Abstract
The structures of polio-, coxsackie-, and rhinovirus polymerases have revealed a conserved yet unusual protein conformation surrounding their buried N termini where a β-strand distortion results in a solvent-exposed hydrophobic amino acid at residue 5. In a previous study, we found that coxsackievirus polymerase activity increased or decreased depending on the size of the amino acid at residue 5 and proposed that this residue becomes buried during the catalytic cycle. In this work, we extend our studies to show that poliovirus polymerase activity is also dependent on the nature of residue 5 and further elucidate which aspects of polymerase function are affected. Poliovirus polymerases with mutations of tryptophan 5 retain wild-type elongation rates, RNA binding affinities, and elongation complex formation rates but form unstable elongation complexes. A large hydrophobic residue is required to maintain the polymerase in an elongation-competent conformation, and smaller hydrophobic residues at position 5 progressively decrease the stability of elongation complexes and their processivity on genome-length templates. Consistent with this, the mutations also reduced viral RNA production in a cell-free replication system. In vivo, viruses containing residue 5 mutants produce viable virus, and an aromatic phenylalanine was maintained with only a slightly decreased virus growth rate. However, nonaromatic amino acids resulted in slow-growing viruses that reverted to wild type. The structural basis for this polymerase phenotype is yet to be determined, and we speculate that amino acid residue 5 interacts directly with template RNA or is involved in a protein structural interaction that stabilizes the elongation complex.
Members of the Picornaviridae family of small RNA viruses cause a wide range of diseases in humans, including liver disease, heart disease, aseptic meningitis, the common cold, and poliomyelitis. The picornaviruses include the most common human viruses, which are the rhinoviruses that spread through respiratory pathways, and the second most common viruses, which are enteroviruses that spread by fecal-oral transmission. These viruses have ≈7.5-kb positive-sense genomes containing a single large open reading frame encoding a ≈250-kDa polyprotein that is cleaved into about a dozen different proteins by viral proteases (20). Their genome life cycle is completely RNA based, with replication being driven by the viral 3Dpol protein, an RNA-dependent RNA polymerase (RdRP).
After viral RNA translation and polyprotein processing, 3Dpol replicates the infecting positive-strand RNA template into a negative-strand intermediate that is subsequently used as a template for positive-strand synthesis. During these processes, 3Dpol interacts with multiple templates, substrates, and other viral proteins; however, many aspects of these events remain obscure. The crystal structures of several picornaviral 3Dpol enzymes have been solved, and these all conform to the “right hand” analogy commonly used to describe polymerases as having palm, thumb, and finger domains (10, 14, 18, 22, 27, 29). Based on homology to other polymerases and the structures of 3Dpol-RNA complexes with foot-and-mouth disease and Norwalk virus polymerases, the finger domain interacts with the template RNA, the palm domain contains the active-site aspartate residues that coordinate the metals needed for catalysis, and the thumb domain contacts the exiting duplex RNA product (13, 26, 30).
Poliovirus is among the most-studied picornaviruses, and its polymerase has been thoroughly characterized biochemically (3, 15) and structurally (28, 29). Processive RNA synthesis requires the formation of a stable 3Dpol elongation complex through a multistep process involving at least two conformational changes (2). First, following RNA binding, there is a slow (t1/2 ≈ 12 s) conformational change that results in a 3Dpol-RNA complex poised for nucleoside triphosphate (NTP) incorporation. Second, following the addition of the first nucleotide to the primer, there is another conformational change to produce a very stable elongation complex with an in vitro half-life on the order of several hours. The polymerase begins processive elongation after the formation of this stable elongation complex, and each nucleotide addition cycle involves a five-step mechanism, of which NTP repositioning and NTP catalysis are rate limiting (3). Similar experiments using the homologous foot-and-mouth disease virus polymerase reveal an analogous set of complexes resulting in an elongation complex with a half-life of 27 h (1). Although these viral polymerase complexes have been well characterized biochemically, there is relatively little known about any structural changes involved in elongation complex formation or the catalytic cycle itself; all the 3Dpol structures solved thus far show essentially the same conformation, with no evidence for significant conformational changes upon RNA or NTP binding.
The activation of several picornaviral polymerases is dependent upon correct cleavage of 3Dpol from the viral 3CDpro precursor protein in order to create a new N terminus that can be buried in a pocket at the base of the 3Dpol finger domain. This buried N terminus has been observed in poliovirus, coxsackievirus, rhinovirus, and foot-and-mouth disease virus polymerases (10, 14, 22, 29). In solving the structure of poliovirus polymerase, we observed that burying the N terminus resulted in a subtle but important conformational change in the active site whereby Asp238 was repositioned to make a key hydrogen bond with the 2′ hydroxyl of a bound NTP (29). Addition or deletion of a single residue at the N terminus abolished enzyme activity, and mutation of Gly1 to alanine resulted in a partially active enzyme with slightly altered positioning of Asp238. Further data from coxsackievirus polymerase showed that the addition of a second N-terminal glycine also inactivated the enzyme, but activity could be restored by also deleting Glu2, indicating that there is a specific length requirement in the N-terminal sequence of the enzyme (10). A prime candidate for involvement in such a length requirement is residue Phe5 of coxsackievirus 3Dpol that corresponds to Trp5 in poliovirus 3Dpol. In the 3Dpol structures, there is a backbone distortion in the β-strand composed of residues 1 to 9 that results in this large hydrophobic amino acid being solvent exposed rather than buried in an adjacent hydrophobic pocket (Fig. 1 A). This unusual conformation at residue 5 is conserved among picornaviral polymerase structures, and substitution mutations at this residue had significant effects on coxsackievirus polymerase activity (10). Large hydrophobic amino acids at residue 5 increased 3Dpol activity, while small amino acids at residue 5 decreased 3Dpol activity (10). Based on these data, we previously proposed that the 3Dpol catalytic cycle involves a conformational change wherein residue 5 flips into an adjacent hydrophobic patch on the polymerase to aid in NTP positioning, and such a conformational change would require the N terminus to be correctly buried to act as a stable pivot for the rotational movement.
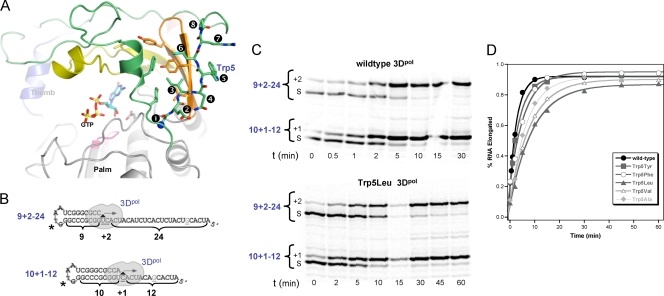
FIG. 1.
Elongation complex formation. (A) Structure of poliovirus polymerase showing the distortion of the β-sheet conformation between residues 3 and 4 that results in Trp5 being solvent exposed adjacent to a large hydrophobic patch composed of residues from the index (green) and middle (orange) fingers. (B) Cartoon of the PETE (polymerase elongation template element) RNAs used in complex formation assays. Both RNAs are G-less until the sixth nucleotide from the end, which limits secondary structure and stops elongation before the 5′ end to avoid possible end effects. The asterisk indicates the position of the amino-modified deoxythymidine where the IRDye label is covalently attached. (C) Denaturing PAGE showing the time course for formation of the +1 and +2 products from the two RNAs. Elongation complexes were formed by incubating 1 μM each PETE RNA, 15 μM 3Dpol, and 40 μM ATP and GTP at 22°C for various times as indicated. (D) Kinetics of +2 complex formation rates obtained from band intensity data curve fit to a single exponential. The resulting formation time constants are listed in Table 1.
In this work, we have investigated the role of residue 5 in 3Dpol in further detail by examining how a series of mutations in poliovirus 3Dpol affects RNA binding, elongation complex formation, elongation rate, and elongation complex stability. The data show that residue 5 mutations have major effects on the stability of the elongation complex, with minor effects on elongation complex formation and no effect on RNA binding affinities and elongation rates. Replication defects are also observed in the context of viral replication centers where residue 5 mutations significantly reduce RNA synthesis in cell-free coupled translation-replication reactions and slow the growth of infectious virus in cells.
MATERIALS AND METHODS
Protein expression and purification.
