Abstract
Interferons (IFNs) are key mediators of the host innate antiviral immune response. To identify IFN-stimulated genes (ISGs) that instigate an antiviral state against two medically important flaviviruses, West Nile virus (WNV) and dengue virus (DENV), we tested 36 ISGs that are commonly induced by IFN-α for antiviral activity against the two viruses. We discovered that five ISGs efficiently suppressed WNV and/or DENV infection when they were individually expressed in HEK293 cells. Mechanistic analyses revealed that two structurally related cell plasma membrane proteins, IFITM2 and IFITM3, disrupted early steps (entry and/or uncoating) of the viral infection. In contrast, three IFN-induced cellular enzymes, viperin, ISG20, and double-stranded-RNA-activated protein kinase, inhibited steps in viral proteins and/or RNA biosynthesis. Our results thus imply that the antiviral activity of IFN-α is collectively mediated by a panel of ISGs that disrupt multiple steps of the DENV and WNV life cycles.
West Nile virus (WNV) and dengue virus (DENV) are mosquito-borne flaviviruses that cause invasive neurological diseases and lethal hemorrhagic fever in humans, respectively (6, 32). Since its first incursion into New York City in 1999, WNV has rapidly spread throughout the continental United States and has recently reached South America (29, 34). In most cases, WNV infection of people resolves as an asymptomatic or a mild febrile illness. However, approximately 1% of infections result in severe neurological disorders, such as encephalitis and meningitis (27). Unlike WNV, for which people are only accidental hosts, DENV has fully adapted to humans (32). It has apparently lost the need for an enzootic cycle and causes a range of diseases in people, from acute febrile illness to life-threatening dengue hemorrhagic fever/dengue shock syndrome (6). Four distinct serotypes of DENV have spread throughout the tropical and subtropical parts of the world, with an estimated 50 to 100 million human cases annually and about 2.5 billion people worldwide being at risk of infection (32). Effective antiviral therapies and vaccines to treat or prevent WNV and DENV infections in humans are not yet available.
Type I interferons (IFNs), represented by IFN-α and IFN-β, have been demonstrated to play an essential role in defending against WNV and DENV infections. For example, mice with deficiencies in the induction of type I IFNs and the receptor or JAK-STAT signal transduction pathway of the cytokines are vulnerable to WNV and DENV infections (7, 38, 42, 49-51). In addition, a strain of WNV that fails to block the type I IFN signal transduction pathway is phenotypically attenuated in mice (23, 50). Clinically, during acute DENV infection, innate immune responses play a key role in determining disease outcome (35), and resolution of WNV infection requires effective IFN-mediated innate host responses (23, 43, 53). Therefore, understanding how the IFN-mediated innate immune response functions is one of the critical frontiers in the molecular biology of WNV and DENV pathogenesis (1, 44).
IFNs inhibit virus infection by induction of IFN-stimulated genes (ISGs) that disrupt distinct steps of the viral replication cycle (47). However, although IFN treatment of cells induces the expression of hundreds of cellular genes (9), only approximately a dozen ISGs have been experimentally demonstrated to instigate an antiviral state against selected viruses (41). As mentioned above, although there is ample evidence suggesting that IFN-mediated innate immunity plays a critical role in defending against WNV and DENV infections, the underlying antiviral mechanism of the cytokines remains to be understood (6, 16, 31). With WNV, previous studies suggested that mice lacking double-stranded-RNA-activated protein kinase (PKR) and RNase L were more susceptible to the virus infection and had increased viral loads in multiple peripheral organs and neuronal tissues, in comparison with congenic wild-type mice (43). In addition, genetic studies showed that a nonsense mutation in the gene encoding the 2′,5′-oligoadenylate synthetase 1b (OAS1b) isoform was associated with WNV susceptibility in mice, and expression of wild-type OAS1b in mouse fibroblasts efficiently inhibited WNV infection (22, 33, 37, 45). For DENV, it was reported recently that viperin was among the highly induced ISGs in DENV-infected cells and overexpression of viperin in A549 cells significantly reduced DENV replication (13).
In principle, to understand how IFNs inhibit DENV and WNV infections, it is essential to know the repertoire of ISGs that are directly implicated in antiviral action and understand how these antiviral ISGs work individually and coordinately to limit virus replication. To achieve this goal, we set out to systematically identify the ISGs that are able to inhibit infection with the two viruses and elucidate their antiviral mechanisms.
Establishment and characterization of cell lines inducibly expressing ISGs.
Overexpression and small interfering RNA (siRNA)-mediated knockdown of ISG expression are two common approaches to identify ISGs involved in the induction of an antiviral state by IFNs (52, 56, 59). However, the antiviral effects of the cytokines are most likely mediated by multiple pathways (59, 60), and hence, ablation of a single ISG's function by siRNA could be compensated for by other antiviral pathways. We therefore decided to take the approach of establishing stable cell lines that can express individual ISGs in a tetracycline-inducible manner. These cell lines allowed the systematic identification of antiviral ISGs and a detailed characterization of their antiviral mechanism (21). We initially chose the HEK293-derived cell line FLP-IN T Rex (Invitrogen) as the “platform” for our ISG expression system. The FLP-IN T Rex genome contains stable integrations of a single FRT site and a gene that expresses a TET repressor (3). FLP-IN T Rex cells were therefore cotransfected with FRT site-containing plasmids that encode a desired ISG (pcDNA5/FRT/ISG) and plasmid pOG44, which expresses Flp IN recombinase (21). The transfected cells were selected with blasticidin and hygromycin, and the antibiotic-resistant cells were pooled and expanded into cell lines (3). The resulting cell line contains a single integration of an ISG cDNA through the FRT site with its expression under the control of a TET-on promoter.
