Abstract
A novel tightly regulated gene expression system was developed for Escherichia coli by applying the regulatory elements of the Pseudomonas putida F1 cym and cmt operons to control target gene expression at the transcriptional level by using p-isopropylbenzoate (cumate) as an inducer. This novel expression system, referred to as the cumate gene switch, includes a specific expression vector, pNEW, that contains a partial T5 phage promoter combined with the Pseudomonas-based synthetic operator and the cymR repressor protein-encoding gene designed to express constitutively in the host strain. The induction of transcription relies on the addition of the exogenous inducer (cumate), which is nontoxic to the culture, water soluble, and inexpensive. The characteristics and potential of the expression system were determined. Using flow cytometry and fed-batch fermentations, we have shown that, with the newly developed cumate-regulated system, (i) higher recombinant product yields can be obtained than with the pET (isopropyl-β-d-thiogalactopyranoside [IPTG])-induced expression system, (ii) expression is tightly regulated, (iii) addition of cumate quickly results in a fully induced and homogenous protein-expressing population in contrast to the bimodal expression profile of an IPTG-induced population, (iv) expression can be modulated by varying the cumate concentration, and (v) the cumate-induced population remains induced and fully expressing even at 8 h following induction, resulting in high yields of the target protein Furthermore, the cumate gene switch described in this article is applicable to a wide range of E. coli strains.
A variety of gene expression systems exist for the production of recombinant proteins and as a tool in metabolic engineering. These systems differ in the hosts, plasmids, and promoters being utilized. The variety of existing expression systems reflects the diversity, complexity, and toxicity of the proteins being produced, requiring in certain instances that the product be expressed at various concentrations (14) or at different phases of cell growth (14) and be soluble (4), secreted (13) or compartmentalized (40, 45). In an attempt to meet these challenges, the search for or the development of new hosts and expression vectors is ongoing.
Gene transcription can be enhanced by replacing a promoter sequence naturally associated with that gene with a sequence of a stronger promoter (17, 42). The nature of the bioprocess will dictate the promoter of choice. A process that requires high-level expression will necessitate a strong constitutive or regulated promoter. A process requiring the expression of a product with toxicity issues for the cell will benefit from a regulated expression system that does not possess basal expression under repressed conditions (26, 37). The promoters Plac, Ptrp, Ptac, λPL, PT7, PBAD, and PlacUV5 are commonly utilized for the construction of expression vectors (1, 18). Among these, PlacUV5, Ptac, and the combined system of PT7 with PlacUV5 are widely used, because of their reportedly isopropyl-β-d-thiogalactopyranoside (IPTG) dose-dependent expression (rheostatic control) (14, 38). Although T7 expression systems are known to express high levels of recombinant proteins, they are susceptible to leaky expression (14, 34, 38a). The high cost and potential toxicity of IPTG can preclude the use of these expression systems in large-scale production of recombinant proteins (2, 12, 22, 27, 28, 46, 47).
The PBAD expression system possesses the nutritionally inducible arabinose promoter (PBAD), which uses the sugar l-arabinose as an inducer. Although the PBAD system, which relies on catabolite repression, shows a more stringent regulation of target gene expression than do other expression systems, the reported achievable expression yields are relatively low. The strength of this system is its excellent inducer (arabinose) dose-response characteristic. However, subsaturating inducer concentrations promote extensive expression heterogeneity within the microbial population (39).
Notwithstanding the leakiness issues with the T7 expression system, Escherichia coli BL21(DE3) harboring pET-derived plasmids has become the industry's preferred expression system due mainly to its high expression capabilities. An expression system that combines tight expression regulation with the favorable T7 expression capabilities is being actively pursued (36, 41). Versatile and tightly regulated expression systems are also a requirement of the rapidly growing field of metabolic engineering. Coordinated and modulated expression of multiple genes may require the availability of different promoters (9, 25). It is therefore necessary for the biotechnology industry in general to continue developing new expression systems and promoters, especially systems that are independent of the host strain, cultivation medium, and growth rate. We have embarked on the development of a novel expression system for E. coli possessing the following salient traits: (i) a strong and tightly regulated inducible promoter; (ii) modulated and consistent expression in all cells within a culture; and (iii) applicability to a wide range of host strains without prior genetic modification, unlike the pET system, which requires the E. coli host strains to be genetically modified in order to express a T7 RNA polymerase for transcription to occur. To do so, we have made use of the cumate (p-isopropylbenzoate)-regulated expression system (cumate gene switch), firstly developed for mammalian cells and, shortly after, for methylotrophic bacteria (7, 31).
