Abstract
The p6 region of HIV-1 Gag contains two late (L) domains, PTAP and LYPXnL, that bind Tsg101 and Alix, respectively. Interactions with these two cellular proteins recruit members of the host's fission machinery (ESCRT) to facilitate HIV-1 release. Other retroviruses gain access to the host ESCRT components by utilizing a PPXY-type L domain that interacts with cellular Nedd4-like ubiquitin ligases. Despite the absence of a PPXY motif in HIV-1 Gag, interaction with the ubiquitin ligase Nedd4-2 was recently shown to stimulate HIV-1 release. We show here that another Nedd4-like ubiquitin ligase, Nedd4-1, corrected release defects resulting from the disruption of PTAP (PTAP−), suggesting that HIV-1 Gag also recruits Nedd4-1 to facilitate virus release. Notably, Nedd4-1 remediation of HIV-1 PTAP− budding defects is independent of cellular Tsg101, implying that Nedd4-1's function in HIV-1 release does not involve ESCRT-I components and is therefore distinct from that of Nedd4-2. Consistent with this finding, deletion of the p6 region decreased Nedd4-1-Gag interaction, and disruption of the LYPXnL motif eliminated Nedd4-1-mediated restoration of HIV-1 PTAP−. This result indicated that both Nedd4-1 interaction with Gag and function in virus release occur through the Alix-binding LYPXnL motif. Mutations of basic residues located in the NC domain of Gag that are critical for Alix's facilitation of HIV-1 release, also disrupted release mediated by Nedd4-1, further confirming a Nedd4-1-Alix functional interdependence. In fact we found that Nedd4-1 binds Alix in both immunoprecipitation and yeast-two-hybrid assays. In addition, Nedd4-1 requires its catalytic activity to promote virus release. Remarkably, RNAi knockdown of cellular Nedd4-1 eliminated Alix ubiquitination in the cell and impeded its ability to function in HIV-1 release. Together our data support a model in which Alix recruits Nedd4-1 to facilitate HIV-1 release mediated through the LYPXnL/Alix budding pathway via a mechanism that involves Alix ubiquitination.
Retroviral Gag polyproteins bear short conserved sequences that control virus budding and release. As such, these motifs have been dubbed late or L domains (49). Three types of L domains have thus far been characterized: PT/SAP, LYPXnL, and PPPY motifs (5, 9, 32). They recruit host proteins known to function in the vacuolar protein sorting (vps) of cargo proteins and the generation of multivesicular bodies (MVB) compartments (2). It is currently accepted that budding of vesicles into MVB involves the sequential recruitment of endosomal sorting complexes required for transport (ESCRT-I, -II, and -III) and the activity of the VPS4 AAA-ATPase (22). These sorting events are believed to be triggered by recognition of ubiquitin molecules conjugated to cargo proteins (20, 24, 41). For retrovirus budding, L domain motifs are the primary signals in Gag that elicit the recruitment of ESCRT components to facilitate viral budding. Consequently, mutations in L domain motifs or dominant-negative interference with the function of ESCRT-III members or the VPS4 ATPase adversely affect virus release. This indicates that Gag interactions with the ESCRT machinery are necessary for virus budding and separation from the cell (7, 10, 15, 16, 21, 28, 44).
Two late domains have been identified within the p6 region of human immunodeficiency virus type 1 (HIV-1) Gag protein: the PTAP and LYPXnL motifs. The PTAP motif binds the cellular protein Tsg101 (15, 39, 40, 47), whereas the LYPXnL motif is the docking site for Alix (44). Tsg101 functions in HIV-1 budding (15) as a member of ESCRT-I (30, 48), a soluble complex required for the generation of MVB. This process is topologically similar to HIV-1 budding and requires the recruitment of ESCRT-III members called the charged-multivesicular body proteins (3, 29, 48) and the activity of the VPS4 AAA-ATPase (4, 48). In addition to binding the LYPXnL motif, Alix also interacts with the nucleocapsid (NC) domain of HIV-1 Gag (13, 38), thus linking Gag to components of ESCRT-III that are critical for virus release (13).
Other retroviruses, including the human T-cell leukemia virus (HTLV) and the Moloney murine leukemia virus (MoMLV), utilize the PPPY-type L domain to efficiently release virus (7, 26, 51). The PPPY motif binds members of the Nedd4-like ubiquitin ligase family (6, 7, 16, 19, 25, 43), whose normal cellular function is to ubiquitinate cargo proteins and target them into the MVB sorting pathway (11, 12, 20). Members of the Nedd4-like ubiquitin ligase family include Nedd4-1, Nedd4-2 (also known as Nedd4L), WWP-1/2, and Itch. They contain three distinct domains: an N-terminal membrane binding C2 domain (12), a central PPPY-interacting WW domain (43), and a C-terminal HECT domain that contains the ubiquitin ligase active site (42). The functional requirement for the binding of Nedd4-like ubiquitin ligases to the PPPY motif in virus budding has been demonstrated (7, 16, 18, 19, 25, 26, 28, 50, 51). Overexpression of dominant-negative mutants of Nedd4-like ligases, ESCRT-III components, or VPS4 cause a potent inhibition of PPPY-dependent virus release (7, 19, 29, 31, 52) and induce assembly and budding defects similar to those observed after perturbation of the PPPY motif (26, 51). These observations demonstrated that Nedd4-like ligases connect Gag encoding PPPY motif to ESCRT-III and VPS4 proteins to facilitate virus release.
Whereas the role of Nedd4-like ubiquitin ligases in virus budding has been established, the protein interactions that link them to the cell's ESCRT-III pathway are still unknown. Evidence for associations of Nedd4-like ligases with ESCRT proteins have been previously reported and include: the binding of Nedd4-like ubiquitin ligases LD1 and Nedd4-1 to ESCRT-I member Tsg101 (6, 31), the colocalization of multiple Nedd4-like ubiquitin ligases with endosomal compartments (1, 28), the requirement of the cell's ESCRT pathway for Itch mediated L domain independent stimulation of MoMLV release (23), and the ubiquitination of ESCRT-I components with a shorter isoform, Nedd4-2s (8). Therefore, Nedd4-like ubiquitin ligase interactions with members of the cell's ESCRT pathway may provide retroviral Gag with access to the host budding machinery required for virus release.
Although HIV-1 Gag does not carry the PPPY canonical sequence known to interact with Nedd4-like ubiquitin ligases, both Nedd4-1 and Nedd4-2 were shown to restore the release of the HIV-1 PTAP− mutant, albeit Nedd4-1 with less efficiency than Nedd4-2 (8, 46). These findings suggested that HIV-1 might utilize cellular Nedd4-like ubiquitin ligases to increase virus release. We present here evidence demonstrating that Nedd4-1 interacts with Gag and enhances HIV-1 PTAP− virus release. Furthermore, we show that Nedd4-1's function in HIV-1 release is distinct from that of Nedd4-2 in both its viral and cellular requirements. Notably, we found that Nedd4-1 enhancement of HIV-1 release requires the Alix-binding LYPXnL L domain motif in the p6 region and basic residues in the NC domain. In addition, Alix's facilitation of HIV-1 release requires cellular Nedd4-1, since mutations in NC that prevented Alix-mediated HIV-1 release also eliminated release by overexpression of Nedd4-1. This suggested a Nedd4-1-Alix physical and functional interdependence. In agreement with this, we found Nedd4-1 to bind and ubiquitinate Alix in the cell. Taken together, these results support a model in which Alix recruits Nedd4-1 to facilitate late steps of HIV-1 release through the LYPXnL L domain motif via a mechanism that involves Alix ubiquitination.