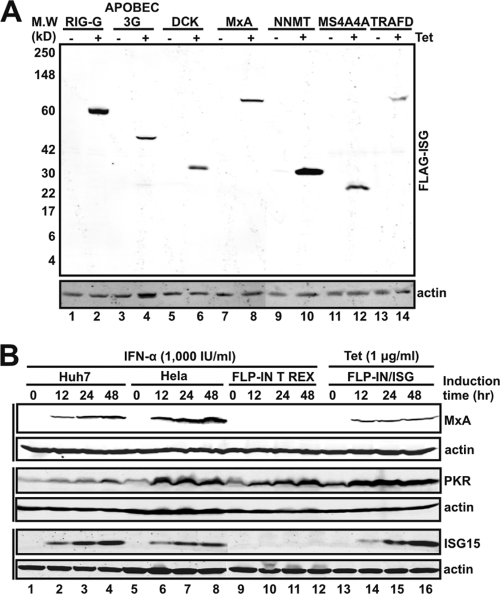
Following an approach similar to that used to identify ISGs that inhibit hepatitis C virus (HCV) replication (21), we expanded our FLP-IN T Rex-derived ISG-expressing cell line library to include 36 ISGs that are commonly induced by IFN-α in human hepatocytes (17), neuronal cells (55), macrophages, and dendritic cells (25, 61), which are the pathologically relevant host cells of WNV and/or DENV infection in vivo (20, 26, 28) (Table 1). Moreover, the majority of the 36 ISGs have been shown by microarray studies to be induced in WNV- and/or DENV-infected cells (13-15, 46, 58). Tetracycline-inducible expression of the desired ISG in each of the 36 cell lines was confirmed by Western blot assay with a mouse monoclonal antibody against FLAG tag (21) (Fig. 1 A). Furthermore, the kinetics and level of ISG expression by the cell lines upon induction by the addition of tetracycline to the culture medium, as demonstrated with three ISG-expressing cell lines (MxA, PKR, and ISG15), are similar to those produced by 1,000-IU/ml IFN-α treatment in Huh7 and HeLa cells (Fig. 1B). Interestingly, due to a low-level expression of signaling components such as STAT2 and IRF9, FLP-IN T Rex cells are partially defective in IFN response and express undetectable levels of MxA and ISG15, but normal level of PKR protein, upon IFN-α treatment.
TABLE 1.
Thirty-six tetracycline-inducible FLP-IN T Rex-derived cell lines that expressed the individual ISGs and their effects on WNV and DENV infection
| ISG | GenBank accession no. | Inhibition of WNV VLP infection | Inhibition of WNV replicon CFEa | Inhibition of DENV-1 VLP or DENV-2 infection |
|---|---|---|---|---|
| ISG56 | NM_001548 | − | − | − |
| RIG-G | NM_001549 | − | − | − |
| GBP1 | NM_002053 | − | − | − |
| MxA | NM_002462 | − | − | − |
| IFITM1 | NM_003641 | − | − | − |
| IFITM2 | NM_006435 | + | − | + |
| IFITM3 | NM_021034 | + | − | + |
| ISG15 | NM_005101 | − | − | − |
| USP18 | NM_017414 | − | − | − |
| UBE2L6 | NM_004223 | − | − | − |
| ISG12 | BN000227 | − | − | − |
| ADAR1 | NM_001111 | − | − | − |
| APOBEC3G | NM_021822 | − | − | − |
| ISG20 | NM_002201 | + | + | + |
| Viperin | AF442151 | + | + | + |
| PKR | AH008429 | + | + | − |
| OAS1 Var1 | NM_016816 | − | − | − |
| OAS1 Var2 | NM_002534 | − | − | − |
| OAS-L Var1 | NM_003733 | − | − | − |
| OAS-L Var2 | NM_198213 | − | − | − |
| MTAP44 | D28915 | − | − | − |
| PLSCR1 | NM_021105 | − | − | − |
| PLSCR2 | NM_020359 | − | − | − |
| MAPK8 | NM_002750 | − | − | − |
| SAMHD1 | NM_015474 | − | − | − |
| IFI44 | NM_006417 | − | − | − |
| IFI44L | NM_006820 | − | − | − |
| BST2 | NM_004335 | − | − | − |
| FLJ20637 | AK000644 | − | − | − |
| FLJ38348 | AK095667 | − | − | − |
| STAF50 | X82200 | − | − | − |
| FLJ20035 | AK000042 | − | − | − |
| NNMT | NM_006169 | − | − | − |
| DCK | NM_000788 | − | − | − |
| MS4A4A | BC020648 | − | − | − |
| TRAFD1 | NM_006700 | − | − | − |
CFE, colony formation efficiency.
FIG. 1.
Characterization of ISG expression following tetracycline (Tet) induction in FLP-IN/ISG cell lines. (A) Demonstration of the desired ISG expression in the seven newly established cell lines. FLP-IN T Rex-derived cell lines that inducibly express individual ISGs were established as described previously (21). The cells were cultured in the absence or presence of tetracycline for 48 h and then harvested. The levels of N-terminally FLAG-tagged ISG protein expression in cell lysates were determined by Western blot analysis with a monoclonal antibody against the FLAG tag as previously described (21). M. W., molecular mass. (B) Comparison of the kinetics and levels of ISG expression upon IFN-α treatment and tetracycline induction. Huh7, HeLa, and parental FLP-IN T Rex cells were left untreated or treated with 1,000 IU/ml IFN-α, and cells were harvested at the indicated times after IFN-α treatment. Three FLP-IN/ISG cell lines that express MxA, PKR, and ISG15, respectively, were cultured in the absence or presence of 1 μg/ml tetracycline, and cells were harvested at the indicated times after the addition of the antibiotic. The levels of the three IFN-induced proteins in cell lysates were determined by Western blot analysis. Briefly, cell monolayers were washed once with phosphate-buffered saline and lysed with 1× Laemmli buffer. A fraction of the cell lysate was separated on SDS-12% polyacrylamide gels and electrophoretically transferred onto polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked with phosphate-buffered saline containing 5% nonfat dry milk and probed with antibodies against MxA (Proteintech Group), PKR (a gift from Pat Romano, Institute for Hepatitis Virus Research, Hepatitis B Foundation, Doylestown, PA), ISG15 (Cell Signaling), and β-actin (Chemicon International). Bound antibody was revealed by IRDye secondary antibodies and visualized by the Li-COR Odyssey system.
Identification of ISGs that inhibit WNV and DENV infections.
