Abstract
Three functional NiFe hydrogenases were previously characterized in Thiocapsa roseopersicina BBS: two of them are attached to the periplasmic membrane (HynSL and HupSL), and one is localized in the cytoplasm (HoxEFUYH). The ongoing genome sequencing project revealed the presence of genes coding for another soluble Hox-type hydrogenase enzyme (hox2FUYH). Hox2 is a heterotetrameric enzyme; no indication for an additional subunit was found. Detailed comparative in vivo and in vitro activity and expression analyses of HoxEFUYH (Hox1) and the newly discovered Hox2 enzyme were performed. Functional differences between the two soluble NiFe hydrogenases were disclosed. Hox1 seems to be connected to both sulfur metabolism and dark/photofermentative processes. The bidirectional Hox2 hydrogenase was shown to be metabolically active under specific conditions: it can evolve hydrogen in the presence of glucose at low sodium thiosulfate concentration. However, under nitrogen-fixing conditions, it can oxidize H2 but less than the other hydrogenases in the cell.
Hydrogenases are metalloenzymes involved in microbial hydrogen metabolism. A great variety of them have been identified and studied in various microorganisms and grouped on the basis of their metal content as NiFe, FeFe, and iron-sulfur cluster free hydrogenases (10, 42, 43). The basic protein structure of NiFe hydrogenases is heterodimeric, while FeFe hydrogenases are mostly composed of a single amino acid chain with multiple iron-sulfur clusters (28, 43, 44). Well-defined maturation proteins assist for the assembly and activation of hydrogenase enzymes; NiFe hydrogenases require a more complex accessory machinery than FeFe enzymes (2, 3, 24).
Thiocapsa roseopersicina BBS is a photosynthetic purple sulfur bacterium belonging to the Chromatiaceae family (4). It prefers to utilize reduced sulfur compounds for anaerobic photochemolithoautotrophic growth, but simple organic substrates such as glucose or acetate can be also used as extra carbon, energy, and electron sources. It can be cultivated under aerobic (nonphotosynthetic) conditions in the presence of organic compounds. In the absence of other nitrogen sources, it is able to fix molecular nitrogen; this process is accompanied by H2 production. T. roseopersicina was earlier shown to possess at least three NiFe hydrogenases varying in their in vivo functions, localizations, and compositions. Hyn and Hup hydrogenases are attached to the membrane facing the periplasmic side (6, 18, 30). Hyn is a bidirectional enzyme with extraordinary stability (17). Recent study has demonstrated that the HynSL subunits are physiologically connected to cellular redox processes via the Isp1 and Isp2 proteins, which play an essential role in electron transfer (27). The second membrane-associated enzyme, Hup, is involved in H2 oxidation and shows homology to uptake hydrogenases, which recycle H2 produced by the nitrogenase enzyme complex or present in the environment. Next to the hydrogenase small and large subunits (HupSL), a b-type cytochrome, HupC, was demonstrated to be part of the in vivo active enzyme as a transmitter of electrons to the quinone pool (27). In several bacteria, e.g., Rhodobacter capsulatus (7) and Ralstonia eutropha (15, 20), the expression of the hydrogenase(s) was shown to be regulated by the hydrogen level in the environment. The genes encoding the hydrogen-sensing system also exist in T. roseopersicina (hupUV, hupT, and hupR), but the hupTUV genes proved to be silent in the wild-type strain—only hupR is expressed—which is why expression of hupSL genes is constitutive (16).
A Hox-type soluble hydrogenase was also identified in T. roseopersicina (31); it is a representative of the bidirectional heteromultimeric cytoplasmic NiFe hydrogenases (37, 39). Enzymes belonging to this group are basically composed of two moieties: hydrogenase (HoxYH) and diaphorase (HoxFU) heterodimers. Additional subunits were identified in few cases. In R. eutropha H16, two HoxI proteins completing the Hox complex were suggested to provide a binding domain for NADPH (5). HoxE has been identified as the fifth subunit of heteropentameric NAD+-reducing Hox hydrogenases in several cyanobacteria, Allochromatium vinosum and T. roseopersicina (21, 31, 37). In-frame deletion of the hoxE gene ceased both the H2-producing and -oxidizing activities of Hox in vivo, but these were not affected in vitro. Consequently, an electron transfer role of the HoxE subunit was suggested (31, 32).
The possibility of the presence of further hydrogenases in T. roseopersicina was noted few years ago (31). In the hynSL hupSL hoxH triple-mutant strain (GB112131), a small in vivo and in vitro hydrogenase activity could be measured under photomixotrophic growth conditions (both CO2 and organic compounds are used for growth) at the late growth phase. This residual activity could not be detected in the hypF mutant strain (M539). Since HypF protein has an essential role in the maturation process of all NiFe hydrogenases (9), these results suggested the presence of a previously unknown hydrogenase. Here we describe the identification and characterization of the second Hox-type hydrogenase, emphasizing the functional similarities and differences between the two soluble enzymes of this bacterium. In order to distinguish between the two Hox-type enzymes unequivocally, the HoxEFUYH complex will be renamed Hox1 and the newly described Hox2FUYH enzyme is called Hox2.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
Strains and plasmids are listed in Table 1; primers can be found in Table 2. T. roseopersicina strains were maintained in Pfennig's mineral medium (29). Pfennig's medium was modified by changing the sodium thiosulfate (Na2S2O3, anhydrous form) concentration (media PC4 and PC2, containing 4 g liter−1 and 2 g liter−1 sodium thiosulfate, respectively) and by supplementing the medium with various organic substrates (glucose [media PC4G and PC2G], sodium pyruvate, sodium acetate, sucrose, sodium formate, sodium succinate, sodium lactate, and sodium fumarate, all added at 5 g liter−1). Under nitrogen-fixing conditions, the NH4Cl was omitted (NC medium and NC medium designated as described above according to the thiosulfate and glucose content). Cells were grown anaerobically in liquid cultures illuminated with continuous light using incandescent light bulbs of 60 W or in the dark at 27 to 30°C. Plates were supplemented with sodium acetate (2 g liter−1) and solidified with Phytagel (7 g liter−1). Plates were incubated in anaerobic jars for 2 weeks. E. coli strains were maintained on LB agar plates (34). Antibiotics were used in the following concentrations: for E. coli, 100 μg ml−1 ampicillin and 25 μg ml−1 kanamycin; for T. roseopersicina, 5 μg ml−1 gentamicin, 25 μg ml−1 kanamycin, 5 μg ml−1 streptomycin, and 50 μg ml−1 erythromycin.
TABLE 1.
Characteristics of the strains and vectors used in this study
| Strain or vector | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| Thiocapsa roseopersicina | ||
| BBS | Wild type | 4 |
| M539 | BBS hypF::miniTn5 Kmr | 9 |
| GB112131 | hynSL::SmrhupSL::GmrhoxH::Emr (Emr) oriented as hox operon | 31 |
| GB1121 | hynSL::SmrhupSL::Gmr | 31 |
| GB11213141 | hynSL::SmrhupSL::GmrhoxH::Emr ΔhoxH2 | This work |
| GB112141 | hynSL::SmrhupSL::Gmr ΔhoxH2 | This work |
| GB11213141/pMHE6hoxH2c | hynSL::SmrhupSL::GmrhoxH::Emr ΔhoxH2 harboring pMHE6hoxH2c | This work |
| E. coli | ||
| S17-1(λpir) | 294 (recA pro res mod) Tpr Smr (pRP4-2-Tc::Mu-Km::Tn7) λpir | 12 |
| XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| Vectors | ||
| pMHE6crtKm | Broad-host-range cloning vector | 8 |
| pBluescript SK(+) | Cloning vector; Ampr | Stratagene |
| pBhox2 | 6,038-bp EcoRV-HindIII fragment cloned into pBluescript SK(+) | This work |
| pK18mobsacB | sacB RP4 oriT ColE1 ori Kmr | 35 |
| phoxH2_2 | Up-and downstream regions of hoxH2 in pK18mobsacB; Kmr | This work |
| pMHE6hoxH2c | 1,484-bp NdeI-HindIII fragment in pMHE6crtKm; Kmr | This work |
TABLE 2.
