studies. In some cases, spa typing could discriminate MRSA isolates of the same PFGE type (29). A combination of PFGE and spa typing was more useful than either method alone in discriminating MRSA isolates.
MRSA isolates with SCCmec type I were obtained only in 2008, and these isolates also had spa type t1767. Isolates with SCCmec type I are not commonly encountered in human hospitals; however, they are sometimes isolated from Japanese hospital inpatients at risk of HA-MRSA infection (36, 54). It has been reported that MRSA isolates harboring SCCmec type I obtained from inpatients belong to ST 8 or ST 30 strains (36, 54). There is little data regarding multilocus sequence analysis of MRSA having spa type t1767; however, the only MRSA with spa type t1767, which was isolated in Hong Kong, belonged to ST8 (http://www.spaserver.ridom.de/).
Although the 10 human MRSA carriers were treated to eradicate colonization, two of these carriers were found to be harboring MRSA isolates even after 1 month of treatment. Recurrent MRSA infection in a patient and healthy MRSA carriers has been reported previously (24, 50). The genotype of MRSA from a recurrent carrier changed from SCCmec type II without kdp gene and spa type t1265 to SCCmec type II harboring the kdp gene and spa type t002. Because the latter genotype was more common among the MRSA isolates obtained from veterinary staff and environmental surfaces, this carrier might be infected from other MRSA carriers or contaminated environmental surfaces. Treatment of MRSA carriers should be undertaken simultaneously with the disinfection of the entire hospital and implementation of hand hygiene practices, which has been reported as a strong protective factor against MRSA colonization (1).
MRSA was not detected in the five buccal mucosa samples of dogs owned by MRCPS-positive individuals. We chose buccal swab and not nasal swab, which is the traditional sample for MRSA isolation, because sampling from dogs was done at home by each owner without enough immobilization. Loeffler et al. reported that the isolation frequency of MRSA from the buccal swabs of dogs was as high as that from the nasal swabs (21). However, in another previous study, the nasal, axillary, and rectal swabs of dogs were tested for isolation MRSA, and the MRSA isolate was obtained only from the rectum (11). Combination sampling of the nasal, buccal, and rectal swabs obtained from dogs may facilitate detection of MRSA carriage.
In this investigation, we used the CHROMagar MRSA medium for the isolation of MRCPS. This medium has been reported to be highly effective in selecting for MRSA (10, 45). In a previous study describing the prevalence of MRSP, samples were directly inoculated onto mannitol-salt agar containing 2 mg/liter oxacillin and after the enrichment of staphylococci in broth containing 7.5% sodium chloride (11). In other studies, MRSP isolates were selected from staphylococcal strains by testing their susceptibility to oxacillin (38, 40). The CHROMagar MRSA medium used in this study contained 6 mg/liter cefoxitin. Although we did not determine the MICs of cefoxitin for the MRSP isolates obtained in this study, we found that approximately half of the MRSP isolates were susceptible to cefazolin. Sasaki et al. also reported that almost all MRSP isolates were susceptible to cefazolin and cefotiame (38). The higher susceptibility of MRSP to cephalosporin than MRSA might explain why the colony size of MRSP on the CHROMagar MRSA medium was considerably smaller than that of MRSA. On the other hand, even cefazolin-susceptible MRSP isolates could be obtained by using the CHROMagar MRSA medium in this study. In the present study, the frequency of MRSP isolated from individuals affiliated with the veterinary hospital (5.9%) was similar to that reported previously (38). Although it was reported that some MRSP isolates from dog patients had low-level resistance to oxacillin (MIC, 2 to 4 μg/ml) (38), all MRSP isolates obtained in this study showed high-level resistance to oxacillin (MIC, >16 μg/ml). The CHROMagar MRSA medium might be unable to detect MRSP that has low-level resistance to oxacillin.