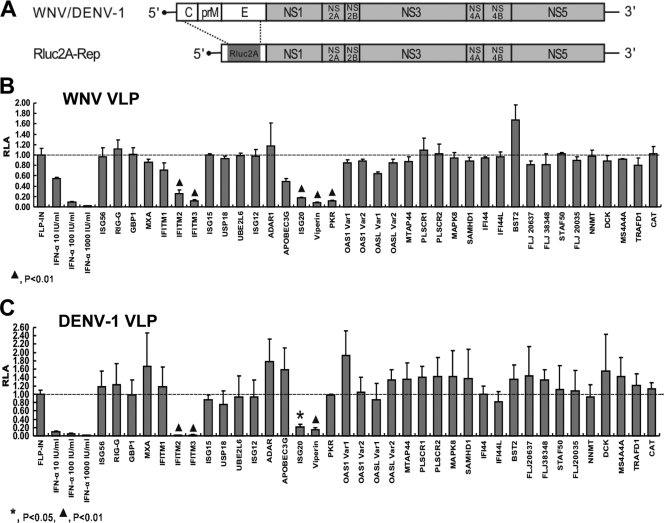
To identify ISGs that are able to inhibit WNV and/or DENV infections, we initially examined the effects of each individual ISG on WNV and DENV-1 replicon-containing virus-like particle (VLP) infections in FLP-IN T Rex-derived ISG-expressing cell lines. As described previously (39), WNV and DENV-1 VLPs were prepared by providing the viral structural proteins in trans in cells containing viral replicons. Like virions, the VLPs are able to infect permissive cells and initiate replicon RNA replication. However, because the replicon Rluc2A-Rep contains a Renilla luciferase gene in lieu of viral structural genes, as depicted in Fig. 2 A, infection of cells with such replicon-containing VLPs fails to produce progeny virions and spread to neighboring cells. Instead, replicon replication results in the expression of Renilla luciferase, which can serve as a convenient and quantitative reporter of virus infection and genome replication. Therefore, the VLP infection assay will provide a robust experimental system for the identification of ISGs that inhibit steps in the early stage (virion uptake and uncoating) and/or biosynthetic stage (viral protein/RNA biosynthesis and turnover) of the viral replication cycle but will not reveal the ISGs that target late-stage events such as virion assembly and secretion.
FIG. 2.
Effects of ISG expression on WNV and DENV-1 VLP infection. (A) Schematic representation of the structures of subgenomic replicons packaged within WNV and DENV VLPs. Compared with the full-length WNV or DENV genome, the VLP-containing reporter replicon Rluc2A-Rep contains an in-frame replacement of the structural genes with the Renilla luciferase gene fused with the 2A fragment from food-and-mouth disease virus (39). (B and C) FLP-IN/ISG and FLP-IN/CAT cells were seeded into the wells of a 96-well plate at a density of 5 × 104 cells per well. At 24 h postseeding, the cells were cultured in the absence or presence of 1 μg/ml tetracycline for 24 h, infected with WNV (panel B) or DENV-1 (panel C) VLPs at a multiplicity of infection of 1, refed with medium with or without 1 μg/ml tetracycline for an additional 24 h, and assayed for luciferase activity with a Renilla luciferase assay kit (Promega). As controls, parental FLP-IN T Rex cells were left untreated or pretreated with the indicated concentrations of IFN-α for 24 h, infected with WNV VLPs, cultured further with or without the indicated concentrations of IFN-α for an additional 24 h, and assayed for luciferase activity as described above. The RLA represents the mean ± the standard deviation (n = 4) of the ratio of light units obtained from wells treated with IFN or cultured in the presence of tetracycline to the light units obtained from wells that were left untreated or cultured in the absence of tetracycline.
FLP-IN/CAT cells, which inducibly express the control protein chloramphenicol acetyltransferase (CAT), and the 36 FLP-IN/ISG cell lines were cultured in the absence or presence of tetracycline for 24 h, followed by infection with WNV or DENV-1 VLPs, and cultured in the absence or presence of tetracycline for an additional 24 h. VLP infection and replicon replication were quantified by measuring Renilla luciferase activity in the cell lysates. As controls, parental FLP-IN T Rex cells were left untreated or pretreated with 10, 100, and 1,000 IU/ml IFN-α, respectively, for 24 h and then infected with WNV or DENV-1 VLPs. The effect of individual ISG expression or IFN-α treatment on VLP infection was expressed as relative luciferase activity (RLA), which is the ratio of luciferase activity (light units) obtained from wells treated with either IFN or tetracycline over that obtained from wells that were left untreated or cultured in the absence of tetracycline. Hence, it is expected that the RLA should be approximately 1 if the ISG does not inhibit VLP infection and less than 1 if the ISG inhibits VLP infection.
As shown in Fig. 2B and C, IFN-α dose dependently inhibited the infection of FLP-IN T Rex cells by both WNV and DENV-1 VLPs. As expected, expression of CAT and the majority of the ISGs affected neither WNV nor DENV-1 VLP infection. However, four ISGs, including IFN-induced transmembrane protein 2 (IFITM2), IFITM3, viperin, and ISG20, significantly reduced the levels of Renilla luciferase activity in both WNV and DENV-1 VLP-infected cells. The antiviral potency of these ISGs is similar to that obtained with 10- to 100-IU/ml IFN-α treatment. Interestingly, expressing one of the best-studied antiviral proteins, PKR, only inhibited infection with WNV, but not DENV-1, VLPs.
Inhibition of WNV and DENV infections by viperin, ISG-20, and PKR requires their enzymatic function.
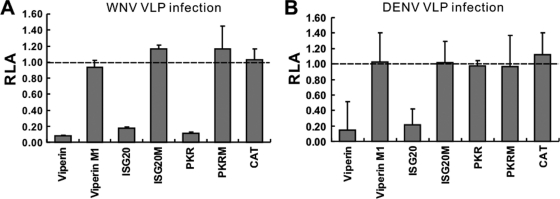
We have previously demonstrated that viperin is a putative radical S-adenosyl-l-methionine (SAM) domain-containing enzyme (21). PKR has already been defined, and ISG20 is a 3′-to-5′ exonuclease. To determine if the inhibitory effects of the three corresponding ISGs on WNV and/or DENV infection depend on their respective enzymatic activities, we tested the antiviral activities of enzymatically inactive mutant viperin, ISG20, and PKR on WNV and DENV VLP infection. As shown in Fig. 3, while the three wild-type ISGs efficiently inhibited WNV and/or DENV-1 VLP infection, expression of a mutant viperin bearing cysteine-to-alanine mutations in its essential motif (CxxxCxxC) for coordination of radical SAM (viperin M1) or enzymatically inactive PKRK296R (PKRM) and ISG20D96G (ISG20M) inhibited neither WNV nor DENV VLP infection. These results thus suggest that inhibition of WNV and DENV infections by the three IFN-induced cellular enzymes requires their respective enzymatic activities.
FIG. 3.
Requirements of the enzymatic activities of viperin, ISG20, and PKR for their antiviral effects against WNV and/or DENV. FLP-IN T Rex-derived cell lines inducibly expressing viperin, ISG20, PKR, the respective enzymatically inactive mutant proteins, and the control protein CAT were seeded into the wells of a 96-well plate at a density of 5 × 104 per well. Tetracycline pretreatment and WNV or DENV VLP infection were as described in the legend to Fig. 2. After infection, cells were cultured with medium with or without tetracycline for 24 h and assayed for luciferase activity. RLA is as defined in the legend to Fig. 2.