Poliovirus polymerase mutations to generate an L446D mutation (29) and add a C-terminal GSSS-His6 tag were introduced into the pET26b-Ub-3D plasmid supplied by Craig E. Cameron, and the resulting plasmid, pET26b-UbDH, was transformed into Escherichia coli BL21 PCG1 cells for expression. By this method, 3Dpol is initially translated as a ubiquitin fusion protein that is then cleaved in vivo by coexpressed ubiquitin-specific carboxyl-terminal protease Ubp1, resulting in a full-length polymerase with the native N-terminal glycine residue (16). Cells were grown overnight at room temperature in NZCYM medium with 25 μg/ml kanamycin, 20 μg/ml chloramphenicol, and 0.4% (wt/vol) d-glucose to reach an optical density at 600 nm (OD600) of ≈1.0, and 5 ml of the overnight culture was then used to inoculate 1 liter of NZCYM medium with 25 μg/ml kanamycin. The cells were grown at 37°C to an OD600 of 0.6 to 0.8 and cooled to room temperature, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, and the cells were grown for an additional 12 to 18 h. The resulting cell pellet was resuspended in a lysis buffer of 50 mM Tris (pH 8.0), 300 mM NaCl, 20% (vol/vol) glycerol, and 0.02% (wt/vol) NaN3 and lysed at 18,000 lb/in2 in a model M-110L Microfluidizer (Microfluidics, Newton, MA). The lysate was centrifuged for 40 min at 17,000 rpm in a Sorvall SS-34 rotor and loaded onto a nickel-charged chelating Sepharose fast-flow column (GE Healthcare), followed by step elution with 350 mM imidazole in 50 mM Tris (pH 8.0), 300 mM NaCl, 20% (vol/vol) glycerol, and 0.02% (wt/vol) NaN3. Fractions containing the polymerase were pooled and diluted to reduce the NaCl concentration to ≈0.11 M prior to loading onto a HiTrap Q HP column (GE Healthcare) and eluting with a linear gradient to 1 M NaCl in 25 mM Tris (pH 8.5), 20% (vol/vol) glycerol, and 0.02% (wt/vol) NaN3. The pooled fractions were concentrated to ≈0.8 ml and run over a Superdex 200 gel filtration column (GE Healthcare) equilibrated in 200 mM NaCl, 20% (vol/vol) glycerol, 5 mM Tris (pH 7.5), and 0.02% (wt/vol) NaN3. Pooled fractions were supplemented with 5 mM Tris (2-carboxyethyl)phosphine (TCEP), concentrated to ≈400 μM, flash frozen with liquid nitrogen, and stored at −80°C in 5- to 10-μl aliquots. Protein concentrations were determined based on absorbance at 280 nm, and extinction coefficients were calculated based on the amino acid sequence by using the ExPASy Proteomics Server (http://www.expasy.ch/tools/protparam.html).
RNA oligonucleotides.
RNA oligonucleotides were synthesized by Integrated DNA Technologies with an amino modifier deoxythymidine residue at the variable N position of the GNRA-type RNA tetraloop (i.e., GdTAA). The oligonucleotides were labeled at this site with IRdye 800RS NHS ester (Li-Cor Biosciences) in a buffer containing 200 mM Na2CO3/NaHCO3 (pH 9.5) and 12.5 mM EDTA, ethanol precipitated, and resuspended in Tris-EDTA (TE) buffer, and any remaining unreacted label was removed via Biospin P6 columns (Bio-Rad). Labeling efficiency was determined by the relative absorbances of the final material at 260 nm and 767 nm. The intramolecular hairpin structure that serves as a 3Dpol primer template was formed by heating the RNA to 95°C for 15 min in 50 mM NaCl, 5 mM MgCl2, 10 mM Tris (pH 8.0), followed by snap cooling on ice.
RNA binding assay.
RNA binding reactions were performed as previously described using 10 nM 8-6 PETE RNA which has an 8-bp hairpin stem with a 6-nucleotide (nt) template in 75 mM NaCl, 50 mM HEPES (pH 6.5), 1.5 mM MgCl2, 0.1% NP-40, 4 mM dithiothreitol (DTT) (24). Fluorescence polarization data were collected in a Perkin-Elmer Victor V multimode microplate reader, using black 384-well polystyrene plates. RNA dissociation constants were determined by curve fitting the data to a single-site binding isotherm with a Hill coefficient to account for the apparent cooperativity of the polarization data using Kaleidagraph (Synergy Software).
Elongation complex formation.
Elongation complex formation assays were performed using two different RNA molecules in the same reaction to allow us to determine the formation rates of the +1 and +2 complexes simultaneously. PETE RNA molecules used in these experiments were 9+2−24 and 10+1−12 (Fig. 1B), which are named using an X+Y−Z convention, where X is the number of base pairs in the priming stem, Y is the number of nucleotides that are incorporated into the RNA to form the “locked” elongation complex, and Z is the number of remaining nucleotides in the single-stranded template. Both RNAs contain a unique guanosine as the sixth nucleotide from the 5′ end, allowing us to pause elongation at this point and avoid any end effects by omitting CTP from the reaction. Elongation complexes were formed by mixing 15 μM 3Dpol, 1 μM each 10+1−12 and 9+2−24 RNA, and 40 μM each ATP and GTP in a reaction buffer of 75 mM NaCl, 50 mM HEPES (pH 6.5), 1.5 mM MgCl2, and 5 mM TCEP. Elongation complex reactions were quenched at the times indicated below by addition of equal volumes of quench buffer consisting of 20 mM EDTA, 50 mM HEPES (pH 6.5), and 75 mM NaCl. Identical reaction mixtures also containing 40 μM UTP were prepared to follow the direct formation of longer +7 or +20 products that stop six nucleotides from the end of the template. Reaction products were separated on 15% polyacrylamide-7 M urea gels that were imaged using a Li-Cor Odyssey infrared imager 9120 and quantified using the manufacturer's Odyssey software. Data from PAGE were plotted as percentages of total RNA that elongated at each time point and fitted to a single exponential curve, A*(1 − exp−t/τ) + C, where τ is time constant of complex formation.
RNA elongation.
Elongation rates were measured at 22.5°C in an Applied Photophysics SX-20 stopped-flow instrument in which equal volumes of preformed 3Dpol-RNA elongation complex with 5′ fluorescein-labeled 10−26 PETE RNA and NTP solutions were mixed to initiate the reaction (17). Fluorescence excitation was at 492 nm from a monochromator source with bandwidth set to 9.3 nm, and emission from fluorescein was detected using 515-nm high-pass filters. Total fluorescent (TF) and fluorescence anisotropy (FA) signals were calculated from the parallel and perpendicular polarization signals after background and G factor correction, per standard fluorescence methods (21). Stable +1 elongation complexes were assembled prior to the elongation reaction, using a 30-min incubation with 15 μM polymerase, 1 μM RNA, and 20 μM ATP, and then diluted 10-fold in reaction buffer and stored on ice until they were loaded into the stopped-flow instrument. Once loaded, the samples were allowed to temperature equilibrate for at least 5 min prior to data collection. The final solution measured in the stopped-flow experiment after rapid mixing contains 50 nM RNA, 750 nM polymerase, and 5 to 120 μM each NTP in reaction buffer. Elongation rates in number of nucleotides per second were calculated from the length of the lag phase observed at each NTP concentration and plotted against substrate concentration to determine Vmax and apparent Km values (17).
Elongation complex stability.
Elongation complexes were formed as described above using the 9+2−24 PETE RNA and RiboLock RNase inhibitor (Fermentas) added to 3 units/μl, with a room temperature incubation period of 5 to 15 min, depending on complex formation rates from Table 1. Reaction mixtures were then diluted up to 10-fold in a reaction buffer consisting of 50 mM HEPES (pH 6.5), 5 mM TCEP, 1.5 mM MgCl2, and either 75 mM or 325 mM NaCl (300 mM NaCl final concentration) to limit polymerase-RNA rebinding and reinitiation. Dilutions were also carried out in 75 mM NaCl with 100 ng/μl heparin to prevent RNA rebinding. An equivalent concentration of the 10+1−12 RNA was added to the reaction mixture immediately after the dilution step to serve as a degradation and reinitiation control. At the time points indicated below, small aliquots of the reaction mixture were chased to longer +20 products by the addition of equal volumes of 40 μM UTP solution, resulting in a 20 μM final concentration of GTP, UTP, and ATP; elongation to +20 product was allowed to proceed for 3 min. CTP was omitted from the chase step to stop the reaction six nucleotides from the end of the template strand and avoid potential end effects. Reaction products were analyzed by denaturing PAGE, and the fraction of active elongation complexes was determined by dividing the amount of elongated (+20) RNA by the sum of the fully elongated (+20) and nonelongated (+2) RNAs. Data analysis by exponential curve fitting was performed using Kaleidagraph (Synergy Software).