Descloux et al. described canine MRSP isolates with a new SCCmec type II-III, resulting from a combination of S. aureus SCCmec type III and Staphylococcus epidermidis SCCmec type II (8). The structure of SCCmec in 23 MRSP isolates, as confirmed by M-PCR 4 and PCR 3, matched the DNA sequence of SCCmec type II-III (GenBank accession no. AM904732); therefore, the SCCmec type of these 23 MRSP isolates was presumed to be type II-III. There are only a limited number of reports pertaining to the SCCmec type among MRSP isolates (8, 30, 38); accordingly, more data regarding the SCCmec types among MRSP isolates are needed in order to understand the reasons for the sudden emergence of MRSP, although canine S. intermedius isolates, which were presumed to be S. pseudintermedius in the previous studies (3, 39), have been reported as being susceptible to β-lactam antibiotics in the past (41, 46).
Although contact with MRSA animal patients has also been recognized as a factor associated with MRSP carriage, the reason for this association is unknown. A previous study has suggested the transmission of multidrug-resistant S. intermedius species among dogs affected by deep pyoderma and their owners (9). Therefore, animal patients could transmit MRSP infection to humans.
Community-acquired MRSA (CA-MRSA) isolates are generally susceptible to most of the antimicrobial agents studied, except β-lactam antibiotics (32, 36, 49). Almost all MRSA and MRSP isolates investigated in this study were multidrug resistant but susceptible to the antibiotics used for treating MRSA infection in medical practice. All of the MRSP isolates were susceptible to imipenem and minocycline, although most of the MRSA isolates were resistant to these antibiotics. On the other hand, MRSS was susceptible to most antibiotics. Hisata et al. reported that MIC to oxacillin and imipenem differed between SCCmec types or subtypes of MRSA (14). The degree of susceptibilities to cefazolin and imipenem differed in the SCCmec types and bacterial species in this study.
It is necessary to obtain information regarding the prevalence of MRSA infection before implementing strategies for MRSA infection control in veterinary medical practice. The molecular and epidemiological analyses undertaken in the present study could provide some information regarding the transmission of MRSA between humans and companion animals in a veterinary hospital. MRSA carriers can be a source of MRSA infection in animals. MRSA animal patients may then, in turn, lead to MRSA infection in other individuals. Moreover, MRSA contamination was detected in the veterinary hospital environment. MRSA contamination in veterinary hospitals has been reported in previous studies (13, 21). These observations suggest that there is a dynamic transmission cycle of MRSA in veterinary hospitals similar to that occurring in human hospital environments.
Acknowledgments
We thank the staff and students of Rakuno Gakuen University for providing samples.
This work was supported in part by a grant-in-aid from the Ministry of Health, Labor and Welfare (H21-Shokuhin-Ippan-013) and the Food Safety Commission of the Cabinet Office (0707).
Footnotes
Published ahead of print on 11 June 2010.
REFERENCES
- 1.Anderson, M. E. C., S. L. Lefebvre, and J. S. Weese. 2008. Evaluation of prevalence and risk factors for methicillin-resistant Staphylococcus aureus colonization in veterinary personnel attending an international equine veterinary conference. Vet. Microbiol. 129:410-417. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannoehr, J., N. L. Ben Zakour, A. S. Waller, L. Guardabassi, K. L. Thoday, A. H. M. van den Broek, and J. R. Fitzgerald. 2007. Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J. Bacteriol. 189:8685-8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannoehr, J., A. Franco, M. Iurescia, A. Battisti, and J. R. Fitzgerald. 2009. Molecular diagnostic identification of Staphylococcus pseudintermedius. J. Clin. Microbiol. 47:469-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boost, M. V., M. M. O'Donoghue, and K. H. G. Siu. 2007. Characterisation of methicillin-resistant Staphylococcus aureus isolates from dogs and their owners. Clin. Microbiol. Infect. 13:731-733. [DOI] [PubMed] [Google Scholar]