Effects of antiviral ISGs on DENV infection.
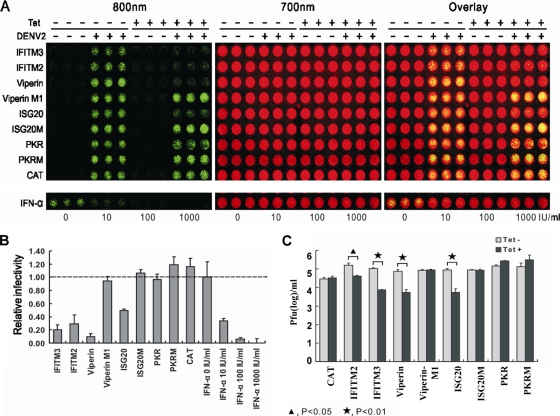
To further validate the results obtained with the VLP infection assay, the effects of the five antiviral ISGs on DENV-2 infection were accessed with two independent assays. First, the effects of the ISGs on DENV infection and spreading were measured with an in-cell Western assay that detected viral envelope (E) protein accumulation in virus-infected cells. As shown in Fig. 4 A, IFN-α dose dependently inhibited DENV-2 infection and its antiviral potency is slightly weaker than that obtained with the DENV-1 VLP assay. Consistent with the results obtained with the VLP assay, PKR did not affect DENV-2 virus infection, but IFITM2, IFITM3, viperin, and ISG20 significantly inhibited DENV-2 infection, as indicated by a 2- to 5-fold reduction of viral E protein accumulation in virally infected cells (Fig. 4B).
FIG. 4.
Effects of ISGs on DENV infection. (A) The stable cell lines that express CAT protein or ISGs and the respective mutant proteins were seeded into the wells of a 96-well plate at a density of 5 × 104 per well. At 24 h postseeding, the cells were cultured in the absence or presence of 1 μg/ml tetracycline for 24 h, infected with serotype 2 DENV at a multiplicity of infection of 0.1, and refed with medium with or without tetracycline. As controls, parental FLP-IN T Rex cells were left untreated or pretreated with the indicated concentrations of IFN-α for 24 h, infected with DENV-2, and cultured further in medium with or without the indicated concentrations of IFN-α. At 48 h after infection, cells were fixed with 3.7% formaldehyde in phosphate-buffered saline, incubated with anti-E antibody 4G2 (1:200; Millipore) for 1 h, and incubated with an anti-mouse IRDye 800CW-labeled secondary antibody together with two reagents for cell staining (DRAQ5 from Biostatus and Sapphire700 from LI-COR). The DENV E protein was visualized in LI-COR Odyssey in the 800-nm channel as green. The cell viability was determined by Sapphire 700 staining (Red color). (B) E protein levels were determined with LI-COR Odyssey and normalized to cell viability. The relative infectivity represents the mean ± the standard deviation (n = 3) of the ratio of the density obtained from wells treated with IFN-α or cultured in the presence of tetracycline to that obtained from wells that were left untreated or cultured in the absence of tetracycline. (C) Effects of ISGs on DENV production. The indicated cell lines were cultured in the absence or presence of 1 μg/ml tetracycline (Tet) for 24 h, infected with DENV-2 at a multiplicity of infection of 0.1, refed with medium with or without tetracycline, and cultured for 3 days. The culture medium was changed every 24 h, and virus yields between 48 and 72 h postinfection were determined by plaque assay and expressed as PFU per milliliter of culture medium (n = 3).
Second, the effects of antiviral ISG expression on DENV-2 yields (titers) were measured by a plaque assay. As shown in Fig. 4C, expression of IFITM3, viperin, and ISG20 reduced the virus yields by approximately 10-fold. However, IFITM2 expression resulted in only a 4-fold reduction of virus titer. In agreement with the results presented above, no significant reduction of virus yield could be detected upon PKR expression. Moreover, in agreement with the results shown in Fig. 3, the results obtained from both the in-cell Western and virus yield reduction assays indicate that the putative radical SAM domain-containing enzymatic activity of viperin and the exonuclease activity of ISG20 are required for their antiviral function against DENV infection.
Identification of viral replication steps disrupted by antiviral ISGs.
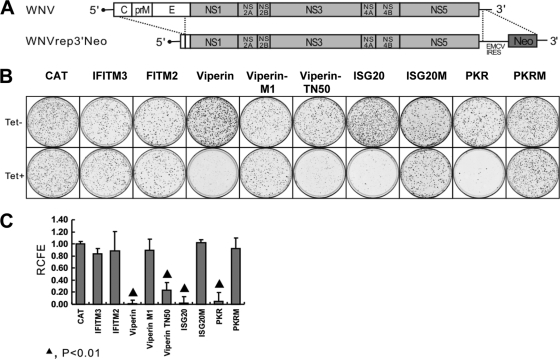
To begin to map the viral replication steps potentially inhibited by each of the antiviral ISGs, we intended first to distinguish whether the ISGs identified inhibit the early events of infection or viral protein/RNA biosynthesis. To achieve this goal, we tested the effects of all of the five antiviral ISGs identified above on WNV replicon RNA replication with a WNV replicon-conferred cell colony formation assay (4). Unlike the VLP infection assay, the colony formation assay identifies only ISGs that target steps in viral protein translation, polyprotein processing, and RNA replication and not ISGs that target steps in virus entry and uncoating, which are bypassed by the direct delivery of replicon RNA into the cells via transfection (48). Briefly, to initiate WNV replicon replication, the individual ISG-expressing cell lines were transfected with total RNA extracted from a FLP-IN T Rex-derived cell line harboring WNV subgenomic replicons (WNVrep3′neo, as depicted in Fig. 5 A) (16, 48). Replicon-replicating cells were selected with 500 μg/ml G418 in the presence or absence of 1 μg/ml tetracycline, respectively. Two weeks after the selection, G418-resistant cell colonies became visible and were stained with crystal violet to facilitate colony counting.
FIG. 5.