Oligonucleotides used in this study
| Primer name | 5′→3′ sequence |
|---|---|
| ohox225 | 5′-GTCTCCAGATTCTTAGTCATG-3′ |
| ohox226 | 5′-CATCCTGCAGCTGGTCGATC-3′ |
| hox2forwSal | 5′-ATCGTATCGTCGACAGTCCATCCGCCGCGTTGCG-3′ |
| hox2revHind | 5′-CAACGTCAAAGCTTTCGGCACCGTCGTCCATAAC-3′ |
| ohoxH2c1 | 5′-GAACGAGCCATATGACTAAGAATCTGGAGACC-3′ |
| ohoxH2c2 | 5′-CGTATATCAAGCTTGGCGTCGATCGAACCGTC-3′ |
| OHOXHQRT | 5′-GTTGTTGTTGGTGGTGGACA-3′ |
| OHOX2FFW | 5′-GGAATACGACCTGAGCGAGATG-3′ |
| OHOX2FREV | 5′-GGAATTTGTCGAGCGTGTTGA-3′ |
| OHOX2UFW | 5′-GCACTTCACCCACTTCTTCC-3′ |
| OHOX2UREV | 5′-GTCGATCACCAGATGGCTCT-3′ |
| OHOX2YFW | 5′-GCCGAGAATGTCGAAGTGTT-3′ |
| OHOX2YREV | 5′-GTAGTCGATCGCAACCACCT-3′ |
| OHOX2HFW | 5′-TCCACGAGGAGATCAAGTCC-3′ |
| OHOX2HREV | 5′-CAGTGCAGCAACTCGATCAT-3′ |
| OHOX1FFW | 5′-GGTGTATGGGCCTATGTTCG-3′ |
| OHOX1FREV | 5′-TGATTGGTCGGACAACGTAA-3′ |
| OHOX1UFW | 5′-GATGCAGATCCAGACCAACA-3′ |
| OHOX1UREV | 5′-GGTAGCTCGCCTGACGATAG-3′ |
| OHOX1YFW | 5′-GGCTGTCACATGTCCTTCCT-3′ |
| OHOX1YREV | 5′-ACCAGGATCTTGCAGTGCTT-3′ |
| OHOX1HFW | 5′-CCGTCGAGGACTTCAGCTAC-3′ |
| OHOX1HREV | 5′-GACCAGTCGGGCATAGTGAT-3′ |
| RRN01 | 5′-GCAACGCGAAGAACCTTACC-3′ |
| RRN02 | 5′-CCAAGGCATCTCTGCCAAGT-3′ |
Isolation of the hox2 gene cluster in T. roseopersicina.
A BLAST search was performed in our local T. roseopersicina genome data bank using the hox1H gene as the query sequence. A genomic locus was identified which harbored putative genes coding for proteins similar to the HoxFUYH subunits of the soluble NAD+-reducing hydrogenases. No other hydrogenase-related gene could be identified in this locus. The genomic locus of hox2W was found similarly.
Deletion of the hoxH2 gene.
The deletion construct was derived from the pK18mobsacB vector (35). The upstream region of the hox2H gene was amplified with the ohox225 and ohox226 primers, and the 742-bp PCR product was cloned into the SmaI-digested pK18mobsacB (phoxH2_1). The downstream region was amplified with the hox2forwSal and the hox2revHind primers and digested with SalI-HindIII enzymes. The 766-bp fragment was cloned into the SalI-HindIII-digested phoxH2_1, yielding phoxH2_2. The plasmid was transformed into E. coli S17-1(λpir) and then conjugated into the next T. roseopersicina mutant strains: GB1121 and GB112131. Single and double recombinants were selected based on kanamycin resistance and the sacB positive selection system (35), yielding the following mutant strains: GB112141 and GB11213141 (Table 1).
ΔhoxH2 complementation.
The hox2H gene was amplified with the ohoxH2c1 and the ohoxH2c2 primers containing NdeI and HindIII restriction sites. The PCR product was digested with NdeI-HindIII enzymes, and the 1,484-bp fragment was cloned into the corresponding sites of pMHE6crtKm vector, resulting in pMHE6hox2c. The plasmid was transformed into E. coli S17-1(λpir) and conjugated into T. roseopersicina strain GB11213141.
RNA isolation and DNase I treatment.
T. roseopersicina liquid cell cultures (4 ml at different stages of growth phase) were pelleted and resuspended in 200 μl phosphate buffer (10 mM, pH 7.0). RNA was isolated and treated with DNase I using the RiboPure Bacteria kit (Ambion) following the manufacturer's protocol.
RT, RT-PCR, and qPCR.
Reverse transcription (RT) was performed with TaqMan reverse transcription reagents (ABi and Roche) using either specific primers or random hexamers for the cDNA synthesis. RT-coupled PCR experiments were carried out as previously described (9). In order to determine the organization of the hox2 genes, the reverse transcription was initiated at primer OHOXHQRT located at the C terminus of the hox2H gene. For the expression analysis of hox2 genes in GB112131 strain, the reverse transcription was initiated either at OHOXHQRT or by using random hexamers. Amplifications were performed with the following primers (listed in Table 2): OHOX2FFW and OHOX2FREV located in the hox2F gene, OHOX2UFW and OHOX2UREV located in the hox2U gene, OHOX2YFW and OHOX2YREV located in the hox2Y gene, and OHOX2HFW and OHOX2HREV located in the hox2H gene. Quantitative PCR (qPCR) experiments were performed using the ABI Step One Plus PCR machine. The levels of expression of hox2 genes (hox2F with primers OHOX2FFW and OHOX2FREV, hox2U with primers OHOX2UFW and OHOX2UREV, hox2Y with primers OHOX2YFW and OHOX2YREV, and hox2H with primers OHOX2HFW and OHOX2HREV), hox1 genes (hox1F with primers OHOX1FFW and OHOX1FREV, hox1U with primers OHOX1UFW and OHOX1UREV, hox1Y with primers OHOX1YFW and OHOX1YREV, and hox1H with primers OHOX1HFW and OHOX1HREV) and 16S rRNA genes (with primers RRN01 and RRN02 of T. roseopersicina) were analyzed in various samples. For the comparative analysis of hox1 and hox2 operons in strain BBS, random hexamers were used for initiation of reverse transcription. At least three biological and three technical replicates were used for each analysis.
Measurement of in vivo hydrogen evolution activity.
T. roseopersicina cultures (60 ml) were grown in various Pfennig media (PC4, PC2, PC4G, and PC2G) under a nitrogen atmosphere in sealed 100-ml Hypo-Vial glasses. Anaerobiosis was established by flushing the gas phase with N2 for 10 min. H2 production was monitored by injecting 200-μl samples of the headspace into a gas chromatograph (Agilent 6890 thermal conductivity detector [TCD]) on each day of growth, starting on the 5th day and ending on the 15th day. Five replicates were used for each in vivo hydrogen evolution measurement.
In vivo hydrogen uptake activity measurements.
T. roseopersicina cells (60 ml) were cultured in sealed vials with a volume of 100 ml (Pierce) in Pfennig medium containing 2 g liter−1 thiosulfate and supplemented with 5 g liter−1 glucose (NC2G) but lacking ammonium chloride under nitrogen atmosphere. H2 production was monitored by injecting 200-μl samples of the headspace into a gas chromatograph (Agilent 6890 TCD detector) on each day of growth, starting on the 5th day and ending on the 15th day. Hydrogen uptake was calculated from the total amounts of hydrogen produced by the nitrogenase complex in cells containing or lacking the hydrogenase of interest. Three replicates were used for each in vivo hydrogen uptake measurement.
Preparation of membrane-associated and soluble protein fractions of T. roseopersicina.