TABLE 1.
Initiation product formation rates of Trp5 mutantsc
| 3Dpol | Initiation product formation rate |
|||
|---|---|---|---|---|
| 10+1−12 RNA |
9+2−24 RNA |
|||
| +1 producta | +7 productb | +2 producta | +20 productb | |
| Wild type | 2.0 ± 0.1 | 2.5 ± 0.5 | 3.0 ± 0.2 | 3.0 ± 0.5 |
| Trp5Tyr mutant | 4.5 ± 0.5 | 5.5 ± 0.1 | 4.2 ± 0.3 | 4.0 ± 0.5 |
| Trp5Phe mutant | 4.0 ± 0.1 | 7.5 ± 0.5 | 5.9 ± 0.5 | 9.0 ± 1.5 |
| Trp5Leu mutant | 5.5 ± 0.5 | 4.0 ± 0.5 | 9.4 ± 0.6 | 5.0 ± 1.0 |
| Trp5Val mutant | 7.0 ± 0.5 | 5.0 ± 0.5 | 8.5 ± 0.3 | 8.0 ± 0.5 |
| Trp5Ala mutant | 7.5 ± 0.5 | 7.5 ± 0.5 | 6.6 ± 0.7 | 7.5 ± 0.5 |
Product formation in the presence of 40 μM each ATP and GTP only.
Product formation in the presence of 40 μM each ATP, GTP, and UTP.
Time constants in minutes based on single exponential curve fits of integrated band intensities.
3Dpol solubility.
Elongation complexes were assembled as described above in 75 and 300 mM NaCl without heparin, diluted 10-fold, and incubated at either room temperature or 37°C at various time points (0 to 180 min) after the 10-fold dilution samples were taken and centrifuged at 17,000 × g for 10 min at 4°C. The supernatant was removed, and soluble protein content was analyzed by SDS-PAGE stained with Coomassie brilliant blue.
Elongation processivity.
RNA templates were T7 transcribed from the pRNA2 plasmid (see below) and linearized by SpeI, AvrII, or MluI to generate 2,201-, 5,424-, or 5,742-nt RNA products, respectively. These can initiate elongation complex formation via snap-back priming from the 3′ end of the transcripts. Elongation reactions were initiated by incubating 20 μM protein, 0.4 μM RNA, and 500 μM each NTP at room temperature for 7 to 15 min in buffer containing 50 mM NaCl, 1.5 mM MgCl2, 4 mM TCEP, 50 mM HEPES (pH 6.5), and 1 unit/μl RiboLock RNase inhibitor (Fermentas). The resulting elongation complexes were then diluted 5-fold into a high-salt buffer (356 mM NaCl [300 mM final concentration], 200 μM each NTP, 1.5 mM MgCl2, 4 mM TCEP, 50 mM HEPES [pH 6.5]) to allow elongation to the end of the template without further initiation. Reaction mixture samples were quenched with 16 mM ETDA after various times and phenol-chloroform-isoamyl alcohol extracted, and product RNA lengths were analyzed by native gel electrophoresis in an ethidium bromide containing 0.7% agarose gel.
Cell-free replication.
pRNA2, referred to as wild type in this report, encodes a poliovirus RNA replicon in which P1 capsid genes have been deleted (nt 1175 to 2956) (12). The T7 transcription of MluI-linearized pRNA2 cDNA yields positive-sense replicon RNA containing two 5′ nonviral guanosine residues that do not affect negative-sense RNA synthesis but prevent subsequent positive-sense synthesis (8). Trp5 mutations to Phe and Ala were generated by the Quick Change PCR mutagenesis protocol (Stratagene, Inc.) in pRNA2 cDNA. Plasmids were isolated from E. coli and digested with KasI and MluI to produce a minimal fragment containing the desired 3Dpol mutation, and this fragment was subcloned into the full-length pRNA2 vector that has not undergone PCR to decrease the chance of introducing additional point mutations.
Cytoplasmic extracts and translation initiation factors were prepared from HeLa cells as previously described (7). Reaction mixtures contained 50% cytoplasmic extract, 20% initiation factors, 10% 10× nucleotide reaction mix (10 mM ATP, 2.5 mM GTP, and 2.5 mM CTP; 600 mM KCH3CO2; 300 mM creatine phosphate; 4 mg/ml creatine kinase; and 155 mM HEPES-KOH [pH 7.4]), and T7 transcripts of poliovirus replicon RNA at 45 μg/ml. Preinitiation RNA replication complexes were formed in the presence of 2 mM guanidine HCl as previously described (5).
Viral protein synthesis from poliovirus replicon mRNA was monitored by incorporation of [35S]methionine (1.2 mCi/ml), which was quantified by acid precipitation followed by scintillation counting. Preinitiation replication complexes were isolated from HeLa S10 reaction mixtures and resuspended in a [α-32P]NTP-labeling reaction mix {27 mM HEPES-KOH (pH 7.4); 60 mM KCH3CO2; 2.3 Mg(CH3CO2)2, 2.6 mM dithiothreitol, 2.3 mM KCl; 50 μg/ml puromycin; 1 μCi/μl [α-32P]UTP, 1 mM ATP, 250 μM GTP and CTP; with or without 2 mM guanidine HCl} and incubated at 37°C for the time periods indicated below. Replication complexes were isolated by centrifugation at 17,000 × g for 15 min at 4°C, resuspended in 0.5% SDS buffer (0.5% SDS, 1 mM EDTA, 100 mM NaCl, 10 mM Tris, [pH 7.5]), phenol-chloroform extracted, and ethanol precipitated, and the resulting product RNA was analyzed by nondenaturing 1% agarose 1× Tris-borate-EDTA (TBE) gel electrophoresis. The agarose gel was dried, and radiolabeled product RNA was detected by phosphorimaging.
Transfection of HeLa cells and virus quantification.
Residue 5 mutations were introduced into poliovirus cDNA by subcloning from pRNA2 via the KasI and MluI restriction sites. Poliovirus RNA encoding wild-type, Trp5Phe, or Trp5Leu 3Dpol was derived by T7 transcription of MluI linearized cDNAs (type 1 Mahoney). HeLa cells (∼106 cells per 35-mm well) were transfected with 2 μg of poliovirus RNAs by using the TransMessenger transfection reagent (Qiagen) according to the manufacturer's instructions. Transfected HeLa cells were grown in 2 ml of cell culture medium (Dulbecco's modified Eagle's medium [DMEM] containing 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin) and incubated at 37°C. Cells were examined for cytopathic effect (CPE) at 24, 48, and 72 h posttransfection (hpt). At 48 and 72 hpt, the cells were subjected to three cycles of freeze-thawing. Poliovirus titers at 48 and 72 hpt were determined by plaque assay as previously described (19).
Sequence of 3Dpol in virion RNA.
Poliovirus RNA was isolated from purified virus particles and converted into cDNA for sequencing. In brief, poliovirus from transfections and infections was purified from cell culture medium by layering 8 ml of the virus onto 3 ml 30% (wt/vol) sucrose in phosphate-buffered saline (PBS) followed by centrifugation at 36,000 rpm for 4 h at 4°C using a Beckman SW41 rotor. The pelleted virus particles were resuspended in 400 μl 0.5% SDS buffer (0.5% SDS, 10 mM Tris-HCl [pH 7.5], 100 mM NaCl). Poliovirus RNA was isolated by phenol-chloroform-isoamyl alcohol extractions and ethanol precipitation.
Poliovirus RNAs were transcribed into cDNA by using Superscript III reverse transcriptase (Invitrogen) and a primer complementary to nucleotides 6930 to 6947 of the poliovirus open reading frame (5′-6947GGTGGTCTAAATCTATGC6930-3′). cDNA corresponding to the 3Dpol region of poliovirus RNA was amplified by 35 PCR cycles (2×) with high-fidelity Phusion DNA polymerase (New England Biolabs), using a forward primer (5′-5284GGCAATGACAATTCTACAAGC5304-3′) and a reverse primer (5′-6519GCTTCAATTAATCTGGATTTC6497-3′) complementary to the poliovirus RNA sequences indicated below. PCR products were sequenced using the forward primer corresponding to poliovirus nucleotides 5816 to 5838. When mixed populations of virus sequence were detected in the sequence of PCR products (as for Trp5Leu at 48 hpt), PCR products were TOPO-TA cloned (Invitrogen), and the sequences of representative cDNA clones were determined. PCR products and cDNA clones were sequenced in the University of Colorado Cancer Center DNA Sequencing Core Laboratory.
RESULTS
Elongation complex formation.