Effects of ISG expression on WNV replicon-conferred cell colony formation. (A) WNV subgenomic replicon WNVneoRep contains an in-frame deletion of the structural genes and insertion of the neomycin phosphotransferase gene (neo) driven by an internal ribosome entry site derived from encephalomyocarditis virus in the 3′ untranslated region. (B) CAT- and ISG-expressing FLP-IN T Rex stable cell lines were electroporated with WNVrep3′neo derived from HEK293/WNVrep cells and selected with G418 in the absence or presence of 1 μg/ml tetracycline for 2 weeks as described previously (4). Cell foci were stained with crystal violet and photographed. A pair of representative plates of each of the five ISGs and their mutant protein-expressing and control protein CAT-expressing cell lines that were cultured in the absence (top) or presence (bottom) of tetracycline (Tet) is presented. (C) The foci of each of the ISG- and CAT-expressing cell lines cultured in three plates in either the absence or the presence of tetracycline were counted. Relative colony formation efficiency (RCFE) was expressed and plotted as the mean ± the standard deviation (n = 3) of the ratios of the number of foci of cells that were selected in the presence of tetracycline to the number obtained from cells that were cultured in the absence of the antibiotic.
As shown in Fig. 5B and C, among the five ISGs that inhibited WNV VLP infection, wild-type viperin, ISG20, and PKR reduced colony formation efficiency by approximately 100-, 50-, and 20-fold, respectively. Consistent with the results obtained with the VLP infection assay, the three enzymatically inactive mutant ISGs did not affect WNV replicon replication. Moreover, in agreement with what we found with HCV replicons, deletion of the N-terminal 50 amino acids of viperin (viperin TN50) significantly compromised its ability to associate with membrane (data not shown) and also partially impaired its antiviral activity against WNV (Fig. 5). Interestingly, IFITM2 and IFITM3 did not affect the colony formation efficiency of WNV replicons.
The reduced colony formation efficiency in responding to the expression of ISG20, viperin, or PKR could be due to either inhibition of WNV replicon replication or cytotoxic effects induced by the three ISGs. The latter possibility had been ruled out by demonstrating that none of the ISGs significantly affected FLP-IN T Rex cell growth (see reference 21 for details). Failure to induce cell death upon the expression of PKR in FLP-IN T Rex cells is most likely due to its relatively low induction level, which is similar to that obtained with 1,000-IU/ml IFN-α treatment, as demonstrated in Fig. 1B. Hence, our results suggest that while viperin, PKR, and ISG20 most likely inhibit replication steps in viral protein and/or RNA biosynthesis, IFITM2 and IFITM3 most probably target a viral replication step(s) before input virion RNA translation, such as virus binding, entry, and nucleocapsid uncoating.
Effects of antiviral ISGs on transient replication of DENV-1 replicons.
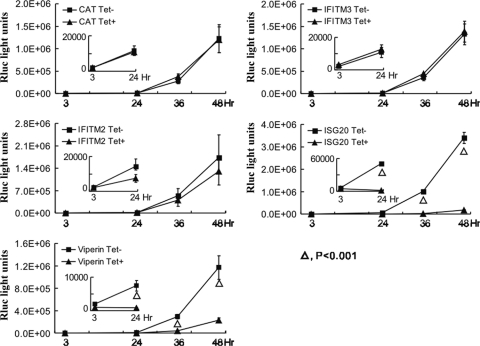
To determine if the antiviral ISGs inhibit DENV infection via the same mechanisms as suggested by the WNV replicon-conferred colony formation assay, we tested the effects of the ISGs on DENV replicon replication with a replicon transient transfection assay. As described previously, transfection of in vitro-transcribed DENV-1 Rluc2A-Rep (depicted in Fig. 2A) into cells resulted in dual-phase luciferase expression. While the first phase of luciferase expression peaked at 3 h posttransfection and declined to the background level at 10 h posttransfection, the second phase of expression started at 24 h posttransfection and increased exponentially (39). It had been demonstrated that the first phase of luciferase expression was translated from the input replicon RNA, but the second phase of luciferase expression required replicon RNA replication (39).
As shown in Fig. 6, while the expression of the control protein CAT and IFN-induced transmembrane proteins (IFITM2 and IFITM3) affected neither phase of Rluc expression upon DENV-1 Rluc2ARep transfection, both viperin and ISG20 significantly reduced the second phase but not the initial phase (3 h posttransfection) of Rluc expression. These results thus indicated that, similar to the results of the WNV replicon assay, viperin and ISG20 inhibited DENV replicon RNA amplification via not-yet-defined mechanisms but IFITM2 and IFITM3 most probably disrupted molecular events before viral RNA translation, i.e., the early events in DENV infection.
FIG. 6.
Effects of antiviral ISGs on DENV-1 replicon replication. FLP-IN/ISG and FLP-IN/CAT cells were cultured in the absence or presence of 1 μg/ml tetracycline (Tet) for 24 h. The cells were electroporated with 10 μg of DENV-1 luciferase replicon RNA (Rluc2A-Rep) per 4 × 106 cells. The transfected cells were seeded into the wells of a 96-well plate at a density of 4 × 105 per well and cultured in the absence or presence of tetracycline. At the indicated times after seeding, cells were washed once with cold phosphate-buffered saline and 250 μl of 1× lysis buffer (Promega) was added. The luciferase activity in the cell lysates were measured in TopCount NXT. Average results and standard deviations (n = 4) are presented. Luciferase activities at the two earlier time points are presented in the inserted graphs on a smaller scale.
IFITM proteins play a role in restriction of DENV infection.
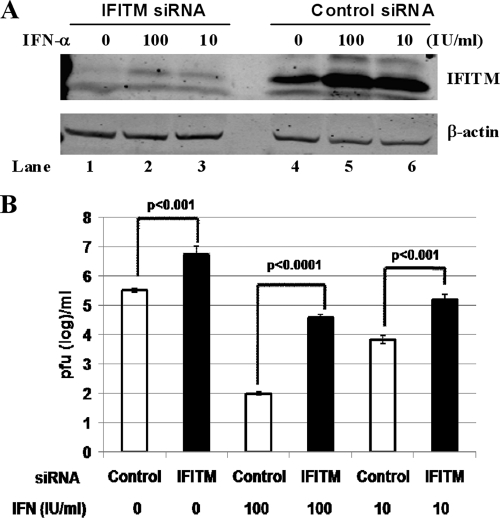
To further confirm the physiological role of the IFITM proteins in controlling DENV infection, we intended to determine the effects of depletion of basal and IFN-induced IFITM expression on DENV infection. As shown in Fig. 7 A, compared with HeLa cells transfected with nontargeting siRNA (control), transfection of SMARTpool siRNA targeting IFITM2 and IFITM3 not only efficiently depleted basal-level IFITM expression (compare lanes 1 and 4) but also significantly reduced IFN-α-induced IFITM expression (compare lanes 2 and 3 with 5 and 6, respectively). Interestingly, depletion of basal IFITM expression dramatically increased the permissiveness of HeLa cells to DENV infection, as indicated by a more-than-10-fold increase in progeny virus production (Fig. 7B). Furthermore, transfection of IFITM siRNA also significantly attenuated the ability of IFN-α to inhibit DENV infection of HeLa cells (Fig. 7B). Similar results were obtained with Huh7 (a hepatoma line) cells upon depletion of IFN-α-induced IFITM expression (data not shown). Hence, our results imply that the IFITM proteins confer basal resistance to DENV but also play an important role in mediating the antiviral response of IFN-α to DENV infection.