- 6.Burstiner, L. C., M. Faires, and J. S. Weese. 2010. Methicillin-resistant Staphylococcus aureus colonization in personnel attending a veterinary surgery conference. Vet. Surg. 39:150-157. [DOI] [PubMed] [Google Scholar]
- 7.CLSI. 2008. Performance standards for antimicrobial disk dilution susceptibility tests for bacteria isolated from animals; approved standard M31-A3, 3rd ed., vol. 28, no. 8. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Descloux, S., A. Rossano, and V. Perreten. 2008. Characterization of new staphylococcal cassette chromosome mec (SCCmec) and topoisomerase genes in fluoroquinolone- and methicillin-resistant Staphylococcus pseudintermedius. J. Clin. Microbiol. 46:1818-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guardabassi, L., M. E. Loeber, and A. Jacobson. 2004. Transmission of multiple antimicrobial-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet. Microbiol. 98:23-27. [DOI] [PubMed] [Google Scholar]
- 10.Han, Z., E. Lautenbach, N. Fishman, and I. Nachamkin. 2007. Evaluation of mannitol salt agar, CHROMagar Staph aureus and CHROMagar MRSA for detection of meticillin-resistant Staphylococcus aureus from nasal swab specimens. J. Med. Microbiol. 56:43-46. [DOI] [PubMed] [Google Scholar]
- 11.Hanselman, B. A., S. Kruth, and J. S. Weese. 2008. Methicillin-resistant staphylococcal colonization in dogs entering a veterinary teaching hospital. Vet. Microbiol. 126:277-281. [DOI] [PubMed] [Google Scholar]
- 12.Hanselman, B. A., S. A. Kruth, J. Rousseau, D. E. Low, B. M. Willey, A. McGeer, and J. S. Weese. 2006. Methicillin-resistant Staphylococcus aureus colonization in veterinary personnel. Emerg. Infect. Dis. 12:1933-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heller, J., S. K. Armstrong, E. K. Girvan, S. W. J. Reid, A. Moodley, and D. J. Mellor. 2009. Prevalence and distribution of meticillin-resistant Staphylococcus aureus within the environment and staff of a university veterinary clinic. J. Small Anim. Pract. 50:168-173. [DOI] [PubMed] [Google Scholar]
- 14.Hisata, K., K. Kuwahara-Arai, M. Yamanoto, T. Ito, Y. Nakatomi, L. Cui, T. Baba, M. Terasawa, C. Sotozono, S. Kinoshita, Y. Yamashiro, and K. Hiramatsu. 2005. Dissemination of methicillin-resistant staphylococci among healthy Japanese children. J. Clin. Microbiol. 43:3364-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hososaka, Y., H. Hanaki, C. Yanagisawa, Y. Yamaguchi, H. Matsui, T. Nakae, S. Iwata, I. Hayashi, and K. Sunakawa. 2006. Nosocomial infection of β-lactam antibiotic-induced vancomycin-resistant Staphylococcus aureus (BIVR). J. Infect. Chemother. 12:181-184. [DOI] [PubMed] [Google Scholar]
- 16.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi, K., N. Takahashi, C. Piao, K. Totsuka, H. Nishida, and T. Uchiyama. 2003. Molecular epidemiology of methicillin-resistant Staphylococcus aureus strains causing neonatal toxic shock syndrome-like exanthematous disease in neonatal and perinatal wards. J. Clin. Microbiol. 41:3001-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwok, A. Y., and A. W. Chow. 2003. Phylogenetic study of Staphylococcus and Macrococcus species based on partial hsp60 gene sequences. Int. J. Syst. Evol. Microbiol. 53:87-92. [DOI] [PubMed] [Google Scholar]
- 20.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 21.Loeffler, A., A. K. Boag, J. Sung, J. A. Lindsay, L. Guardabassi, A. Dalsgaard, H. Smith, K. B. Stevens, and D. H. Lloyd. 2005. Prevalence of methicillin-resistant Staphylococcus aureus among staff and pets in a small animal referral hospital in the UK. J. Antimicrob. Chemother. 56:692-697. [DOI] [PubMed] [Google Scholar]