T. roseopersicina culture (60 ml) was harvested by centrifugation at 7,000 × g. The cells were suspended in 3 ml of 20 mM potassium phosphate buffer (pH 7.0) and sonicated 8 times for 10 s on ice using 15 W of power with a mechanical amplitude of 50 μm. The broken cells were centrifuged at 10,000 × g for 15 min. The debris (remaining whole cells and sulfur crystals) was discarded, and the supernatant was centrifuged at 100,000 × g for 3 h. The pellet was washed twice with 20 mM potassium phosphate buffer (pH 7.0) and used as membrane fraction. The supernatant was considered as the soluble fraction.
Measurement of in vitro MV-dependent hydrogen evolution activity.
T. roseopersicina cultures (60 ml, grown in PC2G medium in sealed 100-ml Hypo-Vials flushed with N2) were harvested, and the soluble fraction was prepared as described above. The soluble fraction (200 μl) was used for the measurement, and 1.76 ml 20 mM potassium phosphate buffer (pH 7.0) and 40 μl 40 mM methyl viologen (MV) were added. The mixture was flushed with nitrogen for 10 min, and the reaction was initiated by injection of 100 μl anaerobic 50 mg ml−1 sodium dithionite. Samples were shaken gently at room temperature for 1 h, and the hydrogen content of the gas phase was determined by gas chromatograph. Three replicates were used for each measurement of in vitro MV-dependent hydrogen evolution.
Measurement of in vitro NADH/NADPH-dependent hydrogen evolution activity.
T. roseopersicina cultures (60 ml, grown in PC2G medium in sealed 100-ml Hypo-Vials flushed with N2) were harvested, and the soluble fraction was prepared as described above. To the soluble fraction (500 μl) used for the measurement, 1.46 ml of 20 mM potassium phosphate buffer (pH 7.0) containing 4 mM dithiothreitol (DTT) and 20 μl 200 mM flavin mononucleotide (FMN) was added. The mixture was flushed with nitrogen for 10 min and incubated at 37°C for 1 h. The reaction was initiated by injecting 20 μl 120 mM anaerobic NADH or NADPH. Samples were shaken gently at 37°C for 6 h, and the hydrogen content of the headspace was tested by injecting 500-μl samples into the gas chromatograph in every 60 min. Three replicates were used for each measurement of in vitro NADH/NADPH-dependent hydrogen evolution.
Measurement of in vitro hydrogen uptake activity.
T. roseopersicina cultures (60 ml, grown in PC2G medium in sealed 100-ml Hypo-Vials flushed with N2) were harvested, and the soluble fraction was prepared as described above. To the soluble fraction (200 μl) used in the measurement, 1.76 ml of 20 mM potassium phosphate buffer (pH 7.0) and 40 μl of 40 mM benzyl viologen (BV) were added. The mixture was flushed with nitrogen for 5 min, followed by flushing with 100% hydrogen for another 5 min. Samples were incubated at 60°C in the spectrophotometer, and measurement of the rate of hydrogenase activity was performed by monitoring the absorbance at 600 nm over time. Three replicates were used for each in vitro hydrogen uptake measurement.
Determination of glucose content.
Changes in glucose concentration were monitored by two approaches. Besides the dinitrosalicylic acid (DNSA) method (25), we applied a simple tool developed for blood sugar tests (GlucoVal). The device was calibrated for Pfennig medium containing glucose (dilution series for glucose), and the measurement proved to be accurate for Pfennig medium containing 0 to 5 g liter−1 glucose. Three parallel samples were measured in each case, and three repetitions were done for each sample.
Determination of thiosulfate content.
Samples with a volume of 1 ml were taken from 60-ml T. roseopersicina cultures on each day of growth. Cell densities were measured at 600 nm spectrophotometrically, and then 1-ml samples were pelleted by centrifugation at 10,000 × g for 5 min. Supernatant was used for thiosulfate determination of the medium. Thiosulfate was identified clearly by UV absorption at 230 nm using quartz cuvettes. The calibration curve for thiosulfate was linear for PCG medium supplemented with 0 to 1.5 g liter−1 thiosulfate. Absorbance values were normalized with cell densities.
Nucleotide sequence accession numbers.
The sequences determined in this study were submitted to GenBank under accession no. GU560006 (hox2FUYH locus) and GU560007 (hox2W locus).
RESULTS
Discovery of the Hox2 hydrogenase in T. roseopersicina.
The triple-mutant T. roseopersicina strain GB112131 retained small but reproducible in vivo hydrogen-producing capability under specific photomixotrophic conditions (PC2G; Pfennig medium containing 2 g liter−1 sodium thiosulfate instead of the standard 4 g liter−1 and 2 g liter−1 sodium hydrogen carbonate and supplemented with glucose). This residual activity could not be observed in the HypF− strain, which implied the presence of an additional functional NiFe hydrogenase in T. roseopersicina. Mining in the genome sequence database of T. roseopersicina (unpublished data) revealed the presence of open reading frames (ORFs) coding for putative proteins showing significant homology to the subunits of bidirectional heteromultimeric cytoplasmic NiFe hydrogenases (42, 43). Since a member of this group has already been identified and characterized in this organism (Hox1) (31), the newly found genes were labeled hox2. The sequence of the deduced Hox2FUYH unambiguously differed from that of the corresponding subunits of the T. roseopersicina Hox1 enzyme: the identity and similarity values between Hox1H and Hox2H subunits were 46% and 63%. The subunits of the Hox2 enzyme show highest similarity to the corresponding subunits of the soluble hydrogenase of Methylococcus capsulatus (Bath) (11, 45) (Table 3). Mining in the genome sequence revealed no coding sequence for additional hydrogenase subunits in the vicinity of the hox2FUYH genes. However, one—so far unknown—ORF resembling the maturation endopeptidase genes was identified in a distant genomic region. Since the coding sequences of all known NiFe hydrogenase-related endopeptidases in T. roseopersicina are well characterized (23), the newly identified sequence was supposed to play a role in the maturation process (cleavage of the C-terminal extension) of the Hox2H subunit and thereby was named “hox2W.”
TABLE 3.
Relative identity values between Hox subunits of various organisms
| Organism | % identity to T. roseopersicina Hox gene subunit: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Hox1 |
Hox2 |
||||||||
| E | F | U | Y | H | 2F | 2U | 2Y | 2H | |
| Ralstonia eutropha H16 | 35 | 33 | 45 | 40 | 44 | 41 | 48 | 47 | |
| M. capsulatus Bath | 38 | 33 | 49 | 46 | 56 | 63 | 65 | 66 | |
| Synechococcus PCC 7002 | 53 | 60 | 57 | 46 | 51 | 28 | 38 | 43 | 42 |
| Nostoc PCC 7422 | 55 | 61 | 58 | 49 | 54 | 28 | 33 | 41 | 43 |
| T. roseopersicina Hox2 strain | 28 | 31 | 49 | 46 | 100 | 100 | 100 | 100 | |
Reverse transcription-coupled PCR was performed to examine whether the hox2FUYH genes form a single operon. Reverse transcription was initiated at hox2H, and specific PCRs on hox2FUYH genes revealed the presence of transcript covering all four genes (data not shown).
Mutant analysis.
Deletion of the hox2H gene was performed in order to examine whether Hox2 is responsible for the hydrogenase activity observed in GB112131. Mutation of hox2H in this strain clearly eliminated the hydrogenase activity; the GB11213141 strain is completely devoid of any hydrogenase activity. Complementation with the hox2H gene using the pMHE6crtKm broad-host-range plasmid restored the hydrogen production; this finally corroborated that Hox2 was the source of the residual hydrogen production in GB112131 (ΔhynSL ΔhupSL Δhox1H) (Table 4).
TABLE 4.
Responsibility of Hox2 for residual hydrogen production observed in T. roseopersicina (ΔhynSL ΔhupSL Δhox1H) strainsa
| Strain genotype | In vivo H2 production (μl H2 mg−1 total protein) |
|---|---|
| ΔhynSL ΔhupSL Δhox1H | 0.49 ± 0.09 |
| ΔhynSL ΔhupSL Δhox1H Δhox2H | 0 ± 0 |
| pMHEhoxH2c/ΔhynSL ΔhupSL Δhox1H Δhox2H | 0.38 ± 0.05 |
| hypF::miniTn5 | 0 ± 0 |
Strains (also listed in Table 1) were cultivated in PC2G medium.