MATERIALS AND METHODS
Proviral constructs.
The wild-type (wt) full-length HIV-1 molecular clone is pNL4-3 and the mutant pNL4-3 PTAP− (PTAP substituted with LIRL) were described previously (21). The generation of HIV-1 PTAP− LYPXnL− (PTAP−/YP−) was described in reference 13. The RKI/PTAP− double mutant was generated by introducing the PTAP to LIRL substitutions into the RKI HIV-1 proviral mutant (13).
Plasmids.
Nedd4-1 cDNA was provided by the Kazusa DNA Institute, Chiba, Japan, and contained the gene accession number KIAA0093. Nedd4-1 vectors encoding the following regions: C2 (1 to 594 bp), C2-WW (1 to 1701 bp), WW (535 to 1701 bp), WW-HECT (535 to 3,003 bp) and either N-terminal tagged hemagglutinin (HA) or 3×Flag were cloned into pHM6 and p3XFLAG mammalian cells expression vectors (Sigma-Aldrich) between the HindIII and KpnI sites. Nedd4-1 mutants C/A, (C894A), LAAL, and AYPA (where either the leucine residues or YP residues were changed to A) were created by site-directed mutagenesis using the QuikChange kit (Stratagene).
Nedd4-2 cDNA (gene accession number KIAA0439) was purchased from Open Biosystems and cloned into pHM6 and p3XFLAG between HindIII and KpnI sites. WWP-1 (NM_007013), a gift from the Miyazano Laboratory (Fred Hutchinson Cancer Research Institute), and Itch gene (NM_031483), a gift from Annie Angers (University of Montreal), were cloned between the NotI and KpnI sites in the pHM6 and p3XFLAG vectors. These vectors were described in Jadwin et al. (23). The pGAGwtPOL-RRE-r, pGAGDΔp6POL-RRE-r constructs, and the pRev1 plasmids were previously described (13). Alix/AIP-1 and derived fragment expression vectors: Broi, Bro1, Bro1-V, delBroV, V-PRD, and PRD were described in reference (13).
Virus release analysis.
293T cells (3 million) were seeded in T25 flasks and transfected the following day using Lipofectamine 2000 (Invitrogen) according to manufacturer instructions. At 24 h after transfection, cell culture medium was filtered through 0.45-μm-pore-size filter and pelleted at 150,000 × g for 1 h on a 20% sucrose cushion. After centrifugation, virus pellets were resuspended in Laemmli buffer containing 5% β-mercaptoethanol (βMe). Cells were washed in phosphate-buffered saline (PBS), pelleted at low speed and resuspended in lysis buffer (1% NP-40, 50 mM Tris [pH 8.0], 150 mM NaCl, and Complete protease inhibitor cocktail [Roche]). Virion pellets and cell lysates were analyzed by SDS-PAGE and Western blotting with an anti-HIV-1 p24 monoclonal antibody (clone 183-H12-5C). Protein expression was detected by using monoclonal anti-HA, anti-Flag, and anti-tubulin antibodies (Sigma).
Coimmunoprecipitation assays.
293T cells (2 million) were seeded into T25 flasks and transfected the following day using Lipofectamine 2000. At 48 h posttransfection, the cells were harvested, washed twice in cold PBS and lysed in RIPA buffer (0.5% NP-40/IGEPAL, 50 mM HEPES [pH 7.3], 150 mM NaCl, 2 mM EDTA, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, and Complete protease inhibitor cocktail). Cell lysates were centrifuged at 16,000 × g, 4°C for 10 min, and supernatants were incubated at 4°C with either anti-HA or anti-Flag mouse monoclonal antibodies conjugated to EZview agarose beads (Sigma). The beads were then washed in radioimmunoprecipitation assay (RIPA) and Tris-buffered saline (TBS) buffers prior to elution with the appropriate peptide(s). Immunoprecipitation eluates and cell lysates (input fractions) were analyzed by SDS-PAGE and Western blot with anti-HA or anti-Flag M2 antibodies.
RNAi knockdown.
293T cells (2 million) were seeded in T25 flasks and transfected with one or a mixture of three stealth Tsg101, Nedd4-1, or Nedd4-2 RNA interference (RNAi) oligonucleotides (Invitrogen) at a 0.2-nmol concentration using Lipofectamine 2000. After 24 h of incubation time, cells were cotransfected with mixture of 0.2 nmol of the RNAi oligonucleotides and proviral DNA in the presence or absence of additional protein expression vectors. Cells and virus were harvested and processed as outlined above. Cellular Tsg101, Nedd4-1, or Nedd4-2 was detected by using mouse monoclonal anti-Tsg101 (BD Biosciences), rabbit anti-Nedd4-1, or anti-Nedd4-2 (Cell Signaling) antibody, respectively. Sequences of RNAi oligonucleotides used for these experiments will be provided upon request.
Alix ubiquitination assay.
293T cells were cotransfected with 1 μg of full-length HIV-1 mutant PTAP− expression plasmid, 2 μg of HA-tagged ubiquitin expression plasmid, 0.5 μg of a Flag-tagged Alix expression plasmid, or the control empty vector, using Lipofectamine 2000. Culture medium was filtered 36 h posttransfection, using 0.45-μm-pore-size filters, and the virions were pelleted at 151,000 × g for 1 h on a 20% sucrose cushion. Viral pellets and cells were lysed in RIPA buffer. Cells lysates were centrifuged at 16,000 × g at 4°C for 10 min; the supernatants and virus lysates were incubated at 4°C with EZview agarose beads covalently attached to anti-Flag mouse monoclonal antibody. The beads were washed in RIPA buffer prior to two successive elutions with the Flag peptide (100 μg/ml; Sigma). Eluates from the immunoprecipitation and cell lysates (input fractions) were analyzed by SDS-PAGE and Western blotting. HIV-1 Gag and p24 proteins were detected by using either a mouse monoclonal anti-HIV-1 p24 antibody (NEA-9306; NEN Life Science, Boston, MA) or an anti-HIV-1 p24 monoclonal antibody (clone 183-H12-5C; NIH AIDS Research and Reference Reagent Program). Alix was detected by using an anti-FLAG M2 antibody and the protein ubiquitin with a mouse anti-HA antibody (1:10,000; Sigma) or anti-ubiquitin antibody (1:100; Sigma).
RESULTS
Overexpression of Nedd4-1 corrects the HIV-1 PTAP− mutant budding defects.