- 22.Ma, X. X., T. Ito, P. Chongtrakool, and K. Hiramatsu. 2006. Predominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J. Clin. Microbiol. 44:4515-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik, S., G. W. Coombs, F. G. O'Brien, H. Peng, and M. D. Barton. 2006. Molecular typing of methicillin-resistant staphylococci isolated from cats and dogs. J. Antimicrob. Chemother. 58:428-431. [DOI] [PubMed] [Google Scholar]
- 24.Manian, F. A. 2003. Asymptomatic nasal carriage of mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MRSA) in a pet dog associated with MRSA infection in household contacts. Clin. Infect. Dis. 36:e26-e28. [DOI] [PubMed] [Google Scholar]
- 25.McLean, C. L., and M. G. Ness. 2008. Meticillin-resistant Staphylococcus aureus in a veterinary orthopaedic referral hospital: staff nasal colonisation and incidence of clinical cases. J. Small Anim. Pract. 49:170-177. [DOI] [PubMed] [Google Scholar]
- 26.Middleton, J. R., W. H. Fales, C. D. Luby, J. L. Oaks, S. Sanchez, J. M. Kinyon, C. C. Wu, C. W. Maddox, R. D. Welsh, and F. Hartmann. 2005. Surveillance of Staphylococcus aureus in veterinary teaching hospitals. J. Clin. Microbiol. 43:2916-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moodley, A., E. C. Nightingale, M. Stegger, S. S. Nielsen, R. L. Skov, and L. Guardabassi. 2008. High risk for nasal carriage of methicillin-resistant Staphylococcus aureus among Danish veterinary practitioners. Scand. J. Work Environ. Health 34:151-157. [DOI] [PubMed] [Google Scholar]
- 29.Moodley, A., M. Stegger, A. F. Bagcigil, K. E. Baptiste, A. Loeffler, D. H. Lloyd, N. J. Williams, N. Leonard, Y. Abbott, R. Skov, and L. Guardabassi. 2006. spa typing of methicillin-resistant Staphylococcus aureus isolated from domestic animals and veterinary staff in the UK and Ireland. J. Antimicrob. Chemother. 58:1118-1123. [DOI] [PubMed] [Google Scholar]
- 30.Moodley, A., M. Stegger, N. L. Ben Zakour, J. R. Fitzgerald, and L. Guardabassi. 2009. Tandem repeat sequence analysis of staphylococcal protein A (spa) gene in methicillin-resistant Staphylococcus pseudintermedius. Vet. Microbiol. 135:320-326. [DOI] [PubMed] [Google Scholar]
- 31.Morris, D. O., K. A. Rook, F. S. Shofer, and S. C. Rankin. 2006. Screening of Staphylococcus aureus, Staphylococcus intermedius, and Staphylococcus schleiferi isolates obtained from small companion animals for antimicrobial resistance: a retrospective review of 749 isolates (2003-04). Vet. Dermatol. 17:332-337. [DOI] [PubMed] [Google Scholar]
- 32.Naimi, T. S., K. H. LeDell, D. J. Boxrud, A. V. Groom, C. D. Steward, S. K. Johnson, J. M. Besser, C. O'Boyle, R. N. Danila, J. E. Cheek, M. T. Osterholm, K. A. Moore, and K. E. Smith. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996-1998. Clin. Infect. Dis. 33:990-996. [DOI] [PubMed] [Google Scholar]
- 33.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Mahony, R., Y. Abbott, F. C. Leonard, B. K. Markey, P. J. Quinn, P. J. Pollock, S. Fanning, and A. S. Rossney. 2005. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from animals and veterinary personnel in Ireland. Vet. Microbiol. 109:285-296. [DOI] [PubMed] [Google Scholar]
- 35.Paule, S. M., A. C. Pasquariello, D. M. Hacek, A. G. Fisher, R. B. Thomson, Jr., K. L. Kaul, and L. R. Peterson. 2004. Direct detection of Staphylococcus aureus from adult and neonate nasal swab specimens using real-time polymerase chain reaction. J. Mol. Diagn. 6:191-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piao, C., T. Karasawa, K. Totsuka, T. Uchiyama, and K. Kikuchi. 2005. Prospective surveillance of community-onset and health care-associated methicillin-resistant Staphylococcus aureus isolated from a university-affiliated hospital in Japan. Microbiol. Immunol. 49:959-970. [DOI] [PubMed] [Google Scholar]