Here we describe the construction of a cumate-inducible expression system (cumate gene switch) developed for E. coli strains, designated pNEW, carrying a synthetic operator and the regulator gene (cymR) of the Pseudomonas putida F1 cmt operon (7, 10, 11, 31). Using several E. coli strains typically utilized in production and expression studies, we demonstrate high expression yields of target protein, tight regulation, rheostatic control, and a homogenous high-expression bacterial culture.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains DH5α, S-17/λpir, K-12, Top10, and BL21(DE3) were used for the heterologous gene expression host. E. coli was cultured in Luria-Bertani broth (LB) at 37°C, and media were solidified with 1.8% agar (Oxoid, Nepean, Canada) when appropriate. Antibiotics were used at the following concentrations (in μg/ml): ampicillin, 100; kanamycin (Km), 50; tetracycline (Tc), 35.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Bacterial strains | ||
| Pseudomonas fluorescens GK13 | Source of phaC1 and phaC2 genes | 21 |
| Pseudomonas putida F1 | Origin of cymR gene and operator sequence in the cmt operon, respectively | 11 |
| E. coli | ||
| S-17/λpir | Tpr Smr, recA thi pro hsdR M+ RP4:2-Tc::Mu::Km Tn7 λpir | 8 |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu)7697 galU galK rpsL (Strr) endA1 nupG | 16 |
| BL21(DE3)pLyS | F−ompT gal dcm lon hsdSB(rB− mB−) λ(DE3) pLysS(Cmr) | EMD Serono Canada |
| DH5α | endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 φ80dlacZΔM15 Δ(lacZYA-argF)U169 λ− | 18 |
| K-12 | F− λ−rph-1 INV(rrnD rrnE) | 23 |
| Plasmids | ||
| pBRI-cymR1 | pBRI80 plasmid containing one copy of cymR expression cassette | 7 |
| pNEW-pre | pET36 plasmid containing Pkm-cymR expression cassettes, lack of T7 promoter and lac operator | This study |
| pCR2.1-TOPO | PCR cloning vector | Invitrogen Inc. |
| pCR-Pkm-cymR | pCR2.1-TOPO plasmid containing Pkm-cymR | This study |
| PCR-T5OP | pCR2.1-TOPO plasmid containing PT5-operator | This study |
| pCR-bgl | pCR2.1-TOPO plasmid containing bgl | This study |
| pCR-est | pCR2.1-TOPO plasmid containing estI | This study |
| pCR-PhaC1 or C2 | pCR2.1-TOPO plasmid containing phaC1 or phaC2 | This study |
| pNEW | Newly constructed regulative expression vector | This study |
| pNEW-MCS | gfp from pNEW-gfp was replaced by MCS from pSL1190 | This study |
| pNEW-mfh | pNEW vector containing mfh fusion peptide expression cassette | This study |
| pNEW-phaC1 or 2 | pNEW vector containing PhaC1 or PhaC2 expression cassette | This study |
| pNEW-bgl | pNEW vector containing lactase expression cassette | This study |
| pNEW-est | pNEW vector containing esterase expression cassette | This study |
| pNEW-gfp | pNEW vector containing gfp expression cassette | This study |
| pET36(b) | T7-based expression vector | EMD Serono Canada |
| pET-gfp | pET36(b) vector containing gfp expression cassette | This study |
| pSL1190 | Source of multiple cloning site for pNEW-MCS | Amersham-Pharmacia |
| pCUM-gfp | gfp gene source | 7 |
| pCESTa | Esterase gene source | 6 |
| pEBIG4 | Lactase gene source | 20 |
| pTSN-6A | Source of mfh fusion peptide | 32 |
Construction of expression vector.
The operator sequence of the cmt operon from P. putida F1 was introduced downstream of the phage T5 promoter (5) by PCR. The pNEW-regulated expression vector was obtained in several steps: first, the PT5-synthetic operator sequence (OP)-green fluorescent protein (GFP) was PCR amplified from pCUM-gfp (7) using primers T5-OP-F-SPH (5′-CGC ATG CAA ATC ATA AAA AAT TTA TTT GCT TTG TGA GCG GAT AAC AAT TAT AAT AGA TTC AAC AAA CAG ACA ATC TGG TCT GTT TGT ATT AT-3′; the SphI site is underlined, the partial T5 promoter is italicized, and the operator site is in bold) and GFP-SAC-R (5′-CGA GCT CTC AGT TGT ACA GTT CAT CCA TGC C-3′; the SacI site is underlined). The 954-bp PCR fragment containing PT5-operator-gfp was cloned into pCR2.1 to create pCR-T5OP. Next, a 954-bp SphI-SacI fragment from pCR-T5OP was then ligated between the SphI and SacI sites of pET36(b) (EMD Serono Canada, Mississauga, Canada) to form pNEW-pre.
Subsequently, the Pkm-cymR (repressor protein-encoding gene under the control of the promoter for the kanamycin resistance gene) was amplified by PCR from pBRI-cymR1 (7) using primers PKM-CYM-MLU-F (5′-CAC GCG TCC GGA ATT GCC AGC TGG GGC GCC CTC TGG TAA GGT TGG GAA GCC CTG CAA AGT AAA CTG GAT GGC TTT CTT GCC GCC AAG GAT CTG ATG GCG CAG GGG ATC AAG ATC TGA TCA AGA GAC AGG ATG AGG ATC GTT TCG CAA GAT GGT GAT CAT GAG TCC AAA GAG AAG AAC ACA G-3′; the MluI site is underlined) and CYM-PCI-R (5′-CAC ATG TCT AGC GCT TGA ATT TCG CGT ACC GCT CTC-3′; the PciI site is underlined). The PCR product containing Pkm-cymR was then cloned into pCR2.1 to create pCR-Pkm-cymR, and the MluI-PciI fragment from pCR-Pkm-cymR was ligated to pNEW-pre digested by the same enzymes to generate pNEW-gfp. Then, the general expression vector pNEW-MCS was constructed as follows. The gfp gene in pNEW-gfp was replaced with the multiple cloning site (MCS) from pSL1190. The MCS was released from pSL1190 by being digested with NheI and SacI and ligated into pNEW-gfp that had been digested with the same restriction enzymes. The MCS sequence from pSL1190 contained two NdeI sites, each possessing ATG as part of its sequence. In order to avoid potential false starts from the MCS, both NdeI sites were removed. Vector pNEW-MCS was digested with AvrII to remove 144 bp containing one NdeI site. The other NdeI site was removed by being digested with NdeI followed by mung bean nuclease (New England Biolabs, Pickering, Canada) treatment and then religated to construct the pNEW expression vector.
Expression modulation in E. coli with different cumate concentrations.
E. coli BL21(DE3) bearing pNEW-gfp was grown in 250-ml baffled shake flasks containing 50 ml LB medium plus 50 μg/ml kanamycin at 37°C and 250 rpm. A 1% overnight culture served as the inoculum. When the cultures reached an optical density (OD) (600 nm) of 0.6 to 0.8, they were induced with cumate at concentrations ranging between 0 and 122 μM. The cultures were harvested after 4 h of induction, and GFP was quantified using a spectrofluorometer (SPECTRAFluor Plus; Tecan Austria Gmbh, Grödig, Austria) under excitation and emission of 485 and 508 nm, respectively. The concentration of GFP was calculated based on a linear relationship between concentration and fluorescence units determined using solutions of purified GFP (Qbiogene Inc., Carlsbad, CA). Biomass was determined from OD readings at 600 nm referenced to dry weight calibration curves.
Flow cytometry analysis.