Many retroviral Gag proteins, including those of the Rous sarcoma virus, MoMLV, and HTLV-1, contain one copy of the PPPY L domain motif that recruits members of Nedd4-like ubiquitin ligase family to facilitate viral release. Although HIV-1 Gag polyprotein contains no obvious Nedd4-like ubiquitin ligase binding site, it was recently described to utilize Nedd4-2s to stimulate virus production (8, 45). This observation led us to speculate that HIV-1 might interact with cellular Nedd4-like ubiquitin ligases to facilitate virus release. Consistent with this view, a correction of HIV-1 PTAP− release defects was observed upon overexpression of Nedd4-1, another member of the Nedd4-like ubiquitin ligase family. However, Nedd4-1's ability to rescue HIV-1 budding defects appeared to be less robust than that obtained after the overexpression of Nedd4-2 and Nedd4-2s (8). Furthermore, we observed that a Flag-tagged Nedd4-1 can also rescue the release of the defective HIV-1 PTAP− (Fig. 1A). In addition, Flag-tagged WWP-1, whose expression in the cell was verified, also exerted a modest stimulatory effect on HIV-1 PTAP−. Conversely, despite robust expression in the cell, Itch had no positive effect on HIV-1 release. These data suggested that HIV-1 Gag might recruit cellular Nedd4-like ubiquitin ligases to facilitate virus release.
FIG. 1.
The ubiquitin ligase Nedd4-1 interacts with HIV-1 Gag and corrects HIV-1 PTAP− virus release defects. (A) Nedd4-1-like ubiquitin ligase's ability to rescue budding defects of HIV-1 PTAP− mutant. 293T cells were transfected with pNL4-3 proviral DNA (HIV-1) or with its PTAP− mutant counterpart, alone (lane 2) or with Flag-tagged Alix (lane 3), Flag-tagged Nedd4-2, Nedd4-1, WWP-1, or Itch (lanes 4 to 7). Cells and viruses were collected 24 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting with an anti-CA antibody. Expression of Alix and Nedd4-like ligases was ascertained by using an anti-Flag antibody. (B) Nedd4-like ligase-Gag binding in immunoprecipitation assays. 293T cells were transfected with the Gag expression vector pGAGwtPOL alone (lane 1) or with Flag-tagged versions of Nedd4-1, Nedd4-2, WWP-1, or Itch (lanes 2 to 5). Cells were lysed in RIPA buffer, and cleared cell lysates were exposed to anti-Flag antibody-conjugated beads. The captured protein complexes were analyzed by SDS-PAGE and Western blotting with the indicated antibodies. (C) Titrations of the amount of Nedd4-1 sufficient to enhance the release of HIV-1 PTAP− mutant. 293T cells were transfected with either proviral pNL4-3 (HIV-1) (lane2) or its PTAP− mutant counterpart, alone (lane 3) or with HA-tagged Alix (lane 4 and 5) or HA-tagged Nedd4-1 (lanes 6 and 7) or Nedd4-2 (lanes 8 and 9). Cells and viruses were collected, and their protein content was analyzed by SDS-PAGE and Western blotting with the indicated antibodies. Relative virus release efficiencies were calculated by densitometry analysis using the following ratio: efficiency = virus-associated CA/cell-associated Gag+CA, and the values obtained (%) are indicated under the panels.
The stimulation of HIV-1 release by overexpression of Nedd4-like ubiquitin ligases suggested that they might interact with Gag in the cell. We therefore examined the ability of Nedd4-1, WWP-1, and Itch to interact with Gag. Flag-tagged versions of the ubiquitin ligases Nedd4-1, WWP-1, and Nedd4-2, were found in complex with Gag in immunoprecipitation assays (Fig. 1B). In contrast, Flag-tagged Itch, as well as the control anti-Flag antibody-coated beads, failed to associate with Gag (Fig. 1B). Together, these results indicate that HIV-1 Gag interact with a subset of Nedd4-like ubiquitin ligases in the cell to enhance virus release and that interaction with Gag is a prerequisite for function in HIV-1 release.
Since we found that Nedd4-1 exerted an efficient and reproducible stimulatory effect on HIV-1 PTAP− release, and WWP-1 displayed only modest stimulation in the virus rescue assay, we conducted a detailed examination of Nedd4-1's cellular and viral functional requirements in HIV-1 release. Careful titrations determined the amount of Nedd4-1 necessary to achieve the optimal stimulatory effect on HIV-1 release (Fig. 1C). These conditions were used to perform the experiments described below.
Nedd4-1-mediated rescue of HIV-1 PTAP− requires the ubiquitin ligase activity but not cellular Tsg101.
The data above provided evidence that HIV-1 can utilize the Nedd4-like ubiquitin ligase Nedd4-1 to correct budding defects resulting from the disruption of the L domain PTAP−. To determine the functional requirements of Nedd4-1 in enhancing HIV-1 release, we first tested whether the ubiquitin ligase activity is essential. We observed that HA-Nedd4-1 rescued the release of HIV-1 PTAP−. However, the enzymatically inactive HA-Nedd4-1 C/A mutant failed to function in the virus rescue assay, despite comparable expression level to wild-type HA-Nedd4-1 (Fig. 2A). Therefore, Nedd4-1-mediated enhancement of HIV-1 PTAP− budding requires the catalytic cysteine residue in the HECT domain.
FIG. 2.

Nedd4-1-mediated rescue of HIV-1 PTAP− release requires its catalytic activity but not the cellular ESCRT-I-associated protein Tsg101. (A) Nedd4-1 stimulation of HIV-1 release requires its catalytic cysteine in the HECT domain. 293T cells were transfected with the HIV-1 PTAP− mutant alone (lane 1), with HA-Alix (lane 2), with HA-Nedd4-1 (lane 2), or with its HA-Nedd4-1C/A mutant (lanes 4). The protein content of cells and viruses was analyzed by SDS-PAGE and Western blotting with the indicated antibodies. (B) Nedd4-1 stimulation of HIV-1 PTAP− virus release is less dependent on cellular Tsg101. 293T cells were transfected with HIV-1 PTAP− mutant proviral DNA either alone (lane 1), with Alix (lanes 2 and 3), with Nedd4-1 (lanes 4 and 5), or HA-Nedd4-2 (lanes 6 and 7) in the presence of cellular Tsg101 (lanes 1, 2, 4, and 6) or Tsg101 RNAi (lanes 3, 5, and 7). The protein content of cells and viruses was analyzed by SDS-PAGE and Western blotting with the indicated antibodies. The virus release efficiency (%) is indicated under the two first panels.
Next, we assessed the effect of Tsg101 RNAi knockdown on the ability of Nedd4-1 to rescue HIV-1 PTAP− release and compared it to that of Nedd4-2 and Alix (Fig. 2B). Nedd4-1 and Alix retained the ability to promote HIV-1 PTAP− release in the absence of cellular Tsg101 (∼60%) (Fig. 2B, lanes 3 and 4). In contrast, Nedd4-2 and Nedd4-2s (data not shown) exhibited at least 5-fold reduction in activity under similar conditions (Fig. 2B, lanes 5 and 6).