Characterization of Hox2 in vivo activity.
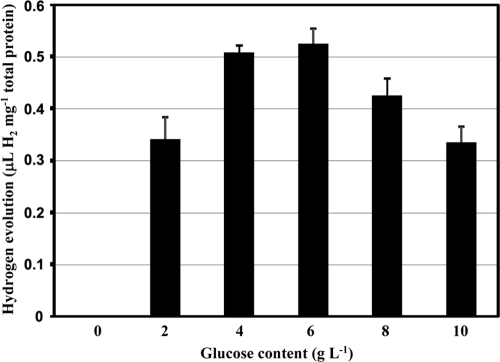
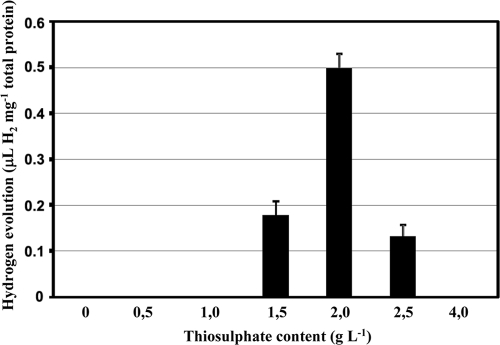
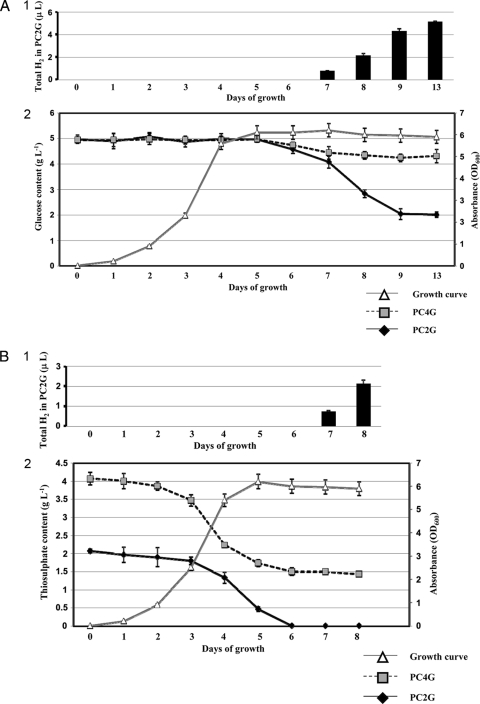
Hox2 activity was not detectable under the conditions which were generally used in previous studies. Our previous investigations indicated that the in vivo activities of the Hox1, Hyn, and Hup hydrogenases were dependent on the nature and quantity of the electron sources used in the growth medium (19, 27). Thiosulfate—being the primary energy and electron source—is one of the key factors connected to the hydrogen metabolism of T. roseopersicina (19, 32). However, changing only the thiosulfate concentration did not induce Hox2 activity, although in vivo hydrogen production by Hox1 can be apparently driven by this compound. We concluded that Hox2 may utilize electrons deriving from components distinct from thiosulfate. Glucose proved to be the inducer of the in vivo hydrogen production by Hox2, but the GB112131 strain was able to evolve hydrogen using glucose only when the thiosulfate concentration was lower than the 4 g liter−1 generally used. The optimal glucose concentration was determined, and a range from 4 to 8 g liter−1 glucose was found to be ideal for in vivo hydrogen production (Fig. 1). Consequently, 5 g liter−1 glucose was used in all experiments hereafter. The thiosulfate concentration of the growth medium was also optimized for Hox2-based hydrogen production: 2 g liter−1 thiosulfate proved to yield maximum hydrogen production in the presence of glucose (Fig. 2). Cell growth was strongly inhibited below 1 g liter−1 thiosulfate, even in the presence of glucose. Thus, Pfennig medium supplemented with 5 g liter−1 glucose and containing 2 g liter−1 thiosulfate (PC2G medium) represented the optimal condition for Hox2-mediated hydrogen production. The level of in vivo hydrogen production by Hox2 is low: approximately 0.45 to 0.55 μl H2 mg−1 total protein. This is the total amount that accumulated in 3 to 4 days. Interestingly, in batch cultures, in vivo hydrogen production by Hox2 starts on the 7th day of growth, and hydrogen production ceases on the 10th day, regardless of the timing of glucose addition (at the culture inoculation time or on the 5th day of growth). Thus, Hox2 becomes physiologically active only in the stationary phase of photomixotrophic growth (the medium contains both carbonate [CO2] and organic substrate). The glucose concentration was monitored continuously during the life cycle of the cultures (Fig. 3 A). A concentration of 5 g liter−1 was added initially, and the same concentration was present in the medium on the 5th day of growth. However, by the 9th day of growth, only about 40% of the original glucose could be detected, regardless of the time of glucose addition (at the culture inoculation time or on the 5th day of growth), indicating that Hox2-mediated in vivo hydrogen production and glucose consumption are linked. No significant glucose consumption could be observed between the 9th and 13th days; this is also in accordance with the graduated cessation of hydrogen-producing activity around the 10th day (Fig. 3A). When GB112131, harboring only Hox2, was grown in a medium containing 4 g liter−1 thiosulfate and 5 g liter−1 glucose, only minimal glucose consumption was observed (Fig. 3A).
FIG. 1.
Determination of the optimal glucose content for Hox2-based hydrogen evolution. Strain GB112131 was cultivated in PC2 supplemented with 0 to 10 g liter−1 glucose. In vivo hydrogen production was measured on each day of growth. Hydrogen that accumulated between the 7th and 10th days of growth is shown.
FIG. 2.
Determination of the optimal thiosulfate content for Hox2-based hydrogen evolution. Strain GB112131 was cultivated in PCG supplemented with 0 to 4 g liter−1 thiosulfate. In vivo hydrogen production was measured on each day of growth. Hydrogen that accumulated between the 7th and 10th days of growth is shown.
FIG. 3.
(A) Glucose utilization and hydrogen production by strain GB112131 as a function of time. (Panel 1) Total hydrogen that accumulated during growth. Hydrogen production could be observed in GB112131 cultures only when grown in PC2G. (Panel 2) Growth curve and glucose content. Samples were taken on each day of growth, and glucose content was determined as described in Materials and Methods. No significant difference was observed in the growth of strain GB112131 in PC2G and PC4G media; thus, results for both media are shown on the same growth curve. OD600, optical density at 600 nm. (B) Thiosulfate consumption and hydrogen production of strain GB112131 as a function of time. (Panel 1) Total hydrogen that accumulated during growth. Hydrogen production could be observed in GB112131 cultures only when grown in PC2G. (Panel 2) Growth curve and thiosulfate content. Samples were taken on each day of growth, and thiosulfate content was determined as described in Materials and Methods. No significant difference was observed in the growth of strain GB112131 in PC2G and PC4G media; thus, results for both media are shown on the same growth curve.
Thiosulfate consumption of GB112131 PC2G cultures was also monitored in line with glucose measurement; rapid and complete thiosulfate consumption was observed, and no thiosulfate remained in the medium by the 6th day of growth (Fig. 3B). This result correlates well with the appearance of hydrogen production on the 7th day in GB112131 grown on PC2G and with the glucose consumption data as well. No hydrogen production could be observed when cultures were supplemented with an additional 2 g liter−1 thiosulfate on the 5th day of growth.
Several organic substrates other than glucose were tested as potential inducers of hydrogenase activity. Only the addition of sodium pyruvate resulted in hydrogen production catalyzed by Hox2; the pyruvate-dependent H2 production was approximately 45% of the amount of hydrogen produced in the presence of 5 g liter−1 glucose.
Light dependence of in vivo Hox2-mediated hydrogen production was also tested: the samples were illuminated until the beginning of the stationary growth phase (5th day), and then half of the samples were wrapped in aluminum foil and were cultivated further in the dark. GB112131 evolves hydrogen only when grown under continuous illumination; no hydrogen production was observed in the dark.