E. coli BL21(DE3) bearing either pNEW-gfp or pET-gfp was grown as described above. At an OD (600 nm) of 0.6 to 0.8, pNEW-gfp- and pET-gfp-bearing cultures were induced with 100 μM cumate and 100 μM IPTG, respectively. At different hours after induction, the cultures were harvested for flow cytometry analysis. Flow cytometry analysis for GFP expression was performed on a Coulter Epics XL-MCL flow cytometry system (Beckman-Coulter, Mississauga, Canada) equipped with an argon ion laser of 15 mW at 488 nm as an excitation source. The green fluorescence emission was detected using a 530/30-nm band pass filter set. Cells were diluted with phosphate-buffered saline (PBS) to an OD of 0.03 at 600 nm and fixed with 2% formaldehyde.
Time course analysis of GFP expression.
Fresh cultures of the recombinant strains harboring pNEW-gfp and pET-gfp in E. coli BL21(DE3) were transferred to 24-well plates in the spectrofluorometer at 37°C. When the OD of cultures reached 0.6 to 0.8 at 600 nm, cultures were induced with cumate or IPTG at two different concentrations (100 μM and 1,000 μM), and the fluorescence was read at 1-min intervals for 60 min. GFP was quantified using a spectrofluorometer under excitation and emission of 485 and 508 nm, respectively. The fluorescence reading was corrected with the relative fluorescent unit (RFU) value at time zero.
Fed-batch fermentations.
GFP production in E. coli BL21(DE3) bearing either pNEW-gfp or pET-gfp was determined in fed-batch fermentation experiments using both rich and chemically defined media in 14-liter bioreactors (BioFlo 110; New Brunswick Scientific, Edison, NJ). Chemically defined medium consisted of R modified medium (48) supplemented with 2% glucose and 50 μg/ml kanamycin. Rich medium consisted of 12 g phytone peptone, 24 g yeast extract, 16.9 mM KH2PO4, 33.1 mM Na2HPO4·H2O, 18.9 mM (NH4)2SO4, 0.08 mM CaCl2, 0.046 mM FeCl2, and 2 mM MgSO4·7H2O per liter. Kanamycin was added at a concentration of 50 μg/ml. For experiments in defined medium, the inoculum was prepared in 2YP medium (12 g phytone peptone, 24 g yeast extract, 40 g glucose per liter, and 50 μg/ml of kanamycin) with 200 μl from glycerol stock. After an incubation period of at least 6 h (280 rpm, 37°C), the culture was centrifuged and the pellet was resuspended in 50 ml of fresh chemically defined medium and aseptically transferred to the fermentors to reach a starting OD (600 nm) of 0.1.
For experiments in rich medium, the inoculum was prepared in the rich medium described above. After an incubation period of 13 h (280 rpm, 37°C), the culture was aseptically transferred to the fermentors to reach a starting OD (600 nm) of 0.1.
The fermentations with defined or rich medium were carried out at 37°C with an aeration and agitation profile of 3.5 liters per minute of air and 250 to 1,000 rpm, respectively. pH was controlled automatically at 6.95 with 28 to 30% NH3 and 28% H3PO4. Dissolved oxygen was controlled at 25% by an agitation/oxygen enrichment cascade, i.e., when the agitation level reached maximum, the gas mixer fed oxygen-enriched air automatically. Undiluted Mazu DF 204 (BASF, Florham Park, NJ) antifoam was injected manually when excess foaming occurred. The depletion of glucose marked the end of the batch phase of the fermentation and coincided with an OD at 600 nm of 20 to 24.
For both rich and defined medium fermentations, the fed-batch phase was initiated by feeding a 50% glucose solution with 50 μg/ml kanamycin. The fed-batch strategy was the fixed growth rate method where the feed pump followed an exponential flow rate increase to support a growth rate of 0.14 h−1.
At an OD at 600 nm ranging between 38 and 42, the fed-batch cultures were induced with the appropriate inducer at two different concentrations (100 and 1,000 μM). Subsequently, on an hourly basis, samples were taken in order to determine GFP and biomass yields. Each sample was aliquoted for dry weight measurement in triplicate and diluted in 0.85% NaCl for OD measurement at 600 nm. The remaining volume of diluted sample was frozen until total measurement of GFP. The samples were compared for the same induction times, which spanned between 1 and 8 h and up to 21 h in some cases.
Western blotting.
Expression of repressor protein (CymR) was determined by Western blotting using standard protocols. CymR was detected with rabbit anti-bCymR antibodies (in-house) and a goat anti-rabbit IgG(H+L) horseradish peroxidase (HRP) conjugate (Pierce Inc., West Grove, PA).
RESULTS AND DISCUSSION
Construction of the cumate-regulated/inducible system in E. coli.
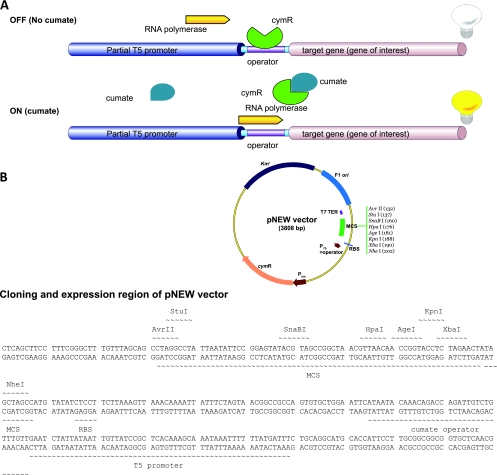
The basic mechanism by which the cumate-regulated expression functions in the native P. putida F1 and how it was applied to mammalian and methylotrophic bacterial cells have been previously described (7, 10, 11, 31). Briefly, the cumate switch requires four major components: a strong promoter, a repressor-binding DNA sequence or operator, expression of a repressor-encoding gene (cymR), and cumate as the inducer (7, 31). The addition of the inducer, cumate, rapidly alters the binding of the repressor CymR to the operator, thereby dislodging the repressor from the operator, culminating in the formation of the CymR-cumate complex and the expression of the target gene. The basic mechanism of this system is summarized in Fig. 1A .