Together, these results demonstrate that Nedd4-1-mediated rescue of HIV-1 PTAP− requires its ubiquitin ligase activity. Interestingly, Tsg101, and therefore ESCRT-I, appears to be dispensable (or less important) for Nedd4-1 function in HIV-1 release, a property that distinguishes Nedd4-1 function in HIV-1 release from that of Nedd4-2 (8).
The p6 region of Gag is involved in the recruitment of the ubiquitin ligase Nedd4-1.
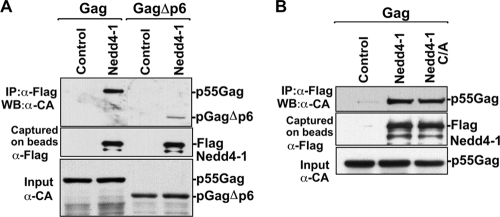
We observed that HIV-1 Gag recruits Nedd4-1 in the cell to enhance virus release. Consequently, to map the regions in Gag that are important for recruitment, interactions between Flag-Nedd4-1 and either Gag or Gag mutant (GagΔp6) were examined in immunoprecipitation assays. The Flag-Nedd4-1 proteins were found to capture Gag (Fig. 3A, lane 2). However, when the p6 region was deleted, a significant decrease of Nedd4-1's ability to interact with Gag was observed (Fig. 3A, lane 4). These results demonstrate that regions within p6 mediate interactions with HIV-1 Gag. Deletion of p6 did not completely eliminate Nedd4-1's interaction with Gag, suggesting the existence of p6-independent binding regions within Gag.
FIG. 3.
Nedd4-1 interaction with HIV-1 Gag involves regions within the p6 domain but not the catalytic cysteine residue. (A) 293T cells were transfected with either pGAGwtPOL alone (lane 1) or its pGAGΔp6POL mutant counterpart (lane 3) and with Flag-Nedd4-1 (lanes 2 and 4). After cell lysis, protein complexes were captured on anti-Flag antibody-coated beads and analyzed by SDS-PAGE and Western blotting with the indicated antibodies. (B) The cysteine residue in the HECT domain of Nedd4-1 is dispensable for binding HIV-1 Gag. 293T cells were transfected with either pGAGwtPOL alone (lane 1), with Flag-Nedd4-1 (lane 2) or with its enzymatically inactive Nedd4-1 C/A mutant (lane 3). Cell lysates were mixed with anti-Flag antibody-conjugated beads, and captured protein complexes were analyzed by SDS-PAGE and Western blotting. An anti-CA antibody was used to detect Gag products, and an anti-Flag antibody was used to detect Flag-Nedd4-1.
To determine whether Nedd4-1 ubiquitin ligase activity is involved in binding Gag, we next examined the binding of the enzymatically inactive Nedd4-1 C/A mutant to Gag. We found that the C/A mutation had no effect on Flag-Nedd4-1-Gag interaction(s) (Fig. 3B). Therefore, the ability of Nedd4-1 to conjugate ubiquitin is not required for its interaction with HIV-1 Gag.
Regions within the C2 domain of Nedd4-1 mediate binding to Gag and function in HIV-1 release.
To determine the regions in Nedd4-1 involved in interactions with HIV-1 Gag, various HA-tagged fragments of Nedd4-1 were tested for their ability to bind Gag in immunoprecipitation assays. A fragment carrying both the C2 and WW domains displayed a binding ability similar to that of the full-length HA-Nedd4-1 (Fig. 4A). This result indicated that that the two N-terminal domains, C2 and WW, were sufficient for binding Gag, whereas the HECT domain was dispensable. In addition, both C2 alone or WW alone retained binding to Gag, albeit with less efficiency than the C2-WW fragment (Fig. 4A), implying that sequences carried within the junction regions located in both the C2 and WW domains contributes to interaction(s). These data suggest that Nedd4-1 binding to Gag maps to regions located within the juxtaposed junction sequences located within the C2 and WW domains of Nedd4-1.
FIG. 4.
Role of Nedd4-1's C2 domain in HIV-1 release. (A) Regions within the Nedd4-1 C2-WW junction are involved in interactions with Gag. 293T cells were transfected with either pGAGwtPOL (lane 1), with the HA-Nedd4-1 (lane 2), or the various Nedd4-1 fragments C2, C2-WW, WW, WW-HECT, and HECT as indicated (lanes 3 to 7). Immunoprecipitation complexes were analyzed by SDS-PAGE and Western blotting with the indicated antibodies. (B) A short leucine-rich motif in the C2 domain is critical for Nedd4-1-mediated enhancement of HIV-1 release. (Upper) Leucine-rich motifs in the C2 domains of Nedd4-1 and Nedd4-2s are shown; the numbers indicate the residue positions in the full-length version of the proteins. (Lower) 293T cells were transfected with pNL4-3 proviral DNA or with its PTAP− mutant counterpart, alone (lane 3) or with HA-tagged wt Nedd4-1 (lane 4), or its mutant counterpart HA-Nedd4-1 LAAL (lane 5) or HA-Nedd4-1 AYPA (lanes 6). The protein content of cells and viruses was analyzed by SDS-PAGE and Western blotting with the indicated antibodies. An asterisk (*) indicates the positions of full-length Nedd4-1 and C2-WW fragment.
Next we sought to determine whether the Nedd4-1 C2-WW fragment played additional role(s) in Nedd4-1 driven HIV-1 release. Knowing that removal of the N-terminal 120 residues of Nedd4-1 C2 domain (Nedd4-1s) increased its stimulatory effect on HIV-1 release (46), we focused our examination on sequences that are located beyond the N-terminal 120 dispensable residues. Similarly, the N terminus of Nedd4-2's C2 domain is not required for its function in enhancing HIV-1 release (8, 46). However, regions contained within the C-terminal end of the C2 domain, present in the shorter isoform Nedd4-2s, were found to be critical. Removal of as little as 34 residues from this region eliminated Nedd4-2s stimulatory activity of HIV-1 release (46). Sequence comparison of the short regions remaining in the N-terminally truncated C2 domains of Nedd4-1s and Nedd4-2s (Fig. 4B) identified conserved leucine residues (L211/L214 and L122/L125, respectively) within both ubiquitin ligases. These leucine residues are found in the 34 residue region essential in Nedd4-2s-mediated stimulation of virus release (46). Substitutions of L211 and L214 residues to alanines (AYPA mutant) caused a near elimination of Nedd4-1 activity in HIV-1 PTAP− rescue assay (Fig. 4B). In contrast, substitution of the proline P213 and tyrosine Y213 residues (mutant LAAL), as well as of P213 and P215 (data not shown), had no effect on Nedd4-1 ability to rescue HIV-1 PTAP− (Fig. 4B). Together, these data demonstrate that the C2 domain of Nedd4-1 contains in its C-terminal region leucine residues (211LYPL214 motif) that play a critical role in Nedd4-1-mediated rescue of HIV-1 PTAP−.