The effects of residue 5 mutations on polymerase RNA binding affinity were determined using a planar aromatic phenylalanine and a smaller nonplanar valine residue. Neither mutation had a significant effect on the affinity for a PETE 8−10 RNA (Table 2), indicating that residue 5 does not play a direct role in RNA binding in the absence of nucleotides and elongation. We then examined what effects the mutations have on the formation of stable 3Dpol elongation complexes as measured by the incorporation rate for the initial nucleotides on a self-priming PETE RNA. Two PETE RNAs were used simultaneously in these experiments; one has a 10-bp hairpin followed by a 13-nt template that allowed the incorporation of only a guanosine when incubated with GTP and ATP, and the other has a 9-bp hairpin with a 26-nt template that allowed the incorporation of both adenosine and guanosine (Fig. 1B). This RNA design allowed us to compare the two RNAs in the same reaction to determine if there are differences between the +1 and +2 product formation rates. The final products of the elongation complex formation reaction have identical structures and differ only in the length of their remaining templating regions. Product formation rates were determined by polyacrylamide gel analysis of the quenched samples followed by band quantitation of the IRdye-labeled RNAs to determine the relative amounts of starting and elongated materials (Fig. 1C).
TABLE 2.
Enzymatic parameters of 3Dpol elongation complexes
| 3Dpol | RNA Kd (μM) | Elongation rate Vmax (nt/s) | NTP Km (μM) | Elongation complex stability (min) |
|---|---|---|---|---|
| Wild type | 8.6 ± 0.6 | 2.1 ± 0.1 | 18 ± 3 | 186 ± 4 |
| Trp5Tyr mutant | NDa | ND | ND | 56 ± 3 |
| Trp5Phe mutant | 9.7 ± 2.4 | 1.8 ± 0.2 | 10 ± 3 | 26 ± 1 |
| Trp5Leu mutant | ND | 1.9 ± 0.2 | 17 ± 5 | 4.4 ± 0.4 |
| Trp5Val mutant | 7.2 ± 0.5 | 3.0 ± 0.3 | 22 ± 7 | 5.3 ± 0.5 |
| Trp5Ala mutant | ND | 2.0 ± 0.1 | 17 ± 2 | 5.3 ± 0.2 |
ND, not done.
The formation rates for the +1 and +2 products by wild-type 3Dpol and the various Trp5 mutants are shown in Fig. 1D and Table 1. All mutants exhibited single exponential kinetics for +1 and +2 product formation, and the rates were determined by numerical curve fitting. The Trp5 mutations reduced both the +1 and +2 complex formation rates compared to those of the wild type, but the effects were fairly small with only a ≈3-fold reduction at most (Table 1). The rate reduction also correlated with the size of the replacement residue, with tyrosine and phenylalanine having fairly small effects, while leucine, valine, and alanine had greater effects. Prior studies of 3Dpol enzymology have shown that the incorporation of the first nucleotide is the rate-limiting step during initiation that is followed by the fast incorporation of additional nucleotides (4). Consistent with this, the rates for formation of the longer +7 and +20 products in the presence of ATP, GTP, and UTP are comparable to those of the +1 and +2 products for each polymerase (Table 1).
Elongation rates.
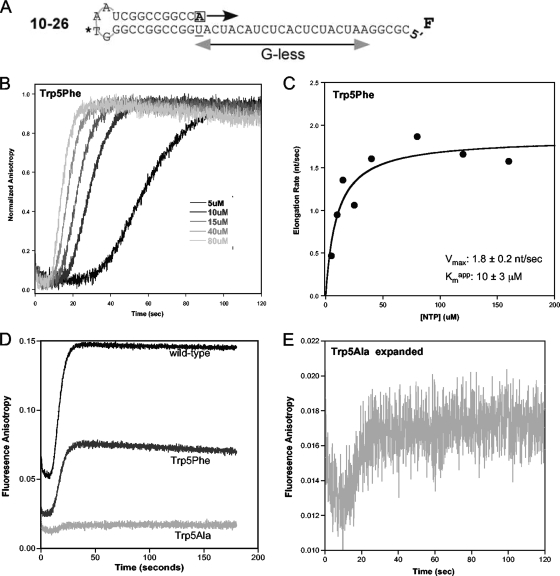
To determine 3Dpol elongation rates, we used the PETE assay in a stopped-flow kinetic format where we detect how long it takes 3Dpol to reach the end of a 5′ fluorescein-labeled template RNA (17). The experiments utilized a 10−26 PETE RNA with a 10-bp priming hairpin and a 26-nt template sequence designed to minimize the possibility of secondary structure (Fig. 2 A). For these experiments, stalled +1 elongation complexes were generated prior to the stopped-flow experiment. The stopped-flow data traces show an initial lag phase that is followed by an increase in anisotropy as the polymerase reaches the end of the template and immobilizes the fluorescein label (Fig. 2B) (17). The length of the lag phase is indicative of the time required for the polymerase to elongate through the 20-nt portion of the template, allowing for the calculation of an elongation rate in the number of nucleotides per second (nt/s). 3Dpol elongation rates were measured at various NTP concentrations and plotted on a Michaelis-Menten curve from which Vmax and an apparent Km value were determined (Fig. 2C and Table 2). The results indicate that all of the mutants exhibited essentially wild-type maximal elongation rates (Vmax) and apparent Km values of 2 nt/s and 20 μM, respectively, indicating that the nature of residue 5 does not significantly affect the 3Dpol elongation rate. Note that these data were obtained at 22.5°C and pH 6.5 to minimize thermal inactivation of the polymerase, but higher rates of ≈70 nt/s are obtained at higher temperatures and pH values (17).
FIG. 2.
3Dpol elongation rates. (A) Schematic of 5′ fluorescein (F) end-labeled PETE RNA used to determine elongation rate by stopped-flow fluorescence anisotropy. (B) Normalized anisotropy data showing the signal increase during the elongation of 10−26 PETE RNA by Trp5Phe 3Dpol at various concentrations of NTPs. The lag phase represents elongation through the 20-nt G-less segment of the template, which shortens with increasing NTP concentrations due to faster elongation rates (17). The increase in anisotropy signal is associated with the addition of the terminal five nucleotides as the polymerase contacts the fluorescein at the 5′ end of the RNA. (C) Michaelis-Menten plot showing the maximum elongation rate (Vmax) and apparent Km for Trp5Phe 3Dpol. (D) Raw stopped-flow data showing the elongation curves of wild-type, Trp5Phe, and Trp5Ala 3Dpol at 40 μM NTPs. The difference in amplitude change and the decrease in starting baseline value indicate that there are fewer complexes being elongated for Trp5Phe and Trp5Ala mutants compared to the wild-type enzyme. (E) Expansion of Trp5Ala data from panel D, illustrating that the signal for Trp5Ala retains the expected shape and an accurate elongation rate can be determined despite its low signal amplitude.
While the Trp5 mutants did not affect the 3Dpol elongation rate, we did observe significant differences in the amplitudes of the elongation-associated fluorescence signals from the various Trp5 mutants (Fig. 2D). The amplitude of the elongation-dependent terminal fluorescence change decreased, particularly for the Trp5Ala mutant, although the data traces retained the expected lag phase behavior and maintained a sufficient signal-to-noise ratio to determine elongation rates (Fig. 2E). During the experiments, we also observed that the changes in anisotropy values consistently decreased during sequential replicate stopped-flow runs from the same preassembled +1 elongation complex mixture, and this effect was most pronounced with mutations to the smaller valine and alanine residues. Because the baseline fluorescence anisotropy signal is representative of the amount of RNA bound to polymerase and the amplitude of the signal change is dependent on the number of RNA molecules being elongated, these findings suggested that the mutations at Trp5 decreased the amount of active 3Dpol elongation complex in the reaction mixtures.
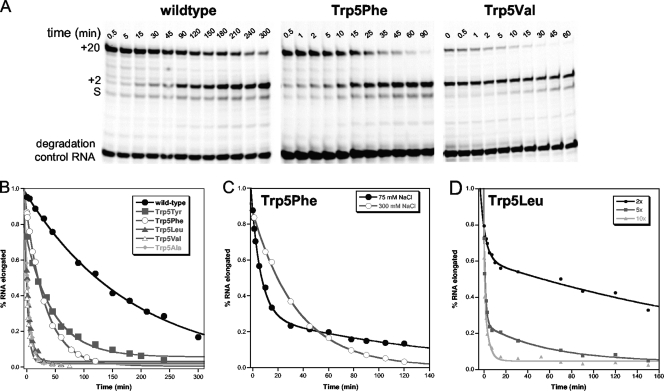
Elongation complex stability.