FIG. 1.
(A) A schematic diagram of the mechanism of action of the cumate gene switch developed for E. coli strains. The mechanism is described in Results and Discussion. (B) Physical map of the pNEW expression vector. RBS, ribosome-binding site.
Since E. coli remains the prokaryotic expression host of choice for the production of many recombinant proteins, it was decided to adapt the cumate-regulated expression system to E. coli strains. The construct of the cumate switch in E. coli described herein is a simplified and more flexible version of the Methylobacterium extorquens cumate switch. All the regulatory elements have been incorporated in the one plasmid named pNEW as shown in Fig. 1B (the complete sequence is available in the supplemental material). In M. extorquens, overexpression of cymR resulted in growth inhibition. In order to avoid cellular toxicity caused by the CymR in M. extorquens, expression levels were reduced by integrating only two copies of cymR in the chromosome (7). In the E. coli cumate switch construct, this cumbersome cloning step was eliminated by putting the 612-bp cymR gene under the control of the weak and constitutive promoter Pkm. CymR expression was confirmed by Western blot analysis (see the supplemental material). Although the level of CymR expression driven by Pkm was not calculated, the important observation was that sufficient CymR was produced, inhibiting transcription of target genes. Within the same plasmid (pNEW) a truncated operator fragment (26 bp) of the cmt operon was inserted downstream of the strong T5 promoter (PT5) which is broadly recognized by RNA polymerases in E. coli strains (3), and upstream of an MCS (Fig. 1B). In order to minimize transcriptional read-through from PT5, the T7 terminator was maintained just downstream of MCS and the kanamycin resistance gene (Kmr) was used for recombinant selection. A variety of restriction sites are available within the MCS to facilitate gene cloning strategies.
Initial evaluation of pNEW as an inducible tightly regulated expression system in E. coli.
To evaluate the performance of the pNEW vector as an inducible system, the expression of the following genes was tested: green fluorescent protein (gfp), polyhydroxyalkanoic acid synthases (phaC1 and phaC2), lactase (bgl), esterase (estI), and synthetic thrombin inhibitory peptide (mfh). These genes were amplified by PCR and individually cloned into pNEW. Each of these recombinant plasmids was individually introduced into various E. coli strains, and the transformants were cultured with or without cumate. Results revealed that the target proteins tested were highly expressed upon cumate induction (see the supplemental material). Target protein activity was not detected in noninduced transformants, demonstrating a tightly regulated expression system.
Applicability of the cumate switch to various E. coli hosts.
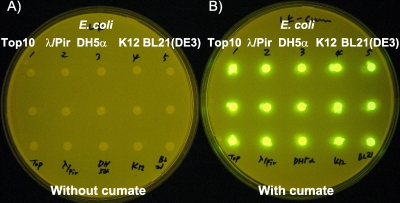
The E. coli strains tested were E. coli DH5α, E. coli Top10, E. coli BL21(DE3), E. coli K-12, and E. coli S-17/λpir. The cumate switch was operational in all strains transformed with pNEW-gfp as shown in Fig. 2. In all cases, cumate-induced shake flask-grown cultures [E. coli DH5α, E. coli Top10, E. coli BL21(DE3), E. coli K-12, and E. coli S-17/λpir] expressed high GFP levels (maximal yields: 355, 375, 380, 424, and 286 mg GFP/liter, respectively). These findings demonstrate that the pNEW system is tightly regulated in different E. coli strains, suggesting its wide applicability. This can be explained by the fact that the cumate switch described herein requires only the interaction of the inducer (cumate) and the constitutively expressed repressor (CymR) on the surface of the PT5-operator DNA fragment of pNEW to alleviate repression. The universal recognition and functionality of PT5 in E. coli strains make it possible for the cumate switch to operate effectively in a wide range of E. coli hosts. It can be said that any E. coli strain that recognizes PT5 can be considered a “pNEW expression-ready” host.
FIG. 2.
Host versatility of the pNEW expression system.
Dose-dependent (rheostat) control of reporter gene expression by cumate.
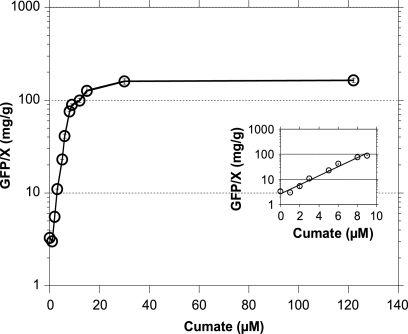
To determine if the cumate switch affords a rheostat mechanism in E. coli, we measured the specific GFP yields of cells transformed with pNEW-gfp induced with various concentrations of cumate, ranging from 0 to 122 μM. Maximal GFP specific yields were obtained at 4 h after induction, and this was achieved with a cumate concentration of 30 μM (Fig. 3). A linear exponential specific GFP yield response (rheostat mechanism) was observed in the presence of subsaturating cumate concentrations in the range of 0 to 10 μM, which culminated in a wide range of specific GFP yields (0 to 90 mg/g [dry weight] of cells). The GFP yield of 90 mg/g, attained at the upper limit of the linear response range, represents approximately 50% of what is maximally achievable in high-cell-density fermentations induced with 100 μM (Table 2). Protein expression driven by PBAD and Plac promoters can also be modulated by varying the inducer concentrations, but the rheostat mechanism functions only at much higher concentrations (≥100 μM) of arabinose and IPTG (14, 19). These two expression systems have been referred to as being functionally “all-or-none” induction systems. With the cumate switch, not only is the rheostat zone achievable with lower and more economical concentrations of inducer (cumate) but it possesses the potential, if desired, to achieve high recombinant protein yields (50% of maximal yields) at the upper inducer concentration levels of the linear rheostat range. This production yield flexibility can be very useful for process development (24, 33) and production strategies and as a tool for metabolic engineering (25).
FIG. 3.
Modulation of gene expression using the cumate gene switch.
TABLE 2.