Nedd4-1 requires the LYPXnL motif to rescue budding defects of HIV-1 PTAP−.
We observed that overexpression of Nedd4-1 corrected virus budding defects resulting from the disruption of the L domain motif PTAP (Fig. 1) and that the p6 region of HIV-1 Gag was involved in interactions with Nedd4-1. To determine whether the remaining LYPXnL motif in HIV-1 Gag PTAP− mutant is involved in Nedd4-1-mediated correction of virus release defects, we tested Nedd4-1's ability to rescue an HIV-1 double mutant lacking both PTAP and LYPXnL L domains (PTAP−/YP−). Mutation of the LYPXnL motif caused a near elimination of Nedd4-1-mediated rescue of HIV-1 PTAP−/YP− (Fig. 5 A, lane 9), even though Nedd4-1 rescue of HIV-1 PTAP− resulted in ∼69% of virus release (Fig. 5A, lane 8). In contrast, full-length Nedd4-2 driven rescue of the HIV-1 PTAP− motif was only minimally affected by the mutation of the LYPXnL motif, since ∼ 90% release of HIV-1 PTAP−/YP− was detected (Fig. 5B, lanes 8 and 9). Together, these results demonstrate that unlike Nedd4-2, Nedd4-1 requires the LYPXnL motif to enhance the release of HIV-1 mutant lacking the PTAP motif.
FIG. 5.
The Alix-binding LYPXnL motif is required for Nedd4-1's remediation of the HIV-1 PTAP− release defects. 293T cells were transfected with pNL4-3 proviral DNA (lane 2), with its LYPXnL− mutant virus (YP−) (lane 3), with the PTAP− mutant virus (lanes 4, 6, and 8), or with the L domain double-mutant PTAP−/YP− virus (lanes 5, 7, and 9), either alone or with HA-Alix (lanes 6 and 7 in panels A and B), HA-tagged Nedd4-1 (lanes 8 and 9 in panel A), or HA-Nedd4-2 (lanes 8 and 9 in panel B). The protein content of cells and viruses was analyzed by SDS-PAGE and Western blotting with an anti-CA antibody. Expression of Alix, Nedd4-1, and Nedd4-2 was checked by using an anti-HA antibody.
Nedd4-1 facilitation of HIV-1 release requires basic residues in NC.
Nedd4-1 remediation of HIV-1 PTAP− release defects requires the Alix-binding LYPXnL L domain motif in Gag, suggesting that Nedd4-1 function is mediated via Alix. We previously showed that intact basic residues (arginine and lysine) in the NC domain of Gag are required for Alix's Bro1 domain-mediated rescue of HIV-1 PTAP− (13). Consistent with this, we found that Alix function in HIV-1 release also requires basic residues in NC to correct HIV-1 budding defects (V. Dussupt and F. Bouamr, unpublished data). We reasoned that if Nedd4-1 functions with Alix, its mediated restoration of HIV-1 PTAP− release should also be sensitive to disruption of the basic residues in NC. To examine this hypothesis, we examined Nedd4-1's ability to restore the release of HIV-1 PTAP− alone or the double mutant RKI/PTAP−, in which the seven arginine (R) and lysine (K) residues in the N-terminal region of NC were substituted to alanines (13). Although both Alix and Nedd4-1 efficiently rescued the release of HIV-1 PTAP− (Fig. 6, left panel), neither protein was able to restore the release of the HIV-1 RKI/PTAP− double mutant (Fig. 6, right panel). In contrast, the ectopic expression of Nedd4-2s, a ubiquitin ligase that functions independently of both the LYPXnL L domain motif and the NC domain (13, 14), was able to efficiently rescue HIV-1 RKI/PTAP−. This result further confirms a functional link between Nedd4-1 and Alix.
FIG. 6.
Mutation of basic residues in NC alter Alix- and Nedd4-1-mediated rescues of HIV-1 PTAP− release. 293T cells were transfected with proviral DNA of either the PTAP− single mutant (HIV-1) (left), or with the PTAP−/RKI double mutant (right), either alone (lane 1), with HA-tagged Alix, with HA-tagged Nedd4-2s, or untagged Nedd4-1 expression vectors (lanes 2 to 4). Cells and viruses were collected 24 h posttransfection, and their protein contents were analyzed by SDS-PAGE and Western blotting with an anti-CA antibody. Expression of Alix, Nedd4-2s and Nedd4-1 was checked using anti-HA and anti-Nedd4-1 antibodies, respectively.
Nedd4-1 interacts with Alix and is involved in its ubiquitin modification in the cell.
Interactions between Alix's yeast counterpart Bro1p and the ubiquitin ligases Nedd4-1 and Rsp5 have been previously reported (33). Since we found that Nedd4-1-driven rescue of HIV-1 PTAP− is dependent on regions in Gag that are also required for Alix's facilitation of HIV-1 release, we investigated whether Nedd4-1 interacted with Alix by examining HA-Alix's ability to bind Flag-Nedd4-1 in coimmunoprecipitiation assays. Indeed, we observed a strong association of Flag-Nedd4-1 with HA-Alix, in stark contrast to the control and despite equal expression of the Flag-Nedd4-1 protein in both samples (Fig. 7A, left). Alix-Nedd4-1 interaction was also confirmed in the reverse immunoprecipitation assay (Fig. 7A, right). A weak interaction between Alix and Nedd4-1 was also detected in yeast two-hybrid (Y2H) assays (data not shown). These results demonstrated a clear interaction between Alix and Nedd4-1.
FIG. 7.
Cellular Nedd4-1 binds Alix and is involved in its ubiquitin modification in the cell. (A) Nedd4-1 binds Alix. 293T cells were transfected with Flag-Nedd4-1 alone (lane 1) or with HA-Alix (lane 2). Anti-HA antibody-conjugated beads were used to capture HA-Alix (left panel) containing immunocomplexes. Their protein content was analyzed by SDS-PAGE and Western blotting with an anti-HA and anti-Flag antibodies to probe for HA-Alix and Flag-Nedd4-1, respectively. The reverse immunoprecipitation experiment was conducted as described above, using HA-Nedd4-1 and Flag-Alix. (B) Alix is ubiquitin modified in the cell. 293T cells were transfected with Flag-Alix and HA-ubiquitin expression vectors (lanes 1, 3, and 5) or with Flag-Alix alone (lanes 2, 4, and 6). Cells were lysed, and the immunocomplexes captured on anti-Flag antibody-conjugated beads were analyzed by SDS-PAGE and Western blotting. Anti-Flag, anti-HA, or anti-ubiquitin antibodies were used to detect Flag-Alix (left panel, lanes 1 and 2), HA-ubiquitin (lanes 3 and 4), or cellular ubiquitin molecules conjugated to Flag-Alix (lanes 5 and 6) (center and right panels), respectively. MW, molecular weight markers; the asterisks (*) mark the positions of ubiquitin-modified Alix. (C) RNAi knockdown of cellular Nedd4-1 inhibits Alix ubiquitination in the cell. 293T cells were transfected with the expression vector of Flag-Alix alone (center panel, lane 1) or with that of HA-ubiquitin (lane 2) in the absence (lanes 1 and 2) or presence of Nedd4-1 RNAi oligonucleotides (lane 3). Flag-Alix containing immunocomplexes were captured by using anti-Flag antibody-conjugated beads, and their protein content was analyzed by SDS-PAGE and Western blotting with an anti-Flag antibody to detect the expression of Flag-Alix (left [darker exposure] and center panels) or anti-HA antibody to detect ubiquitin molecules conjugated to Flag-Alix (right panel). The arrow points to a darker exposure of the anti-Flag antibody Western blot (center panel) also revealing the ubiquitin-modified Flag-Alix marked with the asterisk (*). The black circle (•) marks the position of Flag-Alix. Anti-Flag and Nedd4-1 antibodies were used to detect Flag-Alix and to verify the RNAi depletion of cellular Nedd4-1, respectively.