To more specifically determine the effects of Trp5 mutations on elongation complex stability, we designed an experiment to first form stalled +2 elongation complexes and then measure, as a function of incubation time, how efficiently the +2 RNA could be elongated to the longer +20 product by the addition of the remaining NTPs. The 9+2−24 RNA was used to generate +2 complexes in the presence of ATP and GTP for 5 to 15 min (to reach >95% complex formation based on the rates listed in Table 1), and the reaction mixtures were then diluted 10-fold with buffer containing either 300 mM NaCl or 75 mM NaCl. The 10+1−12 RNA was also added at this point to serve as a degradation and reinitiation control. At various time points postdilution, small samples of the reaction mixture were tested for their ability to rapidly incorporate additional nucleotides in 3 min, and the products were analyzed by denaturing PAGE and quantitation of the IRdye-labeled RNAs.
As shown by the gels in Fig. 3 A, wild-type 3Dpol forms a very stable elongation complex that efficiently chases the +2 RNA into the +20 product even after several hours of incubation at room temperature in 300 mM NaCl. The various Trp5 mutants, on the other hand, exhibited a rapid loss of elongation ability that ranked according to the size of the mutant residue; tyrosine and phenylalanine had the smallest effects (3- to 7-fold), while leucine, valine, and alanine reduced complex stability more than 35-fold. Quantitation of the gel bands and plotting of the data as a fraction of the +2 RNA that could be elongated, i.e., +20 band ÷ (sum of +2 and +20 bands), showed that the loss of competent elongation complexes exhibited single exponential decay behavior (Fig. 3B). Curve fitting of the data revealed that the wild-type 3Dpol elongation complex has a decay constant of about 3 h; this is reduced to 56 and 26 min for the Trp5Tyr and Trp5Phe mutants and to as little as ≈5 min for the leucine, valine, and alanine mutants (Table 2). No elongation of the 10+1−12 control RNA was observed, confirming that the +20 products arise only from the preformed elongation complexes.
FIG. 3.
Elongation complex stability. (A) Denaturing PAGE showing how efficiently stalled +2 elongation complexes can be chased to the +20 product after various incubation times. Elongation complexes were formed by the preincubation of 1 μM 9+2−24 PETE RNA, 15 μM 3Dpol, and 40 μM ATP and GTP to form the +2 elongation complex from the starting RNA (S), diluted 10× in 300 mM NaCl and then chased at the indicated times (t) by addition of NTPs for 3 min prior to quenching the samples. The stability is determined by calculating the fraction of RNA that is able to chase from the +2 band to the +20 band. (B) Plot of data obtained from PAGE analysis showing single exponential loss of 3Dpol elongation complex stability. (C) Data comparing stability of Trp5Phe elongation complexes in 75 mM and 300 mM NaCl. The 75 mM NaCl decay curve exhibits biphasic behavior where the first phase is indicative of complex stability and the second rate is protein concentration dependent and indicative of 3Dpol rebinding and reinitiating on released RNA. (D) Plot of Trp5Leu elongation complex stability in 75 mM NaCl after 2-, 5-, and 10-fold dilutions of the +2 complex (resulting in 7.5, 3, and 1.5 μM final 3Dpol) to show that the second rate of the biphasic curve is protein concentration dependent.
We did see evidence of RNA release and reinitiation during the incubation period when the elongation complexes were diluted in 75 mM NaCl without heparin, as opposed to 75 mM NaCl with heparin or 300 mM NaCl without heparin. In this case, the elongation complex decay curves showed biphasic characteristics, with a fast phase matching that seen in the presence of higher salt and a slow phase whose amplitude was dependent on the postdilution protein concentration, as determined by changing the dilution factor between the complex formation and incubation steps from 2- to 5- to 10-fold (Fig. 3C and D). We interpret this slow phase formation of the +20 product as being due to reinitiation on RNA that has been released during the extended incubation step. This suggests that the polymerase dissociates from the RNA upon inactivation, allowing the RNA to be rebound and elongated by another polymerase molecule.
To verify that the mutant polymerases remained soluble during the elongation complex stability assays, we repeated the experiments and centrifuged the sample to pellet any precipitated protein and then analyzed the supernatant by SDS-PAGE. These experiments were performed on Trp5Tyr and Trp5Ala mutants, and no loss of soluble 3Dpol was observed for either mutant with assays done at room temperature (data not shown). However, we did observe loss of soluble protein in samples that were incubated at 37°C (≈80% loss in 75 mM NaCl and ≈20% loss in 300 mM NaCl), consistent with the ≈40°C melting point of 3Dpol (28). These results indicate that the polymerase remains soluble throughout the stability assays done at room temperature.
Replication processivity.
Elongation processivity was determined by monitoring the amount of abortive products generated during replication of longer genomic RNAs by 3Dpol. Poliovirus RNA templates were generated by T7 transcription from linearized pRNA2, resulting in 2,201- or 5,424-nt-long RNA templates. These pRNA2-templated T7 transcription reactions also produced an additional 670-nt fragment that corresponds to most of the poliovirus 5′ UTR, including the internal ribosome entry site (IRES). This smaller fragment is due to the presence of a T7 polymerase abortive sequence, 5′-UAUCUGUU-3′ (23), found shortly after the IRES element in the pRNA2 cDNA. This fragment is present in all the 3Dpol elongation reaction mixtures and provides additional elongation data from a shorter RNA template that is rich in secondary structure.
To initiate RNA elongation, we utilized the fact that the 3′ ends of single-stranded RNA transcripts can form localized snap-back structures that create self-priming hairpins for RNA elongation by 3Dpol (11). Experiments were performed by preincubating 0.4 μM RNA, 20 μM polymerase, and 500 μM (each) all four NTPs for 7 to 15 min at room temperature to form elongation complexes, mimicking the locked complex formation step used in the polymerase rate and stability assays. After this preincubation step, the complexes were diluted into a high salt concentration to prevent polymerase rebinding and additional initiation events during the elongation phase of the experiment. At the time points indicated below, the reactions were quenched and extracted with phenol-chloroform, and RNA products were analyzed by nondenaturing agarose gels stained with ethidium bromide.
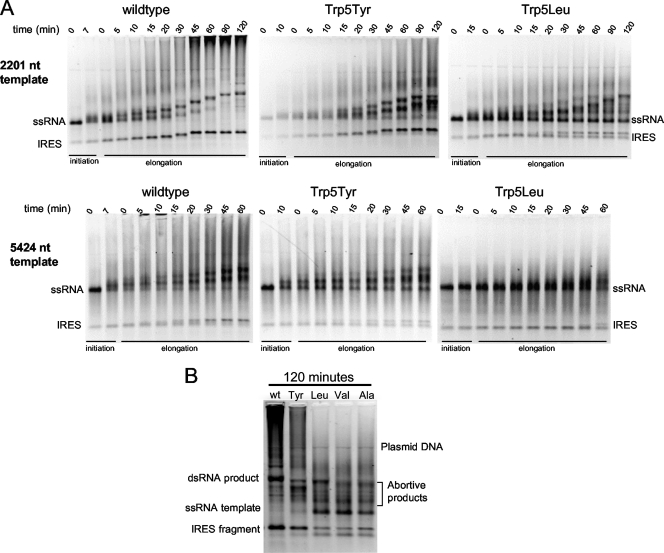
Elongation of the 2,201 (Fig. 4 A, top panel)- and 5,424 (Fig. 4A, bottom panel)-nt-long templates showed that the amount of full-length duplex RNA synthesized by 3Dpol was slightly reduced for the tyrosine and phenylalanine mutants compared to that of the wild type but significantly reduced with the leucine, valine, and alanine mutants. The elongation rate of wild-type and mutant polymerases was calculated to be ∼0.4 nt/s based on the ≈90 min needed to elongate the 2,201-nt template. This is ≈5-fold lower than the rates observed in the PETE assay (Fig. 2 and Table 2) and is likely caused by an increase in the NTP Kmapp due to the increased salt concentration during the elongation phase. In addition to a clear band that reflects full-length duplex RNA, the reactions with wild-type 3Dpol also show the time-dependent accumulation of very large and heterogeneous RNA products just below the sample wells. The amount of these large RNA products is reduced when Trp5 is mutated to phenylalanine or tyrosine and largely eliminated with the smaller leucine, valine, and alanine mutants (Fig. 4B).
FIG. 4.