FIG. 7.
IFITM restricts DENV infection. (A) HeLa cells were transfected with SMARTpool siRNA targeting IFITM2 and IFITM3 or a nontargeting siRNA (control) by following the directions of the manufacturer (Dharmacon). At 72 h posttransfection, cells were retransfected with the same siRNA and left untreated or treated with 100 or 10 IU/ml IFN-α. Cells were harvested at 30 h posttreatment. Levels of IFITM in the cell lysates were detected by Western blot assay with an antibody recognizing IFITM2 and IFITM3 (Proteintech Group). β-Actin served as a loading control. (B) HeLa cells were transfected with SMARTpool siRNA targeting IFITM2 and IFITM3 or a nontargeting siRNA (control) and infected at 72 h posttransfection with DENV-2 at a multiplicity of infection of 1 for 1 h. The infected cells were either left untreated or treated immediately after infection with the indicated concentrations of IFN-α. Virus yields in culture medium harvested at 30 h postinfection were determined with a plaque assay and are expressed as PFU per milliliter of culture medium (n = 3).
Concluding remarks.
The results presented in this report demonstrated that four ISGs, IFITM2, IFITM3, viperin, and ISG20, were able to significantly inhibit both WNV and DENV infections. Considering the similarity in the genome structures and replication strategies of the two viruses, it is not surprising that similar sets of ISGs inhibited the replication of both viruses. However, it is interesting that PKR could inhibit WNV infection but not DENV infection. Although the inability of PKR to inhibit DENV replication is consistent with a previous report demonstrating that IFN-α inhibited DENV infection of cells by preventing translation of viral RNA through a PKR-independent mechanism (10), it will be interesting to know how PKR could selectively recognize and respond to WNV infection but not DENV infection.
Viperin is a putative radical SAM domain-containing enzyme associated with the endoplasmic reticulum through an amphitropic alpha helix structure located within the N-terminal 45 amino acid residues of the protein (18, 21). Viperin had been demonstrated recently to be an important IFN-induced antiviral protein against a variety of viruses, including human cytomegalovirus (5), human immunodeficiency virus (40), alphaviruses (59), HCV (21), and influenza virus (57). Our mutagenesis studies indicated that the putative enzymatic activity is required for viperin to inhibit HCV (21), WNV, and DENV infections (Fig. 3, 4, and 5). In addition, although the N-terminal region was important for viperin to associate with ER membrane and required for its maximum antiviral activity against HCV and WNV, deletion of this region did not completely abolish its antiviral activity (18, 21) (Fig. 5B and C). Hence, viperin association with ER membrane is important but not absolutely required for its antiviral function.
ISG20 is a 3′-to-5′ exonuclease and a member of the DEDD exoribonuclease family (11). It was shown previously by others and by us that ISG20 inhibits infection with several RNA viruses, including vesicular stomatitis virus, influenza virus, encephalomyocarditis virus, HIV, alphavirus, and HCV (11, 12, 21, 59). Although genetic studies strongly suggest that the exonuclease activity of ISG20 is required for its antiviral function (12, 21), it is not yet known whether ISG20 inhibits virus infection by direct cleavage of viral RNA. Interestingly, although ISG20 has been shown to nonspecifically digest single-strand RNA and DNA in vitro (36), expression of ISG20 does not inhibit cell growth and only selectively reduces viral RNA but not host cellular mRNA (21). Furthermore, we demonstrated in this study that ISG20 did not affect the input replicon RNA-directed luciferase expression but potently inhibited replicon RNA replication (Fig. 6). This result may indicate that ISG20 only recognizes viral RNA in a replication complex or associated with certain viral and/or cellular proteins.
One of the most significant discoveries of this study is that the two structurally closely related small ISGs for IFITM2 and IFITM3 efficiently inhibited both WNV and DENV infections, presumably by disrupting the molecular event(s) before translation of input genomic RNA (Fig. 5 and 6). Interestingly, during the revision of the manuscript, Brass and colleagues reported that the IFITM proteins inhibited influenza A H1N1 virus infection by preventing virus entry into cells (2). Moreover, these authors also showed that expression of IFITMs in Vero E6 cells inhibited infection with VLPs that expressed the envelope proteins of three flaviviruses and that siRNA depletion of IFITM3 expression in HeLa cells led to an increase in WNV and DENV infections. Hence, the two lines of independent evidence provided by Brass et al. and by us convincingly demonstrate that IFITMs are novel IFN-induced antiviral proteins that inhibit infection with viruses from at least two distinct families. To our knowledge, the IFITMs are the first IFN-induced antiviral proteins that inhibit virus entry into host cells.
IFITM family genes are typical type I IFN-inducible genes and were upregulated in certain types of tumor cells, but their biological functions remain undefined (19, 54). A previous report suggested that IFITM1 encodes a 17-kDa cell membrane protein that associates with other proteins at the cell surface to relay growth inhibition and aggregation signals in cell lines including K562, Jurkat, and U937 (8). However, expression of none of the three IFITMs in FLP-IN T Rex cells affected cell proliferation (data not shown). Sequence analyses reveal that IFITM2 and IFITM3 differ at only 12 amino acid residues scattered along the 123-amino-acid polypeptides. In contrast, compared with IFITM2 and IFITM3, IFITM1 has a truncation of the N-terminal 20 or 21 amino acid residues and an extended C-terminal region (30). However, the hydropathy plots of all three of the polypeptides are similar and suggest the existence of two transmembrane regions (30). Their membrane association and plasma membrane localization had been demonstrated by cell fractionation and flow cytometry analysis (2, 8; J. M. Weidner and J.-T. Guo, unpublished results). Apparently, the plasma membrane localization of IFITM2 and IFITM3 is consistent with their proposed roles in disrupting the entry and/or other early steps in WNV and DENV infections.