Comparison of GFP yield between the pNEW-GFP and pET-GFP strains in defined medium fed-batch bioreactorsa
| Concn (μM) and inducer | Specific yield (total yield) of GFP at induction time (h)b: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 100 | ||||||||
| Cumate | 30 ± 2 (602 ± 12) | 70 ± 4 (1,644 ± 34) | 105 ± 5 (3,002 ± 25) | 143 ± 7 (4,719 ± 122) | 155 ± 7 (5,443 ± 87) | 149 ± 6 (6,194 ± 119) | 172 ± 7 (6,978 ± 185) | 198 ± 9 (7,838 ± 249) |
| IPTG | 28 ± 2 (554 ± 17) | 60 ± 4 (1,300 ± 33) | 75 ± 5 (1,778 ± 42) | 83 ± 5 (2,486 ± 115) | 73 ± 4 (2,035 ± 70) | 72 ± 4 (2,232 ± 80) | 59 ± 3 (1,948 ± 13) | 59 ± 3 (2,090 ± 33) |
| 1,000 | ||||||||
| Cumate | 28 ± 2 (486 ± 0.4) | 59 ± 3 (1,459 ± 2) | 95 ± 5 (2,851 ± 78) | 125 ± 5 (4,593 ± 44) | 132 ± 6 (4,885 ± 97) | 200 ± 7 (8,569 ± 110) | 224 ± 8 (9,666 ± 153) | 260 ± 9 (11,151 ± 120) |
| IPTG | 30 ± 2 (607 ± 24) | 61 ± 4 (1,578 ± 7) | 88 ± 4 (2,464 ± 9) | 103 ± 5 (3,340 ± 74) | 119 ± 5 (4,238 ± 39) | 127 ± 9 (4,080 ± 219) | − | − |
Triplicate analyses were conducted with samples from the same fermentations.
Specific yield is in mg/g (dry weight [g/liter]); total yield is in mg/liter. −, not measured.
Induction profile of pNEW-GFP and pET-GFP E. coli strains.
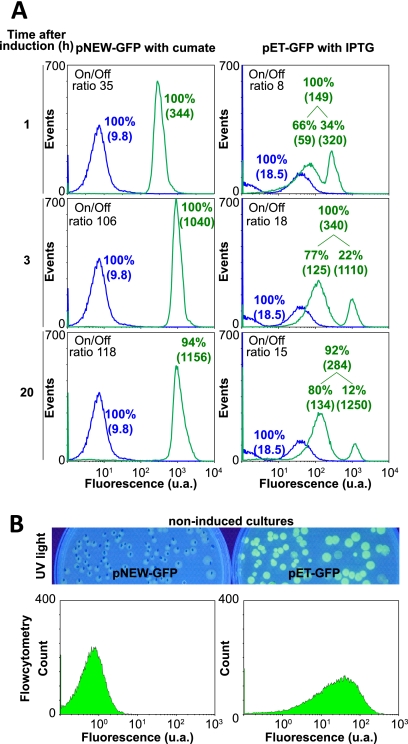
Flow cytometry, which can effectively measure single-cell properties within a microbial population, was used to compare the regulation of the cumate switch and the IPTG-inducible expression systems. E. coli BL21(DE3) cultures bearing either pNEW-gfp (pNEW-GFP strain) or pET-gfp (pET-GFP strain) were grown in shake flasks in the presence or absence of appropriate inducers and prepared for flow cytometry analysis as described in Materials and Methods. Figure 4 reveals that the cumate regulatory elements in pNEW-gfp very tightly regulate the transcription of the heterologous gene (gfp). The noninduced pNEW-GFP strain resulted in very low basal expression or leakiness in comparison to the noninduced pET-GFP strain (mean fluorescence of 9.8 versus 18.5, respectively). Cumate induction culminated in higher induction levels than did the IPTG-regulated pET system. Upon induction, flow cytometry analysis of both expression systems revealed two different expression profiles (Fig. 4A). After 1, 3, and 20 h of induction with cumate, 100% of the pNEW-gfp-bearing population had a homogeneous positive population expressing GFP with a mean fluorescence of 344, 1,040, and 1,156, respectively. In contrast, analysis of IPTG-induced pET-gfp-bearing cultures revealed two distinct populations (bimodal expression) throughout the induction period with respect to gfp expression. At 3 h of induction for example, 77% of the population had a fluorescence mean of 125 and the remaining 22% had a fluorescence mean of 1,100, clearly showing that the induction response is heterogeneous.
FIG. 4.
Flow cytometry analysis of GFP expression. (A) GFP fluorescence intensity peaks from E. coli BL21(DE3) bearing pNEW-gfp and pET-gfp at 1, 3, and 20 h after induction with 100 μM cumate and 100 μM IPTG, respectively. Blue peaks represent noninduced cells; green peaks represent induced cells. (B) Comparison of noninduced GFP expression in E. coli BL21(DE3) bearing pNEW-gfp and pET-gfp grown on solid medium. u.a., arbitrary units.
The on-off ratio, which illustrates the strength of the expression response to an inducer, is calculated by dividing the mean fluorescence of induced cells by the mean fluorescence of the noninduced cells. At 1 h of induction, the on-off ratio for the cumate-induced population was already approximately four times higher than that for the IPTG-induced population (ratio of 35 for the pNEW-GFP strain compared to 8 for the pET-GFP strain). This difference is further accentuated with time. At 3 h of induction, the on-off ratio of cumate-induced culture increases to approximately six times that of the culture induced with IPTG (ratio of 106 for the pNEW-GFP strain compared to 18 for the pET-GFP strain). The GFP yield of both cultures was determined at this point and revealed that the homogenous cumate-induced culture produced approximately 2-fold-higher GFP levels than did the IPTG-induced culture (175 and 92 mg GFP/g [dry weight] of cells, respectively).
However, although the pNEW-gfp-bearing population maintained a single homogenous GFP-expressing profile even up to 20 h of induction, at this time both expression systems revealed the presence of a small number of negative cells, signaling the onset of cell lysis. It should be noted that the homogenous-expression trait of the cumate-induced population is an important feature of gene expression systems, especially when they are to be utilized for carbon flux regulation through metabolically engineered pathways (25).