Elongation processivity of self-primed poliovirus RNA. (A) Ethidium bromide-stained nondenaturing agarose gel electrophoresis showing a time course analysis of RNA elongation through a 2,201-nt template (top) and a 5,424-nt template (bottom). RNA transcripts were preincubated with 20 μM 3Dpol, 0.4 μM RNA, and 0.5 mM NTPs in 75 mM NaCl for 7, 10 or 15 min to allow for initiation, and the reaction mixtures were then diluted 5-fold into 300 mM NaCl and 260 μM NTPs. Reactions were quenched at the indicated times. The IRES band is from a T7 abortive sequence located just after the IRES in the poliovirus genome. (B) Ethidium bromide stain of nondenaturing agarose gel electrophoresis directly comparing elongation of the 2,201-nt RNA template by the different mutant polymerases after 120-min reaction time.
Elongation from self-primed templates also resulted in intermediate RNA bands. Some of these RNAs continue to be elongated, suggesting that there may be sites where the polymerase pauses, while other bands are left behind, indicating that they are abortive products. Figure 4B shows a side-by-side comparison of products formed by wild-type and residue 5 mutant polymerases after 120 min of elongation on the 2,201-nt template. The data show that wild-type 3Dpol produces few abortive products, the tyrosine mutant produced slightly more abortive products, and the leucine, valine, and alanine mutants result in significantly more abortive products. For these smaller mutations, the abortive products represent a significant fraction of the final material when one considers that template utilization is also reduced by less efficient initiation.
The 670-nt band resulting from the T7 abortive sequence in the 5′ untranslated region (UTR) is composed almost entirely of the IRES. This transcript can also self-prime and be elongated by the polymerase, albeit with reduced efficiency for the residue 5 mutants compared to the wild type. Interestingly, the alanine, valine, and leucine mutants are very inefficient at reaching the 5′ end of this short template. Although the gels in Fig. 4 do not have the resolution to show abortive products for the IRES band, it is apparent by the amount of elongation product that the mutant polymerases were extremely inhibited in replication of the IRES, more so than for the full-length RNA elongation. These data suggest that replication through extensive secondary structure is more difficult for residue 5 mutants than for wild-type 3Dpol, significantly slowing the polymerase and giving the unstable mutant polymerases time to dissociate from the template RNA. Consistent with this, the longer subgenomic products generated by the mutants appear slightly shorter than that from the wild-type enzyme (Fig. 4B), suggesting that the mutants may not have replicated through the IRES that would be at the end of the negative-strand transcript.
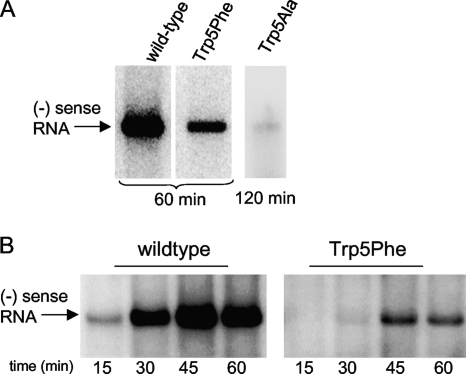
Cell-free replication.
To ascertain the effects of the Trp5 mutations on viral genome synthesis, we carried out cell-free coupled translation-replication reactions. Negative-strand RNA synthesis from subgenomic poliovirus replicons harboring wild-type and Trp5Phe 3Dpol showed replication to the full-length negative strand within 60 min, but the phenylalanine mutant had significantly lower yield than did the wild type (Fig. 5 A). Negative-strand RNA synthesis by the Trp5Ala mutant replicon was too faint to be detected after 60 min of replication, but a small amount of full-length negative-strand RNA product could be detected after 120 min of replication. In a time course experiment with wild-type and Trp5Phe 3Dpol, we see that the two polymerases replicated poliovirus RNA at similar rates, as both reached their maximal yield of negative-strand RNA after ≈45 min of incubation, but the Trp5Phe mutant yielded much less negative-strand RNA product (Fig. 5B). The reduced synthesis of negative-strand RNA by Trp5Phe and Trp5Ala mutants is likely due to instability of the elongation complexes as a result of the residue 5 mutations. Importantly, these results indicate that in authentic RNA replication complexes, the mutations to residue 5 do not affect the elongation rate itself but rather the amount of full-length viral RNA produced.
FIG. 5.
Negative-strand RNA synthesis in a cell-free translation-replication system. (A) Endpoint analysis showing amounts of full-length negative-strand RNA produced by replicons containing wild-type and Trp5Phe 3Dpol in 60 min. Product from the Trp5Ala mutant was barely detectable after 120 min of replication. (B) Time course of product formation showing that both wild-type and Trp5Phe 3Dpol reach maximum RNA quantities after ≈45 min of replication, indicating that the polymerase mutations affect the yield of negative-strand RNA, but not the elongation rate, in the replication complex.
Virus replication.
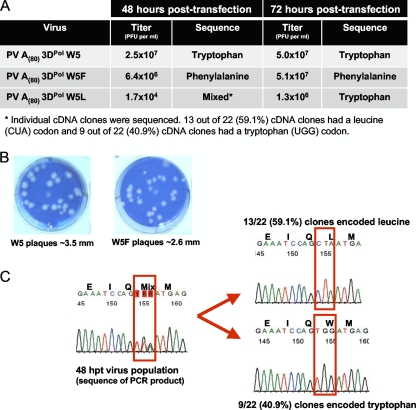
To test the ability of the polymerase mutants to support virus growth, we transfected HeLa cells with positive-strand genomic RNA containing the Trp5Phe and Trp5Leu mutations. Both mutants produced infectious virus (Fig. 6 A), and at 48 hpt, the Trp5Phe mutant had only slightly lower viral titer than did the wild type (6.4 × 106 versus 2.5 × 107 PFU/ml), while the Trp5Leu viral titer was significantly lower, at 1.7 × 104 PFU/ml. After 72 h, the titers of wild-type and Trp5Phe viruses were essentially equivalent at ∼5.0 × 107 PFU/ml, although the phenylalanine mutant produced slightly smaller plaques (Fig. 6B). Sequencing confirmed that the Trp5Phe mutation was stable in the progeny virus population. Growth of the slow Trp5Leu virus, on the other hand, improved dramatically between 48 and 72 h, producing plaques similar to the wild-type virus that suggested a reversion or acquisition of some compensatory mutation. This was confirmed by sequence analysis of the 3Dpol gene from progeny virus showing a mixed population of Leu5 and Trp5 residues at 48 hpt and complete reversion of the virus population to wild-type tryptophan at 72 hpt (Fig. 6C).
FIG. 6.
Viability of poliovirus engineered to express 3Dpol residue 5 mutations. (A) Poliovirus transcripts from cDNAs encoding wild-type, Trp5Phe, and Trp5Leu 3Dpol were transfected into HeLa cells (∼106 cells per 35-mm well). At 48 and 72 hpt, the cells were subjected to three cycles of freeze-thawing. Poliovirus titers at 48 and 72 hpt were determined by plaque assay. Virion RNA from poliovirus recovered at 48 and 72 hpt was converted into cDNA and PCR products sequenced to determine whether the mutations engineered into 3Dpol were maintained within the virus recovered from transfected cells. (B) Representative plaques from wild-type poliovirus and poliovirus with 3Dpol Trp5Phe mutation. (C) Sequence data of Trp5Leu cDNA PCR products from poliovirus at 48 hpt (left) and representative sequence data from individual cDNA clones (right).
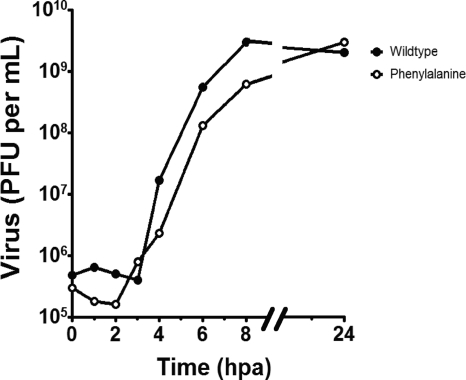
One-step growth curves of the wild type and the Trp5Phe 3Dpol mutant were performed by infection of HeLa cells (∼106 cells per 35-mm well) with a multiplicity of infection (MOI) of 10 viruses per cell. One hour after adsorption, virus inoculum was removed and replaced with 2 ml of culture medium and incubated at 37°C for 1, 2, 3, 4, 6, 8, and 24 h postadsorption (hpa) before undergoing 3 freeze-thaw cycles to release the virus. Growth curves were plotted as virus titer from plaque assays versus time (Fig. 7). In agreement with the prior transfection and plaque assay data, Trp5Phe 3Dpol produced viable virus with a growth rate that was only slightly lower than that of the wild-type virus, and it reached the same endpoint of ∼5.0 × 109 PFU/ml at 24 hpa.