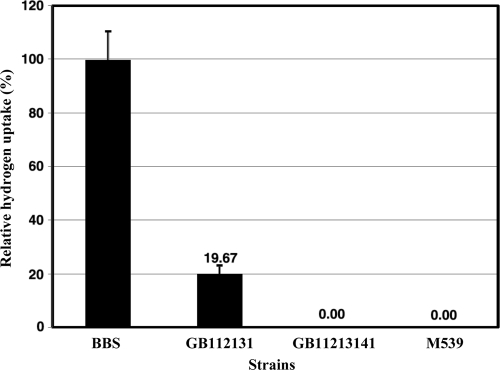
Hox2 was also shown to be a real bidirectional enzyme; in vivo hydrogen uptake mediated by Hox2 could be measured under nitrogen-fixing conditions (NC2G medium, which is PC2G medium lacking ammonium chloride) (Fig. 4). However, this uptake activity was only about 20% of the hydrogen uptake activity of the wild-type strain.
FIG. 4.
In vivo hydrogen uptake mediated by Hox2. NC2G (nitrogen-fixing medium) was used for cell growth, and the hydrogenase uptake activity of Hox2 was calculated as described in Materials and Methods.
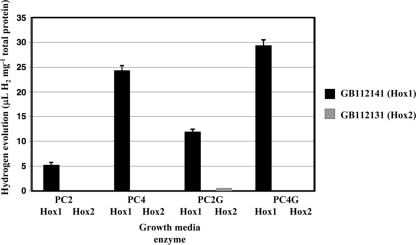
In order to compare the two Hox hydrogenases, Hox1 activity was also examined as a function of thiosulfate and glucose concentrations (Fig. 5). Strain GB112141 (ΔhynSL ΔhupSL Δhox2H) grown under continuous illumination was used to perform these experiments. This strain solely containing Hox1 was able to evolve hydrogen in the original Pfennig medium (PC4), and thiosulfate serves as an electron source (31). As in the case of Hox2, the in vivo hydrogen evolution activity of Hox1 increased in the presence of glucose. However, this induction was independent of the thiosulfate concentration; elevated in vivo hydrogen evolution of Hox1 was observed using either PC2G or PC4G medium. The Hox1 complex showed 20-fold in vivo hydrogen evolution capability relative to Hox2 using PC2G medium. Moreover, Hox1 was able to use glucose—when grown on PC2G—in a slightly earlier growth phase than Hox2. Elevated hydrogen production was observed at the late logarithmic growth phase, around the 6th day of culturing, and the additive effect caused by glucose lasted until the 8th day. No significant difference could be observed in the kinetics of thiosulfate utilization by GB112141 compared to GB112131 (data not shown).
FIG. 5.
Comparison of in vivo hydrogen evolution rates of Hox1 (GB112141) and Hox2 (GB112131) under various growth conditions. Total hydrogen production was measured at the end of the 8th day of growth.
In vitro activity of Hox2.
Besides characterization of in vivo hydrogen production, in vitro activity of Hox2 was examined, as well. Soluble fractions were prepared from GB112131 grown under conditions favoring in vivo H2 production (PC2G). Cells were harvested on the 5th and 10th days of culturing. Hydrogenase activity could be detected in the cytoplasmic fraction regardless of the growth phase, and similar activity levels were found in the samples derived from 5-day-old and 10-day-old cultures. The activity was very low and could only be observed in the H2 evolution direction (0.07 μl H2 h−1 mg−1 total protein); hydrogen uptake could not be detected. The in vitro evolution activity of Hox1 (1.85 μl H2 h−1 mg−1 total protein in the soluble fraction prepared from cells grown under identical conditions) is orders of magnitude higher than that of Hox2. Additionally, the H2 uptake activity of Hox1 enzyme could be easily measured (31, 32). In vitro NAD+/NADP+-reducing and NADH/NADPH-oxidizing activities of the Hox2 complex were also studied; only NADH-dependent hydrogen evolution could be detected (Table 5). No hydrogen-uptake-coupled NAD+/NADP+ reduction could be measured; this can be due to the overall low activity of Hox2 combined with the lower sensitivity of this uptake assay relative to benzyl viologen-mediated hydrogen uptake.
TABLE 5.
Measurement of in vitro NADH/NADPH-dependent hydrogen evolution of Hox2a
| H2 evolution |
In vitro H2 evolution (μl H2 h−1 mg−1 total protein) in T. roseopersicina strain genotype: |
|
|---|---|---|
| ΔhynSL ΔhupSL Δhox1H | ΔhynSL ΔhupSL Δhox2H | |
| NADH dependent | 0.053 ± 0.011 | 0 ± 0 |
| NADPH dependent | 0 ± 0 | 0 ± 0 |
The strains (also listed in Table 1) were cultivated in PC2G medium.
Analysis of expression of hox2 genes.
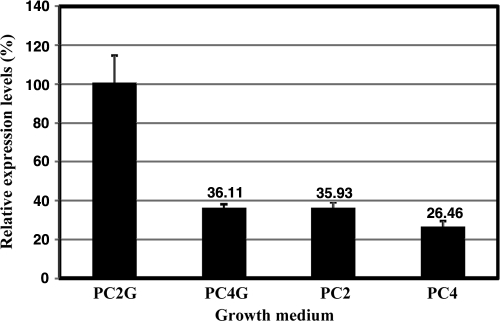
In order to disclose the metabolic routes requiring the Hox2 enzyme, quantifications of hox2 expression (hox2F and hox2H) under various growth conditions were performed. Strain GB112131 was grown under four different conditions (PC2, PC4, PC2G, and PC4G), where thiosulfate and glucose concentrations were used in various combinations (2 g liter−1 and 4 g liter−1 for thiosulfate and 0 g liter−1 and 5 g liter−1 for glucose). Samples were taken from each culture after 5 and 10 days of growth. Expression of the hox2H gene was observed under all conditions applied. The hox2H expression levels were approximately 5 to 6 orders of magnitude lower than those of 16S rRNA genes. In line with the in vivo H2 evolution and hydrogenase activity assays, a significantly higher hox2H expression level could be observed in GB112131 under PC2G conditions compared to the PC2, PC4, and PC4G conditions (Fig. 6). There were no differences between the expression patterns of the samples harvested on the 5th and 10th days of growth. In both time intervals, hox2H had higher expression levels in cells grown on PC2G than in those cultivated on PC4, PC2, or PC4G. These results are in accordance with the data from in vitro activity measurements.
FIG. 6.
Real-time qPCR analysis of hox2H gene expression. Strain GB112131 was grown in various media (PC2G, PC4G, PC2, and PC4), and cells were harvested on the 5th and 10th days of growth.
The level of expression of the hox2 genes was determined by RT-qPCR in cells grown under nitrogen-fixing conditions with and without glucose. No significant difference could be seen in the amount of hox2 mRNA in cells cultivated in the absence and presence of glucose, and the level of expression corresponded to the value obtained for samples grown in nitrogenase-repressed medium in the absence of glucose (basic level; Fig. 6). Consequently, under nitrogen-fixing conditions the hox2 expression was not influenced by the addition of glucose.
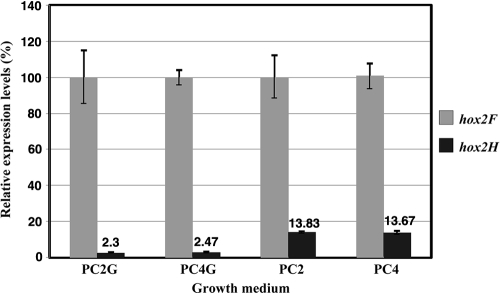
Another interesting phenomenon was the ratio of hox2F to hox2H transcripts when random hexamers were used for cDNA synthesis. The level of hox2F expression was always considerably higher than that of hox2H (40 to 45 times higher in glucose-containing medium [PC2G and PC4G], while 7 to 8 times higher in medium lacking glucose [PC2 and PC4]) (Fig. 7). This result prompted us to perform the same experiments for hox2U and hox2Y genes. The expression level of hox2U was similar to that of hox2F, while hox2Y was expressed at a level almost identical to that of hox2H, pointing toward a dissimilar expression of the hydrogenase and diaphorase dimers. This difference in expression levels could not be detected when reverse transcription was initiated from a hox2H-specific oligonucleotide, which implies the presence of at least two transcriptional units or distinct stability of the mRNA harboring the hoxFU and hoxYH genes. Levels of expression of hox2 in cells growing under illumination and in the dark were also determined. There was no difference in the levels of expression; thus, hox2 expression is not regulated by light, although light is necessary for in vivo activity as was shown above.