These observations led us to examine whether Nedd4-1's interaction with Alix resulted in its ubiquitination. Flag-tagged Alix proteins were expressed in the cell in combination with HA-tagged ubiquitin (named HA-Ub hereafter). Cell extracts were then immunoprecipitated with anti-Flag antibody-coated beads and a portion of the captured Flag-Alix proteins (Fig. 7B, lanes 1 and 2) were separately checked for the presence of HA-Ub molecules. An anti-HA antibody detected a strong band in immunocomplexes purified from cells coexpressing Flag-Alix and HA-Ub but not from those expressing Flag-Alix alone (Fig. 7B, lanes 3 and 4). Based on its size, we concluded that one ubiquitin molecule has been conjugated to Flag-Alix, indicating monoubiquitination. Two faint bands were also detected, suggesting that at least two additional ubiquitin molecules were transferred to Alix. When a portion of the same immunocomplex was separately probed with an anti-ubiquitin antibody, ubiquitinated Alix proteins were also recognized (Fig. 7B, lane 5). Interestingly, the anti-ubiquitin antibody also detected a discrete band migrating at the level of ubiquitin modified Alix in cellular extracts expressing Flag-Alix alone (Fig. 7B, lane 6). This indicated that Flag-Alix was also modified with endogenous cellular ubiquitin.
Ubiquitination of Alix was detected in the cell, suggesting that the host ubiquitin machinery is sufficient to conjugate ubiquitin molecules to Alix. To examine whether Nedd4-1 is involved in this modification, we performed RNAi knockdown assays of cellular Nedd4-1 and examined whether such depletion had any effect on Alix ubiquitination. The data show (Fig. 7C, right panel, lanes 2 and 3) a near obliteration of Alix's ubiquitination when cellular Nedd4-1 was depleted, providing strong evidence of Nedd4-1's involvement in the ubiquitination of Alix.
Ubiquitinated Alix is incorporated HIV-1 PTAP− rescued particles.
It has been previously shown that Alix is incorporated in HIV-1 particles following release (48); thus, we wanted to determine whether ubiquitin modified Alix (Ub-Alix) is present at budding sites at the plasma membrane. If so, we reasoned that ubiquitinated Alix molecules should be incorporated in HIV-1 PTAP− particles, whose release was stimulated with exogenously expressed Flag-Alix. To test this hypothesis, we examined the ubiquitination status of Alix in HIV-1 PTAP− particles that were released upon overexpression of Flag-Alix and HA-Ub proteins (Fig. 8B and C). Flag-Alix was detected in rescued HIV-1 PTAP− particles, whereas very little to no Flag-Alix was released when expressed alone or with HA-Ub (Fig. 8A right, compare lane 6 to lanes 7 and 8). A portion of the same virus-pelleted fraction was analyzed with an anti-HA antibody to probe for the presence of ubiquitin molecules (Fig. 8A, lanes 1 to 4). A band comigrating with Flag-Alix proteins (Fig. 8A, lane 2) was observed, demonstrating that they contain HA-Ub molecules. This band (labeled Ub-Flag-Alix) was slightly higher than Flag-Alix (Fig. 8A, lane 6). Based on its molecular weight and migration pattern, we concluded that virus-associated Flag-Alix proteins carry one ubiquitin molecule (Fig. 8A, left, lane 2). Together, these results indicate that ubiquitin-modified Alix proteins are incorporated in released virus particles, suggesting colocalization with Gag at the plasma membrane in late stages of HIV-1 budding.
FIG. 8.
Ubiquitin-modified Alix is incorporated in HIV-1 particles released via the LYPXnL-driven pathway. 293T cells were transfected with the HIV-1 PTAP− proviral DNA, either alone (lanes 1 and 5) or with the Flag-Alix and HA-tagged ubiquitin (HA-Ub) expression vectors (lanes 2 and 6). Anti-Flag antibody-conjugated beads were used to capture Flag-Alix in HIV-1 PTAP− virions and cell lysates. The immunocomplexes were analyzed by Western blotting with an anti-Flag antibody to detect Flag-Alix in virus particles (panel A, lanes 5 and 6) or proteins pelleted from culture media (panel A, lanes 7 and 8). An anti-HA antibody was used to detect ubiquitin-modified Flag-Alix incorporated within virus (panel A, lanes 1 and 2) or pelleted proteins from culture media (panel A, lanes 3 and 4). Expression of Flag-Alix was checked in captured immnocomplexes using an anti-Flag antibody (panel B, right), and their ubiquitin modification was probed using an anti-HA antibody (panel B, left). Anti-CA antibody detected virus rescue by Flag-Alix (panel C, right lane). The asterisk (*) indicates the position of ubiquitin-modified Flag-Alix.
Alix-mediated rescue of HIV-1 PTAP− release requires cellular Nedd4-1.
To directly examine the role for an Alix-Nedd4-1 complex in HIV-1 release, we RNAi knocked down cellular Nedd4-1 and assessed the effect of such depletion on Alix-mediated rescue of HIV-1 PTAP− release. The results in Fig. 9 show that RNAi knockdown of cellular Nedd4-1 caused a near abrogation of Alix-mediated HIV-1 PTAP− release, whereas depletion of cellular Nedd4-2 did not (Fig. 9, lanes 2 and 3). These findings demonstrated that Alix-dependent rescue of HIV-1 PTAP− requires cellular Nedd4-1 but not Nedd4-2 and that Nedd4-1 plays a specific and critical role in the Alix/LYPXnL-driven HIV-1 release.
FIG. 9.
Endogenous Nedd4-1 is required for Alix's mediated HIV-1 release. 293T cells were transfected twice with the indicated RNAi oligonucleotides at 36-h intervals. At the second transfection with RNAi oligonucleotides, cells were also transfected with the HIV-1 PTAP− mutant proviral DNA in absence (lane 1) or presence of HA-Alix (lanes 2 to 4). After 24 h, the protein content of pelleted virions and cell lysates was analyzed by SDS-PAGE and Western blotting with the indicated antibodies. Relative virus release efficiencies were calculated by densitometry analysis using the following ratio: efficiency = virus-associated CA/cell-associated Gag+CA, and the values obtained (%) are indicated under the panels.