Because the antiviral state induced by IFNs is physiologically mediated by cooperative actions of multiple ISGs, identification of an antiviral ISG based on its ability to inhibit virus infection upon individual expression, as demanded by our assay system, is a high standard. This approach most probably revealed either the most potent antiviral ISGs that directly restrict virus replication or “master” ISGs that lead to the activation of other cellular proteins to control infection with viruses. It is possible that we have missed the ISGs that work cooperatively with other IFN-inducible proteins to achieve their antiviral function. A typical example is ISG15, which requires three IFN-induced conjugating enzymes to be conjugated to its target viral and cellular proteins (24, 62). Nevertheless, the five ISGs identified in this study could represent the major, if not all, cellular genes that mediate the IFN′s antiviral response to WNV and DENV by disrupting multiple steps of the virus life cycle.
Acknowledgments
We thank Andy Cuconati and Pat Romano for critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health (AI061441) and by the Hepatitis B Foundation through an appropriation of the Commonwealth of Pennsylvania. Work in Pei-Yong Shi's laboratory is supported by NIH grants U01 AI061193 and U54-AI057158 (Northeast Biodefense Center). Ju-Tao Guo is the Bruce Witte Scholar of the Hepatitis B Foundation.
Footnotes
Published ahead of print on 9 June 2010.
REFERENCES
- 1.Borden, E. C., G. C. Sen, G. Uze, R. H. Silverman, R. M. Ransohoff, G. R. Foster, and G. R. Stark. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brass, A. L., I. C. Huang, Y. Benita, S. P. John, M. N. Krishnan, E. M. Feeley, B. J. Ryan, J. L. Weyer, L. van der Weyden, E. Fikrig, D. J. Adams, R. J. Xavier, M. Farzan, and S. J. Elledge. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, J., S. O. Gudima, C. Tarn, X. Nie, and J. M. Taylor. 2005. Development of a novel system to study hepatitis delta virus genome replication. J. Virol. 79:8182-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, J., J. T. Guo, D. Jiang, H. Guo, J. M. Taylor, and T. M. Block. 2008. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J. Virol. 82:8215-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin, K. C., and P. Cresswell. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 98:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clyde, K., J. L. Kyle, and E. Harris. 2006. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J. Virol. 80:11418-11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daffis, S., M. A. Samuel, M. S. Suthar, B. C. Keller, M. Gale, Jr., and M. S. Diamond. 2008. Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection. J. Virol. 82:8465-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deblandre, G. A., O. P. Marinx, S. S. Evans, S. Majjaj, O. Leo, D. Caput, G. A. Huez, and M. G. Wathelet. 1995. Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J. Biol. Chem. 270:23860-23866. [DOI] [PubMed] [Google Scholar]
- 9.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamond, M. S., and E. Harris. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289:297-311. [DOI] [PubMed] [Google Scholar]
- 11.Espert, L., G. Degols, C. Gongora, D. Blondel, B. R. Williams, R. H. Silverman, and N. Mechti. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 278:16151-16158. [DOI] [PubMed] [Google Scholar]
- 12.Espert, L., G. Degols, Y. L. Lin, T. Vincent, M. Benkirane, and N. Mechti. 2005. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J. Gen. Virol. 86:2221-2229. [DOI] [PubMed] [Google Scholar]
- 13.Fink, J., F. Gu, L. Ling, T. Tolfvenstam, F. Olfat, K. C. Chin, P. Aw, J. George, V. A. Kuznetsov, M. Schreiber, S. G. Vasudevan, and M. L. Hibberd. 2007. Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl. Trop. Dis. 1:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredericksen, B. L., B. C. Keller, J. Fornek, M. G. Katze, and M. Gale, Jr. 2008. Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 82:609-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredericksen, B. L., M. Smith, M. G. Katze, P. Y. Shi, and M. Gale, Jr. 2004. The host response to West Nile virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737-7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, J., R. Stoyanova, and C. Seeger. 2005. The transcriptome of HCV replicon expressing cell lines in the presence of alpha interferon. Virology 335:264-275. [DOI] [PubMed] [Google Scholar]
- 18.Hinson, E. R., and P. Cresswell. 2009. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 284:4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hisamatsu, T., M. Watanabe, H. Ogata, T. Ezaki, S. Hozawa, H. Ishii, T. Kanai, and T. Hibi. 1999. Interferon-inducible gene family 1-8U expression in colitis-associated colon cancer and severely inflamed mucosa in ulcerative colitis. Cancer Res. 59:5927-5931. [PubMed] [Google Scholar]
- 20.Jessie, K., M. Y. Fong, S. Devi, S. K. Lam, and K. T. Wong. 2004. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J. Infect. Dis. 189:1411-1418. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, D., H. Guo, C. Xu, J. Chang, B. Gu, L. Wang, T. M. Block, and J. T. Guo. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajaste-Rudnitski, A., T. Mashimo, M. P. Frenkiel, J. L. Guenet, M. Lucas, and P. Despres. 2006. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J. Biol. Chem. 281:4624-4637. [DOI] [PubMed] [Google Scholar]
- 23.Keller, B. C., B. L. Fredericksen, M. A. Samuel, R. E. Mock, P. W. Mason, M. S. Diamond, and M. Gale, Jr. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J. Virol. 80:9424-9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, K. I., N. V. Giannakopoulos, H. W. Virgin, and D. E. Zhang. 2004. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 24:9592-9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kota, R. S., J. C. Rutledge, K. Gohil, A. Kumar, R. I. Enelow, and C. V. Ramana. 2006. Regulation of gene expression in RAW 264.7 macrophage cell line by interferon-gamma. Biochem. Biophys. Res. Commun. 342:1137-1146. [DOI] [PubMed] [Google Scholar]
- 26.Kou, Z., M. Quinn, H. Chen, W. W. Rodrigo, R. C. Rose, J. J. Schlesinger, and X. Jin. 2008. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J. Med. Virol. 80:134-146. [DOI] [PubMed] [Google Scholar]
- 27.Kramer, L. D., J. Li, and P. Y. Shi. 2007. West Nile virus. Lancet Neurol. 6:171-181. [DOI] [PubMed] [Google Scholar]
- 28.Kyle, J. L., P. R. Beatty, and E. Harris. 2007. Dengue virus infects macrophages and dendritic cells in a mouse model of infection. J. Infect. Dis. 195:1808-1817. [DOI] [PubMed] [Google Scholar]