FIG. 7.
Ratios of hox2F to hox2H gene expression levels. Strain GB112131 was grown in different media (PC2G, PC4G, PC2, and PC4). Random hexamers were used for reverse transcription reactions.
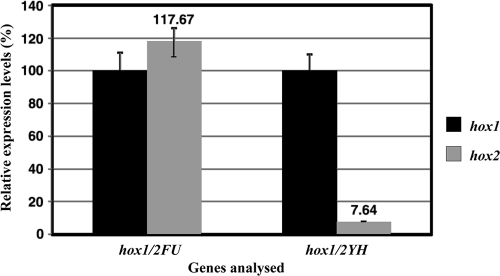
Being relevant for functional studies, the levels of hox1 and hox2 expression were also compared in wild-type cells grown under different conditions (PC2, PC4, PC4G, and PC2G). First of all, it was recognized that the upregulation of the hox2H gene in cells grown in PC2G medium was weaker (by 45 to 50%) in the wild-type strain (BBS) than it was in GB112131. In addition, in PC2G medium, relative to PC4, PC2, and PC4G, expression of the hox1H gene was upregulated in BBS to a similar extent to the hox2H gene. The expression levels of the diaphorase-encoding hox2FU and hox1FU genes were similar, while in the case of the genes coding for hydrogenase subunits, hox1 genes (hox1YH) are expressed at a much higher level than hox2 genes (hox2YH) (Fig. 8), which may explain the remarkable difference between the two Hox-type enzymes in hydrogen-producing capability.
FIG. 8.
Ratios of hox1 to hox2 gene expression levels. Strain BBS was grown in PC2G medium. Random hexamers were used for reverse transcription reactions.
DISCUSSION
T. roseopersicina BBS is suitable for studies of energy metabolism in purple bacteria due to its diverse growth modes, metabolic versatility, and its property of harboring several NiFe hydrogenases. The discovery of hox2 genes was facilitated by the ongoing genome sequencing of T. roseopersicina; preliminary data from the genome annotation suggest that Hox2 is the last hydrogenase identified in this organism. At the genetic level, the hox2YH genes are the coding sequences of the fifth NiFe hydrogenase dimer and Hox2 is the second bidirectional Hox-type hydrogenase in T. roseopersicina. Thus, T. roseopersicina is the first bacterium harboring more than one Hox-type NiFe hydrogenase.
Alignments of deduced sequences showed remarkable variations between the two Hox-type soluble enzymes in T. roseopersicina (Table 3). Hox1 belongs to the cyanobacterium-type Hox enzymes (31), while Hox2 exhibited the highest similarity to the soluble hydrogenase of M. capsulatus (Bath) (11, 45) and resembled the NAD+-reducing hydrogenases of Gammaproteobacteria (e.g., the soluble hydrogenase [SH] of R. eutropha H16) (40) (Table 3). A. vinosum—a close relative of T. roseopersicina—harbors only one Hox-type enzyme, which is very similar to Hox1 (21); the hox2 genes are apparently not present in the genome of this strain (GenBank accession no. CP001896). Hox2 is encoded by four genes (hox2FUYH). Certain members of the Hox enzymes comprise further subunits; HoxE is the fifth subunit besides HoxFUYH in cyanobacteria (37, 39) and in the case of the Hox1 hydrogenase of T. roseopersicina (27, 31), while two HoxI proteins complete the functional soluble hydrogenase of R. eutropha (5). The roles of the additional subunits are distinct. HoxE is essential for the in vivo function of Hox1 but irrelevant for the in vitro activity in T. roseopersicina. Therefore, HoxE was suggested to represent a third redox gate of this enzyme (31). In R. eutropha, the dimeric form of HoxI is attached to the other subunits, leading to a HoxFUYHI2 hexamer (5), which could be activated with both NADH and NADPH. Thus, HoxI might provide an additional nucleotide binding site—for NADPH.
A careful search in the T. roseopersicina genome did not identify any potential gene of either HoxE or HoxI. Therefore, this is the first known organism possessing one four-subunit and one five-subunit NAD+-reducing/NADH-oxidizing NiFe hydrogenase, and both are functional.
There are numerous microorganisms containing heterotetrameric Hox enzymes, such as M. capsulatus Bath (11, 45), Pyrococcus furiosus (Hyh1 and Hyh2) (13, 22, 38), Thermococcus litoralis (Hyh1) (33), and Thermococcus kodakarensis (14). In these organisms, there is no indication for the presence of either HoxE or HoxI. The substrate analysis of P. furiosus Hyh1 and Hyh2 enzymes revealed that Hyh1 was able to utilize NADP+/NADPH only, while Hyh2 could accept both NAD+/NADH and NADP+/NADPH (22). This might be explained by the altered nucleotide binding domain of Hyh2.
In T. roseopersicina, Hox2 was shown to utilize NADH only, similarly to the Hyh1 enzymes in the above-mentioned Archaea or to Hox in M. capsulatus Bath.
Characterization of the in vivo Hox2-related H2-evolving and -oxidizing activities under various growth conditions was performed in order to reveal the in vivo role of this particular NiFe hydrogenase and to highlight the functional similarities and/or differences between the two soluble Hox-type hydrogenases in T. roseopersicina. Genes of the hox2 operon were expressed at a low level when the cells were grown in original Pfennig medium and under nitrogen-fixing conditions. Thiosulfate is the primary electron source for photochemolithoautotrophic growth. It drives the Hox1-mediated hydrogen production under illumination (31, 32), but apparently it cannot serve as an electron source for the Hox2 enzyme in vivo. However, mRNA of the hox2 genes could be detected without doubt in cells grown under this condition. Reduction of the thiosulfate concentration itself did not alter expression of the hox2 genes and did not result in the appearance of the Hox2 enzyme activity; 2 g liter−1 thiosulfate was established as the minimally necessary concentration for normal growth. At this level of thiosulfate, the additional electron-rich compounds may also be metabolized and the inhibiting, masking effects due to the excessive thiosulfate could be avoided. Several organic and inorganic compounds were tested in the Pfennig medium containing 2 g liter−1 thiosulfate, and glucose proved to be an inducer of the Hox2 expression under nitrogenase-repressed conditions. This was demonstrated at the transcript level and supported by the in vivo H2 evolution data. At an elevated (4 g liter−1) thiosulfate concentration, even in the presence of glucose, the expression of the genes dropped to the basic level and no hydrogen production could be detected. This corroborated that the masking phenomenon by thiosulfate indeed took place. Therefore, the low initial thiosulfate concentration and glucose addition were able to render Hox2 functional, and they act together. It was reasonable to assume that thiosulfate is the first electron source utilized by the growing cells, and then, as the culture becomes older, glucose metabolism takes over and donates electrons to the Hox2 hydrogenase. This is compatible with the observation that Hox2-coupled in vivo H2 production appears only in the stationary phase of growth, when thiosulfate content is completely consumed. It is noteworthy that the transcriptional upregulation was independent of growth phase: elevated expression could already be observed at the logarithmic growth phase. The Hox2 hydrogenase complex seems to be continuously upregulated in glucose-containing media but evolves hydrogen only when glucose metabolism starts, indicating that glycolysis provides an elevated metabolic/electron flux toward Hox2 hydrogenase. Therefore, taking into account only the H2-producing role of Hox2, its physiological, phenotypical appearance is related to metabolic shifts rather than to a growth-phase-dependent transcriptional control. One might speculate that the basic amount of Hox2 might not be enough for managing the glucose- and growth-phase-dependent elevated electron flux. A simple way to resolve this problem is to express the enzyme at slightly higher level when the metabolically relevant substrate is present in the medium.