DISCUSSION
Members of the Nedd4-like ubiquitin ligase family have been previously shown to function in HIV-1 budding (8, 45). In the present study we carefully reexamined and compared the role of these ubiquitin ligases in HIV-1 release and used, like our coworkers before us, their ability to correct release defects of HIV-1 PTAP− to gauge function in virus release. When expressed in comparable amounts, Nedd4-1, Nedd4-2, WWP-1, and Itch exhibited various activities in the HIV-1 PTAP− rescue assay, with Nedd4-2 displaying the most effective stimulation. Nedd4-1 exerted an intermediate positive effect, whereas WWP-1 had only a small positive effect on HIV-1 release, possibly due to an inherently low (or tightly regulated) expression level in the cell [the present study and (8)]. In contrast, despite relatively high expression levels, Itch failed to remediate HIV-1 PTAP− release defects, even in conditions similar to those previously shown to robustly rescue budding defects of the MoMLV PPPY− mutant (22). These findings suggested that HIV-1 might recruit a subset of the host Nedd4-1-like ligases to facilitate virus release and thereby increase virus production.
The observation that Itch, which was unable to remediate HIV-1 PTAP− particle release, also failed to bind Gag suggested that Nedd4-like ligase binding to Gag is a prerequisite for function in HIV-1 release. In fact, we established that Nedd4-1 and Gag do interact in the cell. Their binding determinants were found to lie in part within the p6 domain, since deletion of the latter diminished but did not eliminate interaction. It is possible that Nedd4-1 and Gag association involves both p6-dependent and -independent bindings. Nedd4-1's functional requirement in HIV-1 release hinted to the existence of additional binding through the NC domain of Gag. Indeed, like Alix (13, 14, 37, 44), Nedd4-1 required the LYPXnL L domain motif in p6 and basic residues in NC to facilitate HIV-1 release. These results led us to conclude that Gag might recruit Nedd4-1 through interactions with Alix and was confirmed in both cellular extracts and Y2H assays. Together, these findings support a model in which Alix serves as a “bridge protein” that recruits Nedd4-1 to the vicinity of Gag and provide the first evidence of direct interaction between HIV-1 Gag and a cellular Nedd4-1-like ubiquitin ligase.
Analyses of regions in Nedd4-1 involved in interactions with Gag revealed determinants within both the C2 and WW domains. In contrast, the Nedd4-1 HECT domain failed to bind Gag, a finding that is consistent with the ability of the catalytically inactive Nedd4-1 to associate with Gag. Since Alix appears to be the binding intermediary between Gag and Nedd4-1, we speculated that Nedd4-1's C2-WW region might carry interaction sites with Alix. Further studies are under way to confirm these assumptions.
In addition to mediating interactions with Gag, the Nedd4-1 C2 domain plays a key role in the stimulation of HIV-1 release. In fact, mutations of leucine residues in the 211LYPL214 motif of C2 interfered with Nedd4-1's ability to correct HIV-1 PTAP− release defects. Similarly, equivalent leucine residues also found within the short C2 domain of Nedd4-2s were found to be critical for its mediated stimulation of HIV-1 release and incorporation in virus particles (44). However, since mutation of leucine residues in the 211LYPL214 motif (to AYPA) of Nedd4-1 had no effect on its ability to interact with Gag or Alix in the cell (data not shown), we excluded disruption of these interactions as a cause for 211AYPA214's inability to function in HIV-1 release. Importantly, the 211LYPL214 sequence is part of a membrane interacting region in the C2 domain of Nedd4-1 (35, 36). It is thus possible that disruption of the LYPL motif might have prevented Nedd4-1 from engaging in peripheral interactions at the plasma membrane that are necessary for Nedd4-1 facilitation of HIV-1 release. In any case, these data identified the leucine-rich LYPL motif in the C2 region of Nedd4-1 as a critical component of its function in HIV-1 release. Considering its location in the C2 domain (membrane binding) and the hydrophobic nature of the leucine residues required for Nedd4-1 function, a role for the Nedd4-1 LYPL motif in membrane dynamics during virus fission is plausible.
Nedd4-1's ubiquitin-conjugating HECT domain is necessary for its role in HIV-1 release as Nedd4-1's restoration of HIV-1 PTAP− release required its catalytic cysteine residue. Furthermore, overexpression of the C2-WW fragment of Nedd4-1 was not sufficient to restore release of HIV-1 PTAP− (data not shown), demonstrating that ubiquitin conjugation is critical for function. Whether ubiquitin is transferred to Gag (17) or Gag-binding proteins during HIV-1 release has been a matter of debate (53). However, the finding that Nedd4-1 interacts with Alix, suggested that Alix could be the target for Nedd4-1 ubiquitination and that such modification could have a functional significance. Consistent with this, RNAi depletion of cellular Nedd4-1 caused a near elimination of Alix ubiquitination and interfered with its ability to facilitate HIV-1 release. We concluded that the ubiquitin-modification of Alix by Nedd4-1 plays an important role in HIV-1 release. In further support of this notion, we found ubiquitinated Alix in HIV-1 PTAP− particles. Since Alix is known to promote late steps of HIV-1 release, during which it is incorporated in HIV-1 particles (47), the discovery of Ub-Alix in virus particles implied that it colocalizes with Gag in late stages of virus budding. These data suggest a model in which Gag binds Alix, an interaction that recruits Nedd4-1 from the cell's cytoplasm. Alternatively, Gag might recruit a cellular Alix-Nedd4-1 complex to facilitate virus release. Either way, such interactions provide Gag with access to cellular budding complexes containing ubiquitin-modified Alix and possibly Nedd4-1 to facilitate HIV-1 production through the LYPXnL/Alix pathway.
Several lines of evidence in the present study indicated that Nedd4-1 function in HIV-1 release is distinct from that of Nedd4-2 and its shorter isoform Nedd4-2s (8, 45). Unlike Nedd4-2, Nedd4-1 and Alix share functional interdependence in HIV-1 release as the latter's mediated HIV-1 release was inhibited after the RNAi depletion of cellular Nedd4-1. Nedd4-1's facilitation of HIV-1 PTAP− release also required the same binding regions in Gag as Alix and displayed very little to no sensitivity to the depletion of Tsg101 from the cell. This is in stark contrast to Nedd4-2, which exhibited a 5-fold loss in activity under similar circumstances.