- 29.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 30.Lewin, A. R., L. E. Reid, M. McMahon, G. R. Stark, and I. M. Kerr. 1991. Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 199:417-423. [DOI] [PubMed] [Google Scholar]
- 31.Ma, D., D. Jiang, M. Qing, J. M. Weidner, X. Qu, H. Guo, J. Chang, B. Gu, P. Y. Shi, T. M. Block, and J. T. Guo. 2009. Antiviral effect of interferon lambda against West Nile virus. Antiviral Res. 83:53-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackenzie, J. S., D. J. Gubler, and L. R. Petersen. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10:S98-S109. [DOI] [PubMed] [Google Scholar]
- 33.Mashimo, T., M. Lucas, D. Simon-Chazottes, M. P. Frenkiel, X. Montagutelli, P. E. Ceccaldi, V. Deubel, J. L. Guenet, and P. Despres. 2002. A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc. Natl. Acad. Sci. U. S. A. 99:11311-11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morales, M. A., M. Barrandeguy, C. Fabbri, J. B. Garcia, A. Vissani, K. Trono, G. Gutierrez, S. Pigretti, H. Menchaca, N. Garrido, N. Taylor, F. Fernandez, S. Levis, and D. Enria. 2006. West Nile virus isolation from equines in Argentina, 2006. Emerg. Infect. Dis. 12:1559-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro-Sánchez, E., P. Despres, and L. Cedillo-Barron. 2005. Innate immune responses to dengue virus. Arch. Med. Res. 36:425-435. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, L. H., L. Espert, N. Mechti, and D. M. Wilson III. 2001. The human interferon- and estrogen-regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry 40:7174-7179. [DOI] [PubMed] [Google Scholar]
- 37.Perelygin, A. A., S. V. Scherbik, I. B. Zhulin, B. M. Stockman, Y. Li, and M. A. Brinton. 2002. Positional cloning of the murine flavivirus resistance gene. Proc. Natl. Acad. Sci. U. S. A. 99:9322-9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry, S. T., T. R. Prestwood, S. M. Lada, C. A. Benedict, and S. Shresta. 2009. Cardif-mediated signaling controls the initial innate response to dengue virus in vivo. J. Virol. 83:8276-8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puig-Basagoiti, F., T. S. Deas, P. Ren, M. Tilgner, D. M. Ferguson, and P. Y. Shi. 2005. High-throughput assays using a luciferase-expressing replicon, virus-like particles, and full-length virus for West Nile virus drug discovery. Antimicrob. Agents Chemother. 49:4980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivieccio, M. A., H. S. Suh, Y. Zhao, M. L. Zhao, K. C. Chin, S. C. Lee, and C. F. Brosnan. 2006. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J. Immunol. 177:4735-4741. [DOI] [PubMed] [Google Scholar]
- 41.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuel, M. A., and M. S. Diamond. 2005. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J. Virol. 79:13350-13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samuel, M. A., K. Whitby, B. C. Keller, A. Marri, W. Barchet, B. R. Williams, R. H. Silverman, M. Gale, Jr., and M. S. Diamond. 2006. PKR and RNase L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80:7009-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkar, S. N., and G. C. Sen. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245-259. [DOI] [PubMed] [Google Scholar]
- 45.Scherbik, S. V., J. M. Paranjape, B. M. Stockman, R. H. Silverman, and M. A. Brinton. 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80:2987-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scherbik, S. V., B. M. Stockman, and M. A. Brinton. 2007. Differential expression of interferon (IFN) regulatory factors and IFN-stimulated genes at early times after West Nile virus infection of mouse embryo fibroblasts. J. Virol. 81:12005-12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 48.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 49.Shresta, S., J. L. Kyle, H. M. Snider, M. Basavapatna, P. R. Beatty, and E. Harris. 2004. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J. Virol. 78:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shresta, S., K. L. Sharar, D. M. Prigozhin, H. M. Snider, P. R. Beatty, and E. Harris. 2005. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J. Immunol. 175:3946-3954. [DOI] [PubMed] [Google Scholar]
- 51.Shrestha, B., T. Wang, M. A. Samuel, K. Whitby, J. Craft, E. Fikrig, and M. S. Diamond. 2006. Gamma interferon plays a crucial early antiviral role in protection against West Nile virus infection. J. Virol. 80:5338-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor, D. R., M. Puig, M. E. Darnell, K. Mihalik, and S. M. Feinstone. 2005. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J. Virol. 79:6291-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tobler, L. H., M. J. Cameron, M. C. Lanteri, H. E. Prince, A. Danesh, D. Persad, R. S. Lanciotti, P. J. Norris, D. J. Kelvin, and M. P. Busch. 2008. Interferon and interferon-induced chemokine expression is associated with control of acute viremia in West Nile virus-infected blood donors. J. Infect. Dis. 198:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaarala, M. H., K. Porvari, A. Kyllonen, and P. Vihko. 2000. Differentially expressed genes in two LNCaP prostate cancer cell lines reflecting changes during prostate cancer progression. Lab. Invest. 80:1259-1268. [DOI] [PubMed] [Google Scholar]
- 55.Wacher, C., M. Muller, M. J. Hofer, D. R. Getts, R. Zabaras, S. S. Ousman, F. Terenzi, G. C. Sen, N. J. King, and I. L. Campbell. 2007. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J. Virol. 81:860-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, C., J. Pflugheber, R. Sumpter, Jr., D. L. Sodora, D. Hui, G. C. Sen, and M. Gale, Jr. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 77:3898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, X., E. R. Hinson, and P. Cresswell. 2007. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2:96-105. [DOI] [PubMed] [Google Scholar]
- 58.Warke, R. V., K. J. Martin, K. Giaya, S. K. Shaw, A. L. Rothman, and I. Bosch. 2008. TRAIL is a novel antiviral protein against dengue virus. J. Virol. 82:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y., C. W. Burke, K. D. Ryman, and W. B. Klimstra. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 81:11246-11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]
- 61.Zou, W., J. H. Kim, A. Handidu, X. Li, K. I. Kim, M. Yan, J. Li, and D. E. Zhang. 2007. Microarray analysis reveals that type I interferon strongly increases the expression of immune-response related genes in Ubp43 (Usp18) deficient macrophages. Biochem. Biophys. Res. Commun. 356:193-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou, W., and D. E. Zhang. 2006. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J. Biol. Chem. 281:3989-3994. [DOI] [PubMed] [Google Scholar]