The Hox2 hydrogenase is a bidirectonal enzyme; it can oxidize H2, as well. The strain exclusively containing Hox2 had 20% of the uptake activity of the wild-type cells when NC2G medium was used. Similar experiments disclosed that the strain solely possessing Hox1 could take up as much hydrogen as the wild-type strain containing all hydrogenases (31). This means that while Hox1 is able to take over the function of the other hydrogenases, Hox2 can only partially do so. Even the in vitro H2 uptake activity of Hox2 was extremely low.
The level of expression of the hox2 genes under nitrogen-fixing conditions corresponded to the basic level in cells grown in nitrogenase-repressing medium lacking glucose. Not surprisingly, addition of glucose did not alter the expression of hox2 under these circumstances, since catabolism of both glucose and H2 releases electrons.
The in vivo hydrogen evolution by Hox2 requires continuous illumination. Contrary to cyanobacteria, where the hox genes were shown to be regulated by the circadian clock (36), the transcript levels of hox2 genes were not affected by light. The exact role of light is unclear, but it might have an energizing effect in the process.
Glucose is thus an inducer of in vivo Hox2-coupled H2-evolving activity, but this induction is not specific for Hox2: Hox1 was also shown to be able to utilize electrons derived from glucose. The hox1 genes were upregulated in PC2G medium, and hydrogen production by Hox1 could be driven by glucose.
Comparison of the expression levels of the hox2 genes in the wild-type and triple-hydrogenase mutant strains indicated an interplay between the hydrogenases present in the cell. Weaker upregulation of hox2 genes was observed in the wild-type strain (BBS) relative to the triple mutant GB112131, which might be due to the presence and concerted action of three additional functional hydrogenases, with special emphasis on Hox1.
Another interesting observation was the expression ratio of diaphorase- and hydrogenase-encoding genes. Significantly higher expression of hox2FU relative to hox2YH suggests the presence of other, likely shorter, transcripts and/or the distinct stability of the mRNA parts, the hox2YH transcript part being more unstable than the hox2FU mRNA. Various R. eutropha hox transcripts of distinct lengths and stabilities were also observed (26). The expression level of hox1YH hydrogenase genes is much higher than that of hox2YH genes, which may explain the remarkable difference in hydrogen productivities between Hox1 and Hox2.
The question of why two similar Hox-type hydrogenases are preserved by the cells remains open. Two heterotetrameric Hox-type hydrogenases were identified and characterized in the hyperthermophilic archaeon Pyrococcus furiosus: Hyh1 and Hyh2 (13). In this strain, Hyh2 also had a 1-magnitude-lower expression and activity level than Hyh1. Both hyh operons were downregulated by sulfur; consequently, their in vivo function might not be linked to the sulfur metabolism. The physiological role of these hydrogenases is still unclear; they might recycle the hydrogen produced by the energy-conserving membrane-bound hydrogenase (1).
Since the metabolic network in T. roseopersicina obviously differs from that of P. furiosus, the metabolic contexts of their hydrogenases are likely dissimilar. In T. roseopersicina, the two Hox enzymes might link the glucose metabolism to distinct bioenergetic pathways: Hox2 simply connects it to NAD+/NADH housekeeping, while Hox1—possessing an additional redox gate—might have a more complex role which is supposed to be elucidated in the future. From the data presented in this study, we concluded that Hox2 had a physiological role in tuning the NAD+/NADH balance under photomixotrophic conditions at the stage when cells enters into the long-term stationary phase. Under nitrogen-fixing conditions, it can oxidize H2 but less than the other hydrogenases in the cell.
Acknowledgments
This work was supported by EU projects HyVolution FP6-IP-SES6 019825 and FP7 Collaborative Project SOLAR-H2 FP7-Energy-212508 and by domestic funds (NAP-BIO Teller Ede Program OMFB-00441/2007, GOP-1.1.2-07/1-2003 + 8-0007, Asbóth-DAMEC-2007/09, Baross OMFB-00265/2007, and KN-RET-07/2005). This publication was supported by the Dr. Rollin D. Hotchkiss Foundation.
Footnotes
Published ahead of print on 11 June 2010.
REFERENCES
- 1.Adams, M. W., J. F. Holden, A. L. Menon, G. J. Schut, A. M. Grunden, C. Hou, A. M. Hutchins, F. E. Jenney, Jr., C. Kim, K. Ma, G. Pan, R. Roy, R. Sapra, S. V. Story, and M. F. Verhagen. 2001. Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:716-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benoit, S. L., and R. J. Maier. 2008. Hydrogen and nickel metabolism in helicobacter species. Ann. N. Y. Acad. Sci. 1125:242-251. [DOI] [PubMed] [Google Scholar]
- 3.Böck, A., P. W. King, M. Blokesch, and M. C. Posewitz. 2006. Maturation of hydrogenases. Adv. Microb. Physiol. 51:1-71. [DOI] [PubMed] [Google Scholar]
- 4.Bogorov, L. V. 1974. The properties of Thiocapsa roseopersicina, strain BBS, isolated from an estuary of the White Sea. Mikrobiologija 43:326-332. [PubMed] [Google Scholar]
- 5.Burgdorf, T., E. van der Linden, M. Bernhard, Q. Y. Yin, J. W. Back, A. F. Hartog, A. O. Muijsers, C. G. de Koster, S. P. Albracht, and B. Friedrich. 2005. The soluble NAD+-reducing [NiFe]-hydrogenase from Ralstonia eutropha H16 consists of six subunits and can be specifically activated by NADPH. J. Bacteriol. 187:3122-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colbeau, A., K. L. Kovacs, J. Chabert, and P. M. Vignais. 1994. Cloning and sequence of the structural (hupSLC) and accessory (hupDHI) genes for hydrogenase biosynthesis in Thiocapsa roseopersicina. Gene 140:25-31. [DOI] [PubMed] [Google Scholar]
- 7.Elsen, S., A. Colbeau, J. Chabert, and P. M. Vignais. 1996. The hupTUV operon is involved in negative control of hydrogenase synthesis in Rhodobacter capsulatus. J. Bacteriol. 178:5174-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fodor, B. D., Á. T. Kovács, R. Csáki, E. Hunyadi-Gulyás, E. Klement, G. Maróti, L. S. Mészáros, K. F. Medzihradszky, G. Rákhely, and K. L. Kovács. 2004. Modular broad-host-range expression vectors for single-protein and protein complex purification. Appl. Environ. Microbiol. 70:712-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor, B. D., G. Rákhely, Á. T. Kovács, and K. L. Kovács. 2001. Transposon mutagenesis in purple sulfur photosynthetic bacteria: identification of hypF, encoding a protein capable of processing [NiFe] hydrogenases in alpha, beta, and gamma subdivisions of the proteobacteria. Appl. Environ. Microbiol. 67:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghirardi, M. L., M. C. Posewitz, P. C. Maness, A. Dubini, J. Yu, and M. Seibert. 2007. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu. Rev. Plant Biol. 58:71-91. [DOI] [PubMed] [Google Scholar]
- 11.Hanczár, T., R. Csáki, L. Bodrossy, J. C. Murrell, and K. L. Kovács. 2002. Detection and localization of two hydrogenases in Methylococcus capsulatus (Bath) and their potential role in methane metabolism. Arch. Microbiol. 177:167-172. [DOI] [PubMed] [Google Scholar]