In regards to the noninduced (basal) expression and as frequently reported in the literature on this topic (14, 25, 34, 35, 38a), we have observed extensive noninduced GFP expression in the pET-gfp-bearing E. coli strains when grown on solid media. Figure 4B shows the marked difference in noninduced expression between the cumate gene switch and the pET expression system. This leakiness may not be a problem if the expressed protein is not toxic to the cell. However, if the heterologous protein is toxic, leakiness can result in cell metabolic stress, poor culture growth, and lower recombinant protein yields. Using a well-regulated system affords the freedom to choose the time of induction for optimal protein production and optimal culture growth.
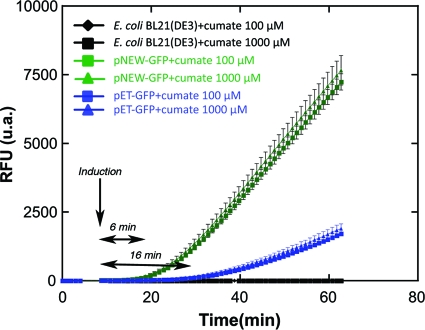
Another important trait of the cumate gene switch is the quickness of response by which it is effectively turned on (Fig. 5). Under the assay conditions utilized, GFP expression in the pNEW-gfp-bearing culture is measurable soon (6 min) after induction with cumate. In contrast, pET-gfp-bearing cultures require a 16-min post-IPTG-induction period before GFP expression can be measured. The increase in the “GFP expression lag time” may be explained by the indirect nature of the IPTG-inducing mechanism in pET systems. IPTG must first induce the transcription of the T7 RNA polymerase in the E. coli host. Only then will transcription of target genes under the control of the T7 promoter occur. In contrast, the induction mechanism of the cumate gene switch is much simpler and direct. It requires only that cumate dislodge the CymR repressor from the (PT5)-operator fragment, permitting the binding of existing E. coli RNA polymerases and expression of target genes.
FIG. 5.
Time course analysis of GFP expression in E. coli BL21(DE3) bearing pNEW-gfp and pET-gfp after induction with cumate and IPTG, respectively. u.a., arbitrary units.
Comparison of GFP yield between the pNEW-GFP and pET-GFP strains in fed-batch bioreactors.
Shake flask and solid medium cultures harboring either the T7 system or the cumate system revealed a higher level of GFP expression in cumate-induced cultures (data not shown). These results were obtained using low-cell-density-growth conditions. Overexpression of heterologous protein is a goal in most industrial protein production processes. However, the high-cell-density bioprocesses often utilized for this purpose may lead to stressful nutrient limitation, thereby culminating in alterations in the organism's physiology, plasmid stability, and ultimately recombinant protein yields (5, 15, 29, 38a, 44). In order to determine the effect of high-cell-density cultivation on GFP yields, high-cell-density fed-batch fermentation experiments were carried out in 14-liter bioreactors. Both rich and chemically defined growth medium formulations were utilized. Cultures were grown to an OD ranging between 38 and 42 at 600 nm. At this point two strategies were used with respect to the concentration of inducer utilized. As was observed and described above in this article, in shake flask cultures, an inducer concentration of 100 μM for both cumate and IPTG was sufficient for maximal GFP expression. However, this IPTG concentration is 10-fold lower than what is normally used in high-cell-density microbial cultivation. Since the inducer concentration of 100 μM may be insufficient to effectively induce the entire high-cell-density microbial population, fed-batch fermentations induced with cumate and IPTG at 1,000 μM concentrations were also tested. Table 2 shows the volumetric and specific GFP yields obtained in E. coli BL21(DE3) harboring either pNEW-gfp or pET-gfp, measured at determined intervals following induction with cumate or IPTG at two different concentrations (100 and 1,000 μM). The cumate-induced cultures consistently outperformed the IPTG-induced cultures with respect to the levels of GFP expression. This difference is accentuated in fed-batch cultures at 8 h after induction with 100 μM respective inducers. At this inducer concentration, cumate-induced cultures produce approximately 2- to 3-fold more GFP (total volumetric and biomass specific) than do IPTG-induced cultures. In contrast to IPTG-induced cultures, the GFP production in cumate-induced cultures continues to increase even up to 8 h of induction, both when cumate is used at 100 μM and when it is used at 1,000 μM. Lengthening the duration of the fermentation process of cultures induced with 1,000 μM cumate from 4 to 8 h results in the doubling of GFP yields. And although prolonging the fermentation process by 4 h in order to achieve a greater recombinant protein yield may not be feasible or practical for certain bioprocesses, it does provide an additional bioprocess option, which is not possible with IPTG-induced cultures. IPTG-induced cultures reach their maximal GFP-producing capacity at 4 to 6 h of induction.
At 6 to 8 h of induction, extensive foaming was observed in IPTG-induced cultures, suggesting cell lysis, an observation also reflected by the decrease in biomass-specific GFP production. The cumate-induced cultures remained healthy, no lysis occurred, and no foaming was observed. There have been numerous reports on the observed toxicity of IPTG to the host, even at concentrations as low as 100 μM (14, 22, 24). We have observed a 10% decrease in the final biomass concentration (dry weight) in high-cell-density cultures induced with 1,000 μM IPTG in comparison to cultures induced with 100 μM IPTG. This observed toxicity effect caused by the inducing agent, contributing to excessive foaming, was not observed in cultures induced with saturating levels of cumate in the range of 100 to 1,000 μM. At identical inducer concentrations, cumate-induced cultures always resulted in higher biomass yields than did IPTG-induced cultures. For example, high-cell-density cultures induced with 1,000 μM cumate reached 33% higher biomass yields than did cultures induced with the same concentration of IPTG (1,000 μM).
It is also important to reiterate that throughout the induction phase, higher GFP yields were obtained in cultures induced with 100 μM cumate than in those induced with 1,000 μM IPTG. This is an important cost-effective advantage, especially for large-scale fermentation processes. Even under high-cell-density conditions, the GFP-producing capacities of cultures after 4 h of induction with 100 μM and with 1,000 μM cumate were similar. The same cannot be said for cultures induced with different IPTG concentrations. Throughout the induction period, the culture induced with 1,000 μM IPTG resulted in greater GFP yields than did the culture induced with 100 μM IPTG (Table 2).