FIG. 7.
One-step growth curve of wild-type and Trp5Phe poliovirus. HeLa cells (∼106 cells per 35-mm well) were infected with 10 PFU per cell of wild-type or Trp5Phe poliovirus. After adsorption for 1 h at room temperature, the virus inoculum was removed, and the cells were given 2 ml of culture medium and incubated at 37°C. Cells were subjected to three cycles of freeze-thawing at 0, 1, 2, 3, 4, 6, 8, and 24 h postadsorption (hpa). Poliovirus titers were determined by plaque assay and plotted versus time.
DISCUSSION
The crystal structures of the poliovirus, coxsackievirus, rhinovirus, and foot-and-mouth disease virus polymerases show a conserved structural motif whereby the very N terminus of the protein is buried in a pocket at the base of the finger domain. This conformation is essential for polymerase activity and can be formed only after the proteolytic processing of 3CDpro to generate the native 3Dpol N terminus. When solving the structure of coxsackievirus B3 polymerase, we also noticed that the structural conservation of a distortion in the β-strand composed of residues 1 to 9 that resulted in the usually hydrophobic residue 5 being exposed on the surface of the protein (Fig. 1A) (10). Through mutational analysis, we found that residue 5 was indeed important for coxsackievirus 3Dpol function; mutations of the native phenylalanine to smaller residues greatly decreased polymerase activity, while mutation to a larger tryptophan increased polymerase activity, as determined by product formation in a poly(A) templated activity assay. The 3Dpol structures lead us to propose that residue 5 could flip into an adjacent hydrophobic pocket during the catalytic cycle, providing a driving force for NTP positioning prior to catalysis.
In this work, we further elucidate the function of residue 5 and the role it plays in the molecular mechanisms of 3Dpol initiation and elongation. Formation of an active 3Dpol elongation complex is a multistep process that has been well characterized for poliovirus (2) and foot-and-mouth disease virus (1) polymerases. Overall, the process consists of (i) RNA binding followed by an initial conformational change to generate a catalytically competent complex, (ii) a second conformational change associated with incorporation of the first NTP to generate the elongation complex, and (iii) processive replication of the entire viral genome. Mutations at residue 5 could affect 3Dpol activity by altering the initiation steps, by altering the elongation rate of the enzyme, or by altering the stability of the elongation complex.
To determine if residue 5 was involved in initial RNA binding, we utilized a solution-based fluorescence polarization assay to measure the affinity of a short hairpin RNA in 75 mM NaCl (24). The data collected from mutations that introduce either a planar phenylalanine or a smaller branched valine showed no significant effects on RNA affinity compared to the wild-type enzyme, indicating that 3Dpol residue 5 is not involved in RNA binding in the absence of nucleotides. To assess initiation and elongation complex formation, we used two different-length hairpin RNA substrates in the same reaction mixture to simultaneously measure +1 product formation due to guanosine addition and +2 product formation due to adenosine and then guanosine addition. The RNAs were designed such that the resulting +1 and +2 complexes had exactly the same RNA sequence in the vicinity of the active site and differed only in the length of the remaining single-stranded template sequence (Fig. 1B). The results show that all the Trp5 mutations slowed the rate of elongation complex formation, with smaller residues having lower complex formation rates (Fig. 1C and D), but the effect of the mutations on the formation rate were at most only ≈3-fold (Table 1). While statistically significant, these changes in rate are not sufficient to be the sole contributor to the changes in activity that we previously observed for the analogous mutations in coxsackievirus 3Dpol (10).
Using our PETE assay in a stopped-flow kinetic format (17), we also established that the mutations at Trp5 do not affect 3Dpol elongation rates, with all the mutations having Vmax and apparent NTP Km values comparable to those of the wild-type enzyme (Table 2). Interestingly, we saw evidence during these elongation experiments suggesting that residue 5 was involved in maintaining a stable elongation complex (Fig. 2D and E). To show that residue 5 mutations have significant effects on the temporal stability of the competent 3Dpol elongation complex, we devised an assay in which the initial elongation complex is stalled after formation of the +2 product by omitting the next nucleotide triphosphate, and the ability of these stalled complexes to rapidly elongate the RNA by another 18 nucleotides was tested as a function of incubation time (Fig. 3).
Wild-type 3Dpol elongation complexes are quite stable and exhibit monoexponential decay with a time constant of about 3 h. The Trp5 mutations, on the other hand, show very rapid loss of elongation ability, with decay time constants of ≈55 and ≈25 min for tyrosine and phenylalanine mutants, respectively, and only 5 to 15 min for the smaller leucine, valine, and alanine mutants (Table 2). All the proteins show monoexponential loss of elongation ability in 300 mM NaCl or in 75 mM NaCl in the presence of heparin. However, in 75 mM NaCl without heparin, we observe a biphasic decay curve whose second phase is dependent on protein concentration (Fig. 3D). The latter observation indicates that the loss of elongation complex competency is associated with a release of the RNA from the polymerase, allowing it to be rebound by another 3Dpol molecule for reinitiation in a secondary reaction whose rate is 3Dpol concentration dependent. Furthermore, the elongation complexes are slightly more stable in 300 mM NaCl than in 75 mM NaCl (Fig. 3C), suggesting that maintaining the proper conformation of the complex may involve hydrophobic interactions that would be favored at the higher salt concentrations.
The decreased elongation complex stability also reduced polymerase processivity during replication of longer RNA templates. Elongation of self-primed 2,201- and 5,424-nt RNA templates showed that mutation of Trp5 to smaller residues resulted in the accumulation of abortive intermediate length products and very little full-length product. These effects were dependent upon the mutation, with the smaller valine and alanine mutants having the most deleterious effects (Fig. 4B). The mutants also showed a decrease in template utilization that is attributed to their reduced initiation and elongation complex formation rates (Tables 1 and 2). The abortive product bands, on the other hand, are attributed to elongation complexes that began during the low-salt initiation step but failed to replicate to the very end of the template. These data indicate that the elongation complex stability is a characteristic of not only a stalled polymerase but of also an actively elongating polymerase. The inability of residue 5 mutants to replicate to the 5′ end of the template can be attributed to reduced processivity that causes an increase in the amounts of aborted RNA products. The difference in processivity between the wild-type and mutant polymerases is also apparent from the amount of very large RNA produced in the reactions after long incubation times (Fig. 4A). The composition of these large RNAs was not determined, but they are presumably due to some form of template switching or internal reannealing during the elongation reactions. The lack of distinct initiation events during the elongation step was confirmed by control reactions that did not show any observable product formation under high-salt conditions (data not shown).
The abortive products generated by the mutant polymerases also show several distinct bands that are indicative of specific pause or dissociation sites along the template. Even the wild-type polymerase produces some abortive bands, something that has not been previously observed in studies showing that 3Dpol is quite processive and fully capable of elongation through secondary structures in the template (11). These bands are likely a result of our experimental conditions during the elongation phase of the reaction, where room temperature and high salt concentrations both act to stabilize RNA secondary structures. RNA structures can inhibit or stall polymerase movement along the template, allowing less stable polymerases to dissociate from the RNA template while more processive polymerases eventually elongate through the duplex section. In such a context, the increased banding seen with the smaller Trp5 mutants suggests that reduced elongation complex stability is a particularly prominent defect when elongating through templates with double-stranded RNA structures. This is also consistent with how inefficiently these mutants replicate the separate 5′ IRES fragment that is rich in secondary structure (Fig. 4B). In contrast, the tyrosine mutant and wild-type 3Dpol containing a tryptophan at residue 5 produce few distinct abortive product bands, and they elongated through the IRES template without difficulty. These results indicate that large hydrophobic amino acids at residue 5 may increase processivity by facilitating the unwinding of double-stranded RNA structures in poliovirus RNA templates.
The effects of reduced elongation complex stability are also observed when the mutant polymerases are introduced into viral replication complexes, using cell-free translation-replication reactions. The poliovirus cell-free replication system was used to compare negative-strand RNA synthesis by wild-type and mutant polymerases (Fig. 5). This system depends on cis-active elements within native viral RNA templates (9, 25) and reveals biochemical defects in RNA replication associated with mutations in 3Dpol (6). In these reactions, 3Dpol mutations result in detectable synthesis of full-length negative-strand RNA product at a timescale comparable to that observed for the wild-type enzyme, consistent with our biochemical finding that 3Dpol elongation rates are not affected by these mutations. However, the amount of full-length negative-strand product produced is significantly reduced, consistent with the reduced initiation rates of the mutant polymerases and the formation of unstable elongation complexes that dissociate prior to reaching the end of the ≈6-kb replicon genome.