- 37.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki, T., K. Kikuchi, Y. Tanaka, N. Takahashi, S. Kamata, and K. Hiramatsu. 2007. Methicillin-resistant Staphylococcus pseudintermedius in a veterinary teaching hospital. J. Clin. Microbiol. 45:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki, T., K. Kikuchi, Y. Tanaka, N. Takahashi, S. Kamata, and K. Hiramatsu. 2007. Reclassification of phenotypically identified Staphylococcus intermedius strains. J. Clin. Microbiol. 45:2770-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz, S., K. Kadlec, and B. Strommenger. 2008. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius detected in the BfT-GermVet monitoring programme 2004-2006 in Germany. J. Antimicrob. Chemother. 61:282-285. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu, A., Y. Wakita, S. Nagase, M. Okabe, T. Koji, T. Hayashi, N. Nagase, A. Sasaki, J. Kawano, K. Yamashita, and M. Takagi. 2001. Antimicrobial susceptibility of Staphylococcus intermedius isolated from healthy and diseased dogs. J. Vet. Med. Sci. 63:357-360. [DOI] [PubMed] [Google Scholar]
- 42.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shore, A., A. S. Rossney, C. T. Keane, M. C. Enright, and D. C. Coleman. 2005. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 49:2070-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strommenger, B., C. Kehrenberg, C. Kettlitz, C. Cuny, J. Verspohl, W. Witte, and S. Schwarz. 2006. Molecular characterization of methicillin-resistant Staphylococcus aureus strains from pet animals and their relationship to human isolates. J. Antimicrob. Chemother. 57:461-465. [DOI] [PubMed] [Google Scholar]
- 45.Taguchi, H., T. Kaneko, M. Onozaki, R. Kubo, and S. Kamiya. 2004. Evaluation of a new chromogenic medium for isolation of MRSA. Kansenshogaku Zasshi. 78:54-58. [DOI] [PubMed] [Google Scholar]
- 46.Talan, D. A., D. Staatz, A. Staatz, E. J. Goldstein, K. Singer, and G. D. Overturf. 1989. Staphylococcus intermedius in canine gingiva and canine-inflicted human wound infections: laboratory characterization of a newly recognized zoonotic pathogen. J. Clin. Microbiol. 27:78-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuchizaki, N., J. Ishikawa, and K. Hotta. 2000. Colony PCR for rapid detection of antibiotic resistance genes in MRSA and enterococci. Jpn. J. Antibiot. 53:422-429. [PubMed] [Google Scholar]
- 48.Ubukata, K., R. Nonoguchi, M. Matsuhashi, and M. Konno. 1989. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J. Bacteriol. 171:2882-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Duijkeren, E., M. J. H. M. Wolfhagen, A. T. A. Box, M. E. O. C. Heck, W. J. B. Wannet, and A. C. Fluit. 2004. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 10:2235-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Duijkeren, E., M. J. H. M. Wolfhagen, M. E. O. C. Heck, and W. J. B. Wannet. 2005. Transmission of a Panton-Valentine leucocidin-positive, methicillin-resistant Staphylococcus aureus strain between humans and a dog. J. Clin. Microbiol. 43:6209-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wettstein, K., S. Descloux, A. Rossano, and V. Perreten. 2008. Emergence of methicillin-resistant Staphylococcus pseudintermedius in Switzerland: three cases of urinary tract infections in cats. Schweiz. Arch. Tierheilkd. 150:339-343. [DOI] [PubMed] [Google Scholar]
- 53.Wulf, M., A. van Nes, A. Eikelenboom-Boskamp, J. de Vries, W. Melchers, C. Klaassen, and A. Voss. 2006. Methicillin-resistant Staphylococcus aureus in veterinary doctors and students, the Netherlands. Emerg. Infect. Dis. 12:1939-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaraket, H., T. Otsuka, K. Saito, S. Dohmae, T. Takano, W. Higuchi, T. Ohkubo, K. Ozaki, M. Takano, I. Reva, T. Baranovich, and T. Yamamoto. 2007. Molecular characterization of methicillin-resistant Staphylococcus aureus in hospitals in Niigata, Japan: divergence and transmission. Microbiol. Immunol. 51:171-176. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]