There are bioprocesses that require the utilization of rich medium formulations for optimal growth characteristic and gene expression (30, 43). Consequently, the robustness of the cumate switch was tested and compared to that of IPTG-induced fed-batch cultures grown in rich medium. Although both the biomass and GFP yields obtained were higher, cumate-induced cultures consistently produced more GFP than did IPTG-induced cultures (data not shown). The loss of GFP productivity at the onset of cell lysis in IPTG-induced cells also occurred in rich media, as it had occurred in chemically defined media (data not shown).
Conclusions.
The cumate gene switch described in this paper has several salient features and benefits that render it an attractive expression system for the production of heterologous proteins. The regulatory elements required for a functional cumate gene switch (promoter-operator sequence and the repressor gene) are incorporated within a plasmid (pNEW) that is transferable and applicable to well-characterized E. coli strains [E. coli DH5α, S-17/λpir, K-12, Top10, and BL21(DE3)]. Due to the simplicity of the switch, induction is achieved quickly. This also results in a fully induced and homogenous population expressing similar levels of heterologous protein at the cellular level. Modulated protein expression can be easily achieved by modifying the concentration of cumate. In contrast to IPTG, the inducer used in the well-characterized pET expression systems, cumate, is inexpensive, and maximal expression of heterologous protein can be achieved at 10% of the usual concentration of IPTG. Also, whereas IPTG is known to cause cellular toxicity issues, cumate is noncytotoxic, at least not at the concentrations utilized to achieve maximal product yields. Furthermore, in contrast to the pET system, cells possessing the cumate gene switch remain fully induced, achieving 2- to 3-fold-higher target protein yields.
Supplementary Material
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baneyx, F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10:411-421. [DOI] [PubMed] [Google Scholar]
- 2.Bhandari, P., and J. Gowrishankar. 1997. An Escherichia coli host strain useful for efficient overproduction of cloned gene products with NaCl as the inducer. J. Bacteriol. 179:4403-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bujard, H., R. Gentz, M. Lanzer, D. Stüber, M. Müller, I. Ibrahimi, M. T. Häuptle, and B. Dobberstein. 1987. A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Methods Enzymol. 155:416-433. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee, D. K., and D. Esposito. 2006. Enhanced soluble protein expression using two new fusion tags. Protein Expr. Purif. 46:122-129. [DOI] [PubMed] [Google Scholar]
- 5.Chaves, A. C., F. G. C. Abath, J. L. Lima-Filho, J. M. S. Cabral, and N. Lucena-Silva. 1999. Studies on growth kinetics and plasmid stability of recombinant Escherichia coli expressing a Schistosoma mansoni antigen. Bioprocess Engin. 21:355-361. [Google Scholar]
- 6.Choi, Y. J., C. B. Míguez, and B. H. Lee. 2004. Characterization and heterologous gene expression of a novel esterase from Lactobacillus casei CL96. Appl. Environ. Microbiol. 70:3213-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, Y. J., L. Morel, D. Bourque, A. Mullick, B. Massie, and C. B. Míguez. 2006. Bestowing inducibility on the cloned methanol dehydrogenase promoter (PmxaF) of Methylobacterium extorquens by applying regulatory elements of Pseudomonas putida F1. Appl. Environ. Microbiol. 72:7723-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 9.De Mey, M., J. Maertens, G. J. Lequeux, W. K. Soetaert, and E. J. Vandamme. 2007. Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnol. 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton, R. W. 1996. p-Cumate catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA carrying the cmt operon. J. Bacteriol. 178:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton, R. W. 1997. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J. Bacteriol. 179:3171-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figge, J., C. Wright, C. J. Collins, T. M. Roberts, and D. M. Livingston. 1988. Stringent regulation of stably integrated chloramphenicol acetyl transferase genes by E. coli lac repressor in monkey cells. Cell 52:713-722. [DOI] [PubMed] [Google Scholar]
- 13.Georgiou, G., and L. Segatori. 2005. Preparative expression of secreted proteins in bacteria: status report and future prospects. Curr. Opin. Biotechnol. 16:538-545. [DOI] [PubMed] [Google Scholar]
- 14.Giacalone, M. J., A. M. Gentile, B. T. Lovitt, N. L. Berkley, C. W. Gunderson, and M. W. Suber. 2006. Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. Biotechniques 3:355-366. [DOI] [PubMed] [Google Scholar]
- 15.Glick, B. R. 1995. Metabolic load and heterologous gene expression. Biotechnol. Adv. 13:247-261. [DOI] [PubMed] [Google Scholar]
- 16.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U. S. A. 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, J. C., M. Jaisani, G. Pandey, and K. J. Mukherjee. 1999. Enhancing recombinant protein yields in Escherichia coli using the T7 system under the control of heat inducible λPL promoter. J. Biotechnol. 68:125-134. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1985. Techniques for transformation of Escherichia coli, p. 109-135. In D. M. Glover (ed.), DNA cloning: a practical approach, vol. 1. IRL Press, McLean, VA.