It appears as though HIV-1 Gag makes more than one contact with members of the host cell Nedd4-like ubiquitin ligase family, either to drive virus release or to stimulate existing virus budding pathways. In addition to interactions with Nedd4-2 (and Nedd4-2s) (8, 44), Gag recruits Nedd4-1 through Alix. Stimulation of HIV-1 release by Nedd4-2 was proposed to involve the ubiquitin modification of Tsg101, a central component of ESCRT-I that bind the PTAP L domain motif (8). Similarly, we found Nedd4-1 to be important for Alix's ubiquitination and mediated HIV-1 release, supporting a role for Alix ubiquitin modification in virus release through the Alix/LYPXnL pathway. It is thus possible that Alix's ubiquitin modification augments its propensity to interact with the host ESCRT factors necessary for HIV-1 release. This is consistent with the observation that the majority of Alix captured in the cell and virus was found to be monoubiquitinated, a modification that typically signals the entry of membrane proteins into MVB via interactions with ESCRT and ESCRT-associated proteins. Alternatively, ubiquitination of Alix might also increase its ability to bind Gag, a role that was recently attributed to the ubiquitination of Tsg101 (34). Therefore, we propose a model in which Nedd4-1-mediated monoubiquitination of Alix drives or enhances HIV-1 production by increasing its ability to bind Gag and/or the fission-inducing ESCRT-III members.
In summary, we presented evidence here demonstrating that Nedd4-1 remedies HIV-1 release defects caused by the disruption of the PTAP L domain motif, revealing a role for Nedd4-1 in HIV-1 production. Nedd4-1's facilitation of HIV-1 release requires both the LYPXnL and the NC domains, the two regions in Gag also required for Alix function. Nedd4-1's interaction with Alix resulted in its monoubiquitination, a posttranslational modification known to mediate interactions with components of the host ESCRT pathway. In addition, RNAi depletion of Nedd4-1 resulted in the near elimination of Alix's ubiquitination and mediated HIV-1 release. Together, these findings establish a physical and functional link between Alix and Nedd4-1 and suggest that Alix recruits Nedd4-1 to HIV-1 budding sites to facilitate virus release through the Alix/LYPXnL pathway. Our studies identify Nedd4-1 as the first cellular ubiquitin ligase that associates with Gag through Alix and facilitates HIV-1 release via a mechanism that involves Alix ubiquitination. This is consistent with recent reports revealing new role(s) for cellular ubiquitin ligases in the production of HIV-1, as well as that of other enveloped viruses (26, 27, 33, 34).
Acknowledgments
We thank Arianna Calistri (University of Padova, Padua, Italy) for the HA-ubiquitin expression plasmid and Vicki Rudd, Que Dang (LMM, NIAID), and Bernard Moss (LVD, NIAID) for helpful discussions and critical reading of the manuscript.
This study was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 2 June 2010.
REFERENCES
- 1.Angers, A., A. R. Ramjaun, and P. S. McPherson. 2004. The HECT domain ligase itch ubiquitinates endophilin and localizes to the trans-Golgi network and endosomal system. J. Biol. Chem. 279:11471-11479. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M. 2005. A protein's final ESCRT. Traffic 6:2-9. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. ESCRT-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 6.Blot, V., F. Perugi, B. Gay, M. C. Prevost, L. Briant, F. Tangy, H. Abriel, O. Staub, M. C. Dokhelar, and C. Pique. 2004. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking toward the multivesicular body pathway prior to virus budding. J. Cell Sci. 117:2357-2367. [DOI] [PubMed] [Google Scholar]
- 7.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, H. Y., E. Morita, U. von Schwedler, B. Muller, H. G. Krausslich, and W. I. Sundquist. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 82:4884-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 10.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. U. S. A. 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, R., and L. Hicke. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276:25974-25981. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, R., D. A. Klos, A. S. Adler, and L. Hicke. 2004. The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J. Cell Biol. 165:135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dussupt, V., M. P. Javid, G. Abou-Jaoude, J. A. Jadwin, J. de La Cruz, K. Nagashima, and F. Bouamr. 2009. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 5:e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher, R. D., H. Y. Chung, Q. Zhai, H. Robinson, W. I. Sundquist, and C. P. Hill. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841-852. [DOI] [PubMed] [Google Scholar]
- 15.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 16.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottwein, E., S. Jager, A. Habermann, and H. G. Krausslich. 2006. Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 gag cause a late budding defect. J. Virol. 80:6267-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. U. S. A. 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidecker, G., P. A. Lloyd, K. Fox, K. Nagashima, and D. Derse. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78:6636-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 21.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurley, J. H., and S. D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jadwin, J. A., V. Rudd, P. Sette, S. Challa, and F. Bouamr. 2010. Late domain-independent rescue of a release-deficient Moloney murine leukemia virus by the ubiquitin ligase itch. J. Virol. 84:704-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 25.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. U. S. A. 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Blanc, I., M. C. Prevost, M. C. Dokhelar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malakhova, O. A., and D. E. Zhang. 2008. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 283:8783-8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medina, G., Y. Zhang, Y. Tang, E. Gottwein, M. L. Vana, F. Bouamr, J. Leis, and C. A. Carter. 2005. The functionally exchangeable L domains in RSV and HIV-1 Gag direct particle release through pathways linked by Tsg101. Traffic 6:880-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 33.Nikko, E., and B. Andre. 2007. Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot. Cell 6:1266-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okumura, A., G. Lu, I. Pitha-Rowe, and P. M. Pitha. 2006. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U. S. A. 103:1440-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okumura, A., P. M. Pitha, and R. N. Harty. 2008. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl. Acad. Sci. U. S. A. 105:3974-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plant, P. J., F. Lafont, S. Lecat, P. Verkade, K. Simons, and D. Rotin. 2000. Apical membrane targeting of Nedd4 is mediated by an association of its C2 domain with annexin XIIIb. J. Cell Biol. 149:1473-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plant, P. J., H. Yeger, O. Staub, P. Howard, and D. Rotin. 1997. The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J. Biol. Chem. 272:32329-32336. [DOI] [PubMed] [Google Scholar]
- 38.Popov, S., E. Popova, M. Inoue, and H. G. Gottlinger. 2008. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 82:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 40.Pornillos, O., S. L. Alam, R. L. Rich, D. G. Myszka, D. R. Davis, and W. I. Sundquist. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raiborg, C., T. E. Rusten, and H. Stenmark. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15:446-455. [DOI] [PubMed] [Google Scholar]
- 42.Rotin, D., and S. Kumar. 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 10:398-409. [DOI] [PubMed] [Google Scholar]
- 43.Staub, O., S. Dho, P. Henry, J. Correa, T. Ishikawa, J. McGlade, and D. Rotin. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15:2371-2380. [PMC free article] [PubMed] [Google Scholar]
- 44.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 45.Usami, Y., S. Popov, and H. G. Gottlinger. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 81:6614-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Usami, Y., S. Popov, E. Popova, and H. G. Gottlinger. 2008. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J. Virol. 82:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. U. S. A. 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 49.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 50.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zamborlini, A., Y. Usami, S. R. Radoshitzky, E. Popova, G. Palu, and H. Gottlinger. 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. USA 103:19140-19145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhadina, M., M. O. McClure, M. C. Johnson, and P. D. Bieniasz. 2007. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc. Natl. Acad. Sci. USA 104:20031-20036. [DOI] [PMC free article] [PubMed] [Google Scholar]