- 12.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenney, F. E., Jr., and M. W. W. Adams. 2008. Hydrogenases of the model hyperthermophiles. Ann. N. Y. Acad. Sci. 1125:252-266. [DOI] [PubMed] [Google Scholar]
- 14.Kanai, T., S. Ito, and T. Imanaka. 2003. Characterization of a cytosolic NiFe-hydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:1705-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleihues, L., O. Lenz, M. Bernhard, T. Buhrke, and B. Friedrich. 2000. The H2 sensor of Ralstonia eutropha is a member of the subclass of regulatory [NiFe] hydrogenases. J. Bacteriol. 182:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovács, A. T., G. Rákhely, J. Balogh, G. Maróti, L. Cournac, P. Carrier, L. S. Mészáros, G. Peltier, and K. L. Kovács. 2005. Hydrogen independent expression of hupSL genes in Thiocapsa roseopersicina BBS. FEBS J. 272:4807-4816. [DOI] [PubMed] [Google Scholar]
- 17.Kovács, K. L., and C. Bagyinka. 1990. Structural properties and functional states of hydrogenase from Thiocapsa roseopersicina. FEMS Microbiol. Rev. 87:407-412. [Google Scholar]
- 18.Kovács, K. L., C. Bagyinka, and L. T. Serebriakova. 1983. Distribution and orientation of hydrogenase in various photosynthetic bacteria. Curr. Microbiol. 9:215-218. [Google Scholar]
- 19.Laurinavichene, T. V., G. Rákhely, K. L. Kovács, and A. A. Tsygankov. 2007. The effect of sulfur compounds on H2 evolution/consumption reactions, mediated by various hydrogenases, in the purple sulfur bacterium, Thiocapsa roseopersicina. Arch. Microbiol. 188:403-410. [DOI] [PubMed] [Google Scholar]
- 20.Lenz, O., A. Strack, A. Tran-Betcke, and B. Friedrich. 1997. A hydrogen-sensing system in transcriptional regulation of hydrogenase gene expression in Alcaligenes species. J. Bacteriol. 179:1655-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long, M., J. Liu, Z. Chen, B. Bleijlevens, W. Roseboom, and S. P. Albracht. 2007. Characterization of a HoxEFUYH type of [NiFe] hydrogenase from Allochromatium vinosum and some EPR and IR properties of the hydrogenase module. J. Biol. Inorg. Chem. 12:62-78. [DOI] [PubMed] [Google Scholar]
- 22.Ma, K., R. Weiss, and M. W. Adams. 2000. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182:1864-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maróti, G., G. Rákhely, J. Maróti, E. Dorogházi, E. Klement, F. K. Medzihradszky, and K. L. Kovács. 2010. Specificity and selectivity of HypC chaperonins and endopeptidases in the molecular assembly machinery of [NiFe] hydrogenases of Thiocapsa roseopersicina. Int. J. Hydrogen Energy. doi: 10.1016/j.ijhydene.2009.10.059. [DOI]
- 24.McGlynn, S. E., S. S. Ruebush, A. Naumov, L. E. Nagy, A. Dubini, P. W. King, J. B. Broderick, M. C. Posewitz, and J. W. Peters. 2007. In vitro activation of [FeFe] hydrogenase: new insights into hydrogenase maturation. J. Biol. Inorg. Chem. 12:443-447. [DOI] [PubMed] [Google Scholar]
- 25.Miller, G. L. 1959. Use of dinitro salicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 26.Oelmüller, U., H. G. Schlegel, and C. G. Friedrich. 1990. Differential stability of mRNA species of Alcaligenes eutrophus soluble and particulate hydrogenases. J. Bacteriol. 172:7057-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palágyi-Mészáros, L. S., J. Maróti, D. Latinovics, T. Balogh, E. Klement, K. F. Medzihradszky, G. Rákhely, and K. L. Kovács. 2009. Electron-transfer subunits of the NiFe hydrogenases in Thiocapsa roseopersicina BBS. FEBS J. 276:164-174. [DOI] [PubMed] [Google Scholar]
- 28.Peters, J. W., W. N. Lanzilotta, B. J. Lemon, and L. C. Seefeldt. 1998. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282:1853-1858. (Errata, Science 283:35, 2102, 1999.) [DOI] [PubMed] [Google Scholar]
- 29.Pfennig, N., and H. G. Trüper. 1991. The family Chromatiaceae, p. 3200-3221. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer, Berlin, Germany.
- 30.Rákhely, G., A. Colbeau, J. Garin, P. M. Vignais, and K. L. Kovács. 1998. Unusual organization of the genes coding for HydSL, the stable [NiFe] hydrogenase in the photosynthetic bacterium Thiocapsa roseopersicina BBS. J. Bacteriol. 180:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rákhely, G., Á. T. Kovács, G. Maróti, B. D. Fodor, G. Csanádi, D. Latinovics, and K. L. Kovács. 2004. Cyanobacterial-type, heteropentameric, NAD+-reducing NiFe hydrogenase in the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. Appl. Environ. Microbiol. 70:722-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rákhely, G., T. V. Laurinavichene, A. A. Tsygankov, and K. L. Kovács. 2007. The role of Hox hydrogenase in the H2 metabolism of Thiocapsa roseopersicina. Biochim. Biophys. Acta 1767:671-676. [DOI] [PubMed] [Google Scholar]
- 33.Rákhely, G., Z. H. Zhou, M. W. W. Adams, and K. L. Kovács. 1999. Biochemical and molecular characterization of the [NiFe] hydrogenase from the hyperthermophilic archaeon, Thermococcus litoralis. Eur. J. Biochem. 266:1158-1165. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 36.Schmitz, O., G. Boison, and H. Bothe. 2001. Quantitative analysis of expression of two circadian clock-controlled gene clusters coding for the bidirectional hydrogenase in the cyanobacterium Synechococcus sp. PCC7942. Mol. Microbiol. 41:1409-1417. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz, O., G. Boison, H. Salzmann, H. Bothe, K. Schütz, S. H. Wang, and T. Happe. 2002. HoxE—a subunit specific for the pentameric bidirectional hydrogenase complex (HoxEFUYH) of cyanobacteria. Biochim. Biophys. Acta 1554:66-74. [DOI] [PubMed] [Google Scholar]
- 38.Silva, P. J., E. C. van den Ban, H. Wassink, H. Haaker, B. de Castro, F. T. Robb, and W. R. Hagen. 2000. Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267:6541-6551. [DOI] [PubMed] [Google Scholar]
- 39.Tamagnini, P., E. Leitao, P. Oliviera, D. Ferreira, F. Pinto, D. J. Harris, T. Heidorn, and P. Lindblad. 2007. Cyanobacterial hydrogenases: diversity, regulation, and high value products. FEMS Microbiol. Rev. 31:692-720. [DOI] [PubMed] [Google Scholar]
- 40.Tran-Betcke, A., U. Warnecke, C. Bocker, C. Zaborosch, and B. Friedrich. 1990. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J. Bacteriol. 172:2920-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tye, J. W., M. Y. Darensbourg, and M. B. Hall. 2008. Refining the active site structure of iron-iron hydrogenase using computational infrared spectroscopy. Inorg. Chem. 47:2380-2388. [DOI] [PubMed] [Google Scholar]
- 42.Vignais, P. M., and A. Colbeau. 2004. Molecular biology of microbial hydrogenases. Curr. Issues Mol. Biol. 6:159-188. [PubMed] [Google Scholar]
- 43.Vignais, P. M., and B. Billoud. 2007. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107:4206-4272. [DOI] [PubMed] [Google Scholar]
- 44.Volbeda, A., M. H. Charon, C. Piras, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
- 45.Ward, N., O. Larsen, J. Sakwa, L. Bruseth, H. Khouri, A. S. Durkin, G. Dimitrov, L. Jiang, D. Scanlan, K. H. Kang, M. Lewis, K. E. Nelson, B. Methe, M. Wu, J. F. Heidelberg, I. T. Paulsen, D. Fouts, J. Ravel, H. Tettelin, Q. Ren, T. Read, R. T. DeBoy, R. Seshadri, S. L. Salzberg, H. B. Jensen, N. K. Birkeland, W. C. Nelson, R. J. Dodson, S. H. Grindhaug, I. Holt, I. Eidhammer, I. Jonasen, S. Vanaken, T. Utterback, T. V. Feldblyum, C. M. Fraser, J. R. Lillehaug, and J. A. Eisen. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:E303. [DOI] [PMC free article] [PubMed] [Google Scholar]