- 19.Hashemzadeh-Bonehi, L., F. Mehraein-Ghomi, C. Mitsopoulos, J. P. Jacob, E. S. Hennessy, and J. K. Broome-Smith. 1998. Importance of using lac rather than ara promoter vectors for modulating the levels of toxic gene products in Escherichia coli. Mol. Microbiol. 30:676-678. [DOI] [PubMed] [Google Scholar]
- 20.Hung, M. N., Z. Xia, N. T. Hu, and B. H. Lee. 2001. Molecular and biochemical analysis of two β-galactosidases from Bifidobacterium infantis HL96. Appl. Environ. Microbiol. 67:4256-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaeger, K. E., A. Steinbüchel, and D. Jendrossek. 1995. Substrate specificities of bacterial polyhydroxyalkanoate depolymerases and lipases: bacterial lipases hydrolyze poly(omega-hydroxyalkanoates). Appl. Environ. Microbiol. 61:3113-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jana, S., and J. K. Deb. 2005. Strategies for the production of heterologous proteins in Escherichia coli. Appl. Microbiol. Biotechnol. 67:289-298. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaur, P., S. Agarwal, and S. Datta. 2009. Delineating bacteriostatic and bacteriocidal targets in Mycobacteria using IPTG inducible antisense expression. PLoS One 4:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keasling, J. D. 1999. Gene-expression tools for the metabolic engineering of bacteria. Trends Biotechnol. 17:452-459. [DOI] [PubMed] [Google Scholar]
- 26.Keyes, W., and A. Mills. 2003. Inducible systems see the light. Trends Biotechnol. 21:53-55. [DOI] [PubMed] [Google Scholar]
- 27.Knodler, L. A., E. O. Sekyere, T. S. Stewart, P. J. Schofield, and M. R. Edwards. 1998. Cloning and expression of a prokaryotic enzyme, arginine deiminase, from a primitive eukaryote Giardia intestinalis. J. Biol. Chem. 273:4470-4477. [DOI] [PubMed] [Google Scholar]
- 28.Kosinski, M. J., U. Rinas, and J. E. Bailey. 1992. Isopropyl-beta-D-thiogalactopyranoside influences the metabolism of E. coli. Appl. Microbiol. Biotechnol. 36:782-784. [Google Scholar]
- 29.Lee, S. Y. 1996. High cell-density culture of Escherichia coli. Trends Biotechnol. 14:98-105. [DOI] [PubMed] [Google Scholar]
- 30.Losen, M., B. Frölich, M. Pohl, and J. Büchs. 2004. Effect of oxygen limitation and medium composition on Escherichia coli fermentation in shake-flask cultures. Biotechnol. Prog. 20:1062-1068. [DOI] [PubMed] [Google Scholar]
- 31.Mullick, A., Y. Xu, R. Warren, M. Koutroumanis, C. Guilbault, S. Broussau, F. Malenfant, L. Bourget, L. Lamoureux, R. Lo, A. Caron, A. Pilotte, and B. Massie. 2006. The cumate gene-switch: a system for regulated expression in mammalian cells. BMC Biotechnol. 6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osborne, M. J., Z. Su, V. Sridaran, and F. Ni. 2003. Efficient expression of isotopically labeled peptides for high resolution NMR studies: application to the Cdc42/Rac binding domains of virulent kinases in Candida albicans. J. Biomol. NMR 26:317-326. [DOI] [PubMed] [Google Scholar]
- 33.Otto, C. M., F. Niagro, X. Su, and C. A. Rawlings. 1995. Expression of recombinant tumor necrosis factor is toxic to Escherichia coli. Clin. Diagn. Lab. Immunol. 2:740-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patkar, A., N. Vijayasankaran, D. W. Urry, and F. Srienc. 2002. Flow cytometry as a useful tool for process development: rapid evaluation of expression systems. J. Biotechnol. 93:217-229. [DOI] [PubMed] [Google Scholar]
- 35.Qing, G., L. C. Ma, A. Khorchid, G. V. Swapna, T. K. Mal, M. M. Takayama, B. Xia, S. Phadtare, H. Ke, T. Acton, G. T. Montelione, M. Ikura, and M. Inouye. 2004. Cold-shock induced high-yield protein production in. Escherichia coli. Nat. Biotechnol. 22:826-827. [DOI] [PubMed] [Google Scholar]
- 36.Riesenberg, D., and R. Guthke. 1999. High cell density cultivation of microorganisms. Appl. Microbiol. Biotechnol. 51:422-430. [DOI] [PubMed] [Google Scholar]
- 37.Rossi, F. M., and H. M. Blau. 1998. Recent advances in inducible gene expression systems. Curr. Opin. Biotechnol. 9:451-456. [DOI] [PubMed] [Google Scholar]
- 38.Schein, C. H., and M. H. M. Noteborn. 1988. Formation of soluble recombinant proteins in E. coli is favored by lower growth temperature. Biotechnology (NY) 6:291-294. [Google Scholar]
- 38a.Sevastsyanovich, Y. R., S. N. Alfasi, and J. A. Cole. 2010. Sense and nonsense from a systems biology approach to microbial recombinant protein production. Biotechnol. Appl. Biochem. 55:9-28. [DOI] [PubMed] [Google Scholar]
- 39.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. U. S. A. 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sørensen, H. P., and K. K. Mortensen. 2005. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Fact. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staub, J. M., B. Garcia, J. Graves, P. T. J. Hajdukiewicz, P. Hunter, N. Nehra, V. Paradkar, M. Schlittler, J. B. Carroll, I. Spatola, D. Ward, Y. E. Guangning, and D. A. Russell. 2000. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat. Biotechnol. 18:333-338. [DOI] [PubMed] [Google Scholar]
- 42.Studier, F. W., and B. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 43.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiry, M., and D. Cingolani. 2002. Optimizing scale-up fermentation processes. Trends Biotechnol. 20:103-105. [DOI] [PubMed] [Google Scholar]
- 45.Thomas, J. G., and F. Baneyx. 1996. Protein misfolding and inclusion body formation in recombinant Escherichia coli cells overexpressing heat-shock proteins. J. Biol. Chem. 271:11141-11147. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Z. W., W. S. Law, and Y. P. Chao. 2004. Improvement of the thermoregulated T7 expression system by using the heat-sensitive lacI. Biotechnol. Prog. 20:1352-1358. [DOI] [PubMed] [Google Scholar]
- 47.Yogender, P., J. C. Gupta, and K. J. Mukherjee. 2001. Optimizing recombinant protein expression in the T7 system under the control of the proUp promoter. Biotechnol. Lett. 23:41-46. [Google Scholar]
- 48.Yoon, S. H., M. J. Han, S. Y. Lee, K. J. Jeong, and J. S. Yoo. 2003. Combined transcriptome and proteome analysis of Escherichia coli during high cell density culture. Biotechnol. Bioeng. 81:753-767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.