Introduction of both the Trp5Phe and Trp5Leu 3Dpol mutations into poliovirus genomes resulted in virus growth after transfection into host cells. The phenylalanine mutation was stably maintained in the progeny virus with a slightly reduced plaque size but an essentially wild-type growth rate in a single-step growth curve. In contrast, virus containing the more deleterious leucine mutant grew very slowly with titers at 48 hpt that were ≈1,000-fold less than that of the wild-type virus. The leucine mutation was not stable, and sequencing of the progeny virus pool revealed a mixed population of leucine and tryptophan at residue 5 after 48 h and complete reversion to the wild-type tryptophan by 72 hpt (Fig. 6C). The loss of the leucine codon is not unexpected given the instability of its elongation complexes on the PETE RNAs and its inefficient synthesis of genomic size RNA in vitro, but it is a bit surprising that only a triple mutation (CUA to UGG) reversion to the wild-type tryptophan was observed, rather than two mutations to generate a phenylalanine (CUA to UUU/C) that has essentially wild-type growth characteristics. These results demonstrate that poliovirus propagation is dependent upon processive RNA replication and that poliovirus 3Dpol can tolerate a small decrease in elongation complex stability and processivity and still maintain viability. The emergence of the Leu5Trp reversion and the genetic stability of the Phe5 mutant may also point to a requirement for a planar residue 5 in order to get optimal viral growth.
While our results show that residue 5 is important for the stability of the elongation complex, the molecular and structural details underlying this stabilization are not yet known. It may be that residue 5 interacts directly with the RNA template via stacking interactions that are likely to be disrupted by mutations to small and nonplanar amino acids, as shown by our data. While this could be the case for poliovirus and coxsackievirus polymerases that have tryptophan and phenylalanine residues at position 5, it is less likely for the structurally homologous rhinovirus enzyme having a small hydrophobic residue and foot-and-mount disease virus 3Dpol where residue 5 is an aspartic acid. Alternatively, residue 5 may play a role in maintaining 3Dpol in the proper conformation for processive elongation. For example, residue 5 could move into the hydrophobic pocket of the finger domain as we suggested when describing the coxsackievirus 3Dpol structure (10); however, based on the new data resulting from this work, we now expect this structural rearrangement to occur only once during elongation complex formation, fixing the hydrophobic residue within the nearby pocket rather than reiteratively flipping in and out of the pocket with each cycle of nucleotide addition. Notably, structures of 3Dpol-RNA complexes from the homologous foot-and-mouth disease virus and Norwalk virus polymerases (13, 14, 30) indicate that residue 5 remains solvent exposed, suggesting that a structural rearrangement involving residue 5 may not occur. However, the RNA templates in these structures are not long enough to reveal whether or not the template strand interacts directly with the polymerase in the vicinity of residue 5. Thus, it remains to be determined whether amino acid residue 5 participates in structural rearrangements, interacts with template RNA, plays a role in strand displacement, or has some other function that we have yet to consider.
In conclusion, we have used poliovirus polymerase to further dissect the importance of residue 5 for 3Dpol function by generating a series of point mutations and measuring their effects on RNA binding, elongation complex formation, elongation rate, the temporal stability of the elongation complex, and viral growth. The data show that while residue 5 plays a small role in elongation complex formation, mutations do not affect the elongation rate of the enzyme and instead destabilize the elongation complex. The results indicate that the size and hydrophobicity, and possibly the planarity, of residue 5 are important for maintaining the 3Dpol in an elongation-competent state. Consistent with this, viral genomes containing Trp5 3Dpol mutations show reduced levels of RNA synthesis in cell-free translation-replication reactions and impaired growth of infectious virus that results in the rapid emergence of revertant virus.
Acknowledgments
We thank Peng Gong and Grace Campagnola for helpful comments on the experiments and manuscript.
This work was supported by NIH grants R01-AI059130 to O.B.P. and R01-AI042189 to D.J.B.
Footnotes
Published ahead of print on 9 June 2010.
REFERENCES
- 1.Arias, A., J. J. Arnold, M. Sierra, E. Smidansky, E. Domingo, and C. E. Cameron. 2008. Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J. Virol. 82:12346-12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, J. J., and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275:5329-5336. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, J. J., and C. E. Cameron. 2004. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43:5126-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, J. J., D. W. Gohara, and C. E. Cameron. 2004. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+. Biochemistry 43:5138-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton, D. J., B. J. Morasco, L. Eisner-Smerage, P. S. Collis, S. E. Diamond, M. J. Hewlett, M. A. Merchant, B. J. O'Donnell, and J. B. Flanegan. 1996. Poliovirus RNA polymerase mutation 3D-M394T results in a temperature-sensitive defect in RNA synthesis. Virology 217:459-469. [DOI] [PubMed] [Google Scholar]
- 7.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 8.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ Cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campagnola, G., M. Weygandt, K. Scoggin, and O. Peersen. 2008. Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases. J. Virol. 82:9458-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, M. W., O. C. Richards, T. M. Dmitrieva, V. Agol, and E. Ehrenfeld. 1993. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J. Virol. 67:3010-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collis, P. S., B. J. O'Donnell, D. J. Barton, J. A. Rogers, and J. B. Flanegan. 1992. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J. Virol. 66:6480-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrer-Orta, C., A. Arias, R. Perez-Luque, C. Escarmis, E. Domingo, and N. Verdaguer. 2007. Sequential structures provide insights into the fidelity of RNA replication. Proc. Natl. Acad. Sci. U. S. A. 104:9463-9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrer-Orta, C., A. Arias, R. Perez-Luque, C. Escarmis, E. Domingo, and N. Verdaguer. 2004. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279:47212-47221. [DOI] [PubMed] [Google Scholar]
- 15.Gohara, D. W., J. J. Arnold, and C. E. Cameron. 2004. Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry 43:5149-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gohara, D. W., C. S. Ha, S. Kumar, B. Ghosh, J. J. Arnold, T. J. Wisniewski, and C. E. Cameron. 1999. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17:128-138. [DOI] [PubMed] [Google Scholar]
- 17.Gong, P., G. Campagnola, and O. B. Peersen. 2009. A quantitative stopped-flow fluorescence assay for measuring polymerase elongation rates. Anal. Biochem. 391:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruez, A., B. Selisko, M. Roberts, G. Bricogne, C. Bussetta, I. Jabafi, B. Coutard, A. M. De Palma, J. Neyts, and B. Canard. 2008. The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases. J. Virol. 82:9577-9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, J. Q., H. L. Townsend, B. K. Jha, J. M. Paranjape, R. H. Silverman, and D. J. Barton. 2007. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J. Virol. 81:5561-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Dorner, E. A. Emini, R. Hanecak, J. J. Lee, S. van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547-553. [DOI] [PubMed] [Google Scholar]
- 21.Lakowicz, J. R. 2006. Principles of fluorescence spectroscopy, 3rd ed. Springer Science and Business Media, LLC, New York, NY.
- 22.Love, R. A., K. A. Maegley, X. Yu, R. A. Ferre, L. K. Lingardo, W. Diehl, H. E. Parge, P. S. Dragovich, and S. A. Fuhrman. 2004. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure 12:1533-1544. [DOI] [PubMed] [Google Scholar]
- 23.Ma, K., D. Temiakov, M. Anikin, and W. T. McAllister. 2005. Probing conformational changes in T7 RNA polymerase during initiation and termination by using engineered disulfide linkages. Proc. Natl. Acad. Sci. U. S. A. 102:17612-17617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mestas, S. P., A. J. Sholders, and O. B. Peersen. 2007. A fluorescence polarization-based screening assay for nucleic acid polymerase elongation activity. Anal. Biochem. 365:194-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng, K. K., J. J. Arnold, and C. E. Cameron. 2008. Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 320:137-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng, K. K., N. Pendas-Franco, J. Rojo, J. A. Boga, A. Machin, J. M. Alonso, and F. Parra. 2004. Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 279:16638-16645. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, A. A., R. A. Albertini, and O. B. Peersen. 2007. Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J. Mol. Biol. 366:1459-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, A. A., and O. B. Peersen. 2004. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 23:3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zamyatkin, D. F., F. Parra, J. M. Martin Alonso, D. A. Harki, B. R. Peterson, P. Grochulski, and K. K. Ng. 2008. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 283:7705-7712. [DOI] [PubMed] [Google